Abstract
Cercospora leaf spot (CLS), caused by Cercospora canescens, is a major threat to mungbean production worldwide. The disease is complicated by its wide host range, diverse pathogenic strains, and the influence of environmental factors. Understanding the interplay between the host, pathogen, and environmental conditions is crucial for developing effective control measures. In a bid to identify and validate CLS resistant mungbean genotypes we conducted multi-environment trials; Genomic selection of mungbean for resistance against CLS necessitates pre-identification in diverse environments. Initially, 110 genotypes were screened under controlled conditions in which thirty-day-old mungbean plants were thoroughly sprayed with the spore suspension using a glass atomizer. After three weeks of revalidation under controlled conditions, Koch’s postulates was adopted for disease identification before selecting 16 genotypes for field testing across four different environments over three successive years. The results obtained from the GGE biplot analysis emphasize the importance of taking both genetic and ambient factors in consideration when evaluating the potential of mungbean genotypes for resistance against CLS and therefore two genotypes “SK-89 (15)” and “WMB-9 (14)” was identified as desirable genotypes. The Additive Main Effects and Multiplicative Interaction (AMMI) model and the GGE biplot have emerged as potent tools for unraveling GEI complexities. It has expanded its application to include evaluation of resistant genotypes and to locate “ideal” evaluation sites and “mega environments” linked to resistance against infection. The study provides valuable insights for future breeding programs, allowing researchers to focus on incorporating these resistant traits into future varieties.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-025-98885-1.
Keywords: Mungbean, Cercospora leaf spot, Multienvironment trial, GGE biplot, Resistance
Subject terms: Biochemistry, Biotechnology, Ecology, Genetics, Microbiology, Molecular biology, Physiology, Plant sciences, Environmental sciences
Introduction
Mungbean (Vigna radiata), also referred to as green gram, stands out as a leguminous crop extensively cultivated for its edible seeds and sprouts. Its nutritional richness, coupled with its adaptability to diverse agroclimatic conditions and ease of cultivation, renders it highly esteemed. Notably, mungbean exhibits remarkable resilience in environments characterized by poor soil quality and limited water resources, making it particularly appealing to farmers across a spectrum of regions. Primarily grown in tropical and subtropical areas, key producers such as India, China, Myanmar, and Thailand collectively drive the bulk of global production. Beyond its role as a dietary staple, mungbean is increasingly acknowledged for its potential in functional foods and nutritional supplements. With its protein content ranging from 20 to 30% and abundance in essential amino acids, it serves as crucial provenance of sustenance for both human race and animals1–3. In defiance of notable strides from recent years, mungbean yield potential continues to face constraints imposed by a variety of stress factors4,5.
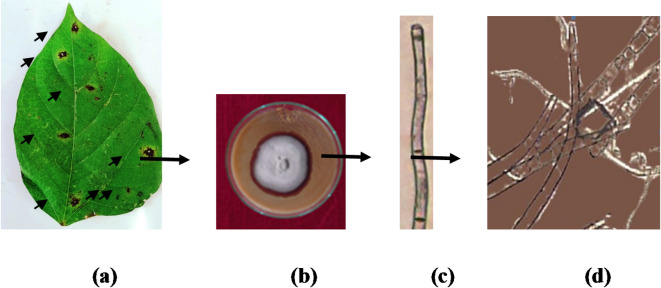
Cercospora leaf spot (CLS) caused by Cercospora canescens, presents significant hazard to global mungbean production, particularly in regions characterized by hot and humid climates6. Typically, the onset of the disease occurs during the flowering stage of mungbean plants, initially manifesting as small necrotic lesions on leaves, eventually progressing to the formation of spots (Fig. 6a). Disease proliferation notably intensifies during the pod filling phase. The genotypes that are vulnerable to CLS experience rapid disease progression, leading to early leaf drop, reduced pod formation, smaller seed size, and significant yield reductions between 46% and 61% 7,8. While the fungus is widespread, the severity of its impact fluctuates with varying climatic pattern, leading to sporadic upsurge9,10. Morphologically, it can be differentiated from other Cercospora species by its distinctive culture, conidiophores and conidia (Fig. 6b–d). Fungicidal approaches for managing the disease are less practicable economically and environmentally. The presence of variability in host plants and pathogens11,12 underscores the urgent necessity of identifying resilient sources of resistance against CLS in mungbean and subsequently integrating these sources into resistance breeding programs to comprehensively address the issue. However, progress in breeding CLS-resistant cultivars has been impeded by challenges in the screening process, the wide host range, and the inherent complexity within the Cercospora genus13–15.
Fig. 6.
Climate factors at experimental sites concerning (a) total rainy days, (b) total Rainfall, and (c) temperature range throughout growth period of mungbean.
The interplay between genotype and environment (GEI) often poses challenges in accurately identifying resilient sources against pathogenic strains and may hinder genetic advancements through selection processes16,17. Moreover, variations in inoculum concentration of C. canescens across diverse settings resulting in varying genotypic reactions and genotype rankings18–20. Hence, it is imperative to locate “optimal testing sites” with high repeatability to screen genotypes exhibiting consistent responses and to precisely evaluate disease dynamics in these critical areas. Minimizing environmental discrepancies during genotype testing is crucial to unveil genuine genotypic reactions18,21–23. Additionally, given the intricacies of field testing against CLS, clustering environments with similar responses (forming “mega environments”) towards host genotype interactions becomes essential. This strategy can streamline screening expenses and provide meaningful insights while upholding trial consistency21,24,25.
Over the past decade, a comprehensive study of different statistical approaches has been conducted to understand genotype by environment interaction (GEI) and to identify ideal genotypes with broad or specific adaptability to different conditions26–28. The Additive Main Effects and Multiplicative Interaction (AMMI) model29–31 and the GGE biplot32–35 have emerged as potent tools for unraveling GEI complexities. Comparative analyses have revealed that the GGE biplot provides a more straightforward and coherent visualization of genotype main effects and GEI compared to the AMMI model, focusing solely on genotypic effects by excluding additive main effects36,37. The GGE biplot has expanded its application to include evaluation of resistant genotypes and to locate “ideal” evaluation sites and “mega environments” linked to resistance against various infections across several leguminous crops18,21,38–40. In mungbean, GGE biplot test has been instrumental in pinpointing resistant genotypes, particularly against YMV resistance18,41–43. It helps to analyse genotypic stability, environmental conditions and subsequent association between genotype and environment (G x E) from assorted conditions39. However, some researchers utilized biplot methodology to evaluate multiple disease resistances in mungbean41,44.
In our study, a GGE biplot approach was adopted to evaluate the influence of genotype, environment; and their interaction (GEI) against CLS, using data from across multiple locations. Furthermore, our objectives encompassed the identification of robust and high-performing mungbean genotypes resistant to CLS and the delineation of “optimal” testing sites based on criteria such as “discrimination power,” “representativeness,” and “desirability index.” Subsequently, we organized diverse testing sites into discrete “mega environments” to offer guidance for future testing initiatives.
Results
Genotypic response towards CLS infestation
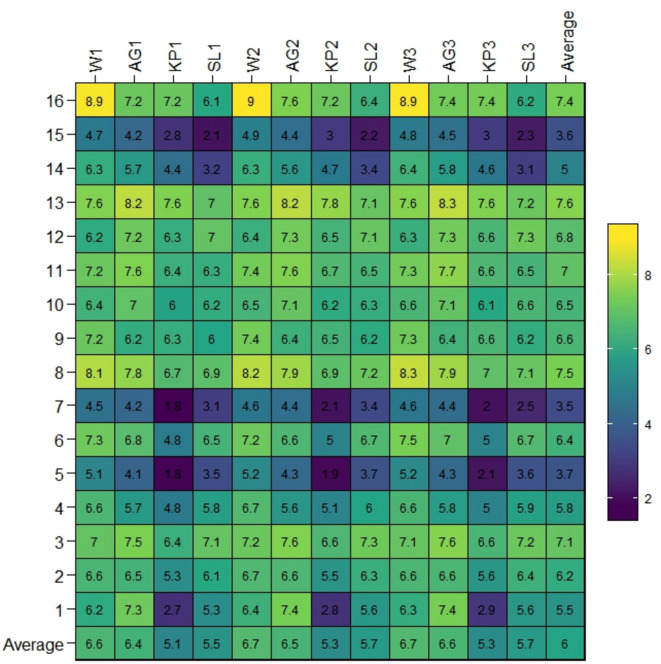
At various tested locations, diverse mungbean genotypes displayed inconsistent reactions to Cercospora leaf spot (CLS) severity. ANOVA conducted on CLS invasion indicated significant impact of genotype, environment, and their interaction (GEI) (Table 1). The proportionate distribution of each source of variation revealed that the environment accounted for 67.40% of overall variation, with genotype × environment interaction contributing 21.20%. The analysis revealed that the environment was responsible for 67.40% of overall variation, while the genotype × environment interaction presented 21.20%. This underscores the substantial impact of the environment on CLS invasion between genotypes across various experimental sites. Figure 1 further demonstrates the varied success of mungbean genotypes and the notable differences in CLS severity among locations. In the initial year (21–23), Kupwara (KP-1) displayed the highest variability, followed by Shalimar (SL-1), whereas Anantnag (AG-1) exhibited the lowest variability followed by Wadura (W-1), signifying notable GEI in relation to the response between host and pathogen. Throughout these years, the inflated prevalence of resistant genotypes was observed in Kupwara during second and third year. In contrast, Anantnag (AG1) recorded the highest frequency of susceptible genotypes during the first year. Additionally, the maximum CLS severity was recorded at Anantnag (8.3) across all the three years (AG-1, AG-2, AG-3), followed by Wadura in the first (W-1) and second year (W-2). Conversely, the lowest severity was noted at Kupwara (2.0) in second (KP-2) and third year (KP-3), followed by Shalimar during second (SL-1) and third year (2.5) (SL-3). For the susceptible check (WMB-1), the severity of CLS varied between 6.1 and 9.0, with an average severity of 7.4 across all locations. This indicates a consistent level of disease pressure over different years and locations. Throughout these years, SK-39, SM-89 and SM-27 displayed moderate resistance, while WMB-9, WMB-78, and SM-6 were identified as moderately susceptible genotypes. The performance of various genotypes showed significant variation across different locations and years (Fig. 2). Based on the average DS score over these locations and years, three genotypes—SM-27, SK-39, and WMB-89—were recognized as moderately resistant, each with mean disease rating below 5. Specifically, SM-27, SK-39, and WMB-89 had average DS scores of 4.07, 3.53, and 3.97, respectively.
Table 1.
ANoVA for CLS intensity in Mungbean tested at 4 different experimental sites.
| Source | Df | Sum Sq | Mean Sq | F value | P value | % contribution |
|---|---|---|---|---|---|---|
| Environment | 11 | 251.87 | 22.8974 | 254.96 | < 0.001 | 67.40% |
| Replication | 24 | 7.99 | 0.3329 | 3.71 | < 0.001 | 2.03% |
| Genotype | 15 | 1034.12 | 5.1686 | 767.66 | < 0.001 | 9.37% |
| Genotype x Environment (G x E) | 165 | 184.79 | 0.0904 | 12.47 | < 0.001 | 21.20% |
| Residuals | 360 | 32.33 | 0.0898 | 1.07 | 0.153 | – |
Significance (p < 0.05).
Fig. 1.
“Heatmap visualization of CLS severity in mungbean across four locations during 3 years. The x-axis shows the tested environments. Locations are denoted as W– (Wadura), AG- (Anantnag), K- (Kupwara), SL- (Shalimar) for year 1 (2019): W1, AG1, KP1, SL1; year 2 (2020): W2, AG2, KP2, SL2; year 3 (2021): W3, AG3, KP3, SL3. The y-axis shows the tested genotypes. The plot legend for disease severity depicts the 1–9 scale for CLS severity rating in color. CLS stands for Cercospora Leaf Spot.”
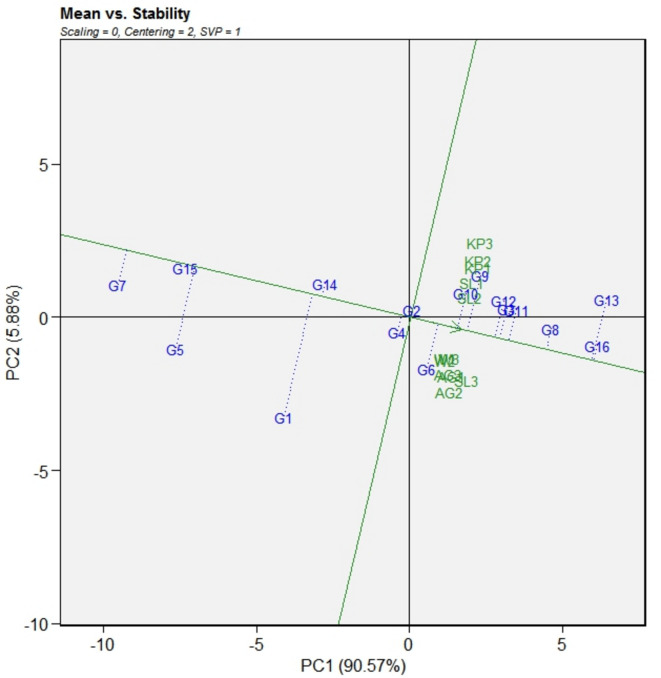
Fig. 2.
The GGE biplot illustrates mean versus stability of 16 mungbean genotypes across four experimental sites. The data were not transformed (transform = 0) and centered by the means values of the environments (centering = 2). The biplot was build upon “row metric preserving.” The testing locations were as follows: year 1 (2021) - W1, AG1, KP1, SL1; year 2 (2022) - W2, AG2, KP2, SL2; year 3 (2023) - W3, AG3, KP3, SL3.
Evaluation of genotypes based on mean performance and stability
The biplot illustrates the ranking of genotypes based on their average performance at different locations and over the year, using the “average environmental coordination” (AEC) axis to visually represent mean and stability (Fig. 2). This biplot test reveals that PC1 and PC2 exhibit 87.58% and 6.8% of the total variation in CLS scoring, respectively, when considering the environment. The AEC axis, represented by a single arrow line extending from the center of the biplot, indicates higher CLS severity in the genotypes. Upon visualization of the figure, it was concluded that SM-6 (2), SM-21 (4), WMB-9 (14), WMB-4 (1), SM-89 (15), SM-27 (5), and SK-39 (7) were placed sinisterly to biplot’s origin, suggesting lower CLS infestation. Conversely, the susceptible check, WMB-1 (16), along with SM-10 (3), WMB-34 (6), SM-66 (8), SK-77 (9), WMB-78 (10), SK-87 (11), SM-92 (12), and SM-107 (13) were situated dextrally to the origin, suggesting higher disease intensity.
Genotype stability was illustrated by a line with two arrows, known as the “AEC ordinate.” The length of genotype projection from this line indicates its stability; longer projections signify less stability. The most desirable genotypes should show low disease intensity and low projection on AEC axis. Consequently, SK-89 (15) evolves as an “ideal” genotype, demonstrating moderate resistance and high stability with a short projection ahead of AEC axis. Genotypes closer to the desirable genotype were appraised as more “desirable.” Thus, SM-21 (4), followed by SM-6 (2), were traced as “desirable” genotypes due to moderately susceptible and consistent responses shown by them across locations. In contrast, genotypes such as SK-39 (7), SM-27 (5) and WMB-4 (1) exhibited moderately susceptible responses but displayed higher projections from the AEC axis, suggesting inconsistency in their reaction to CLS across locations.
Assessment of testing locations: discriminative vs. representativeness and desirability index
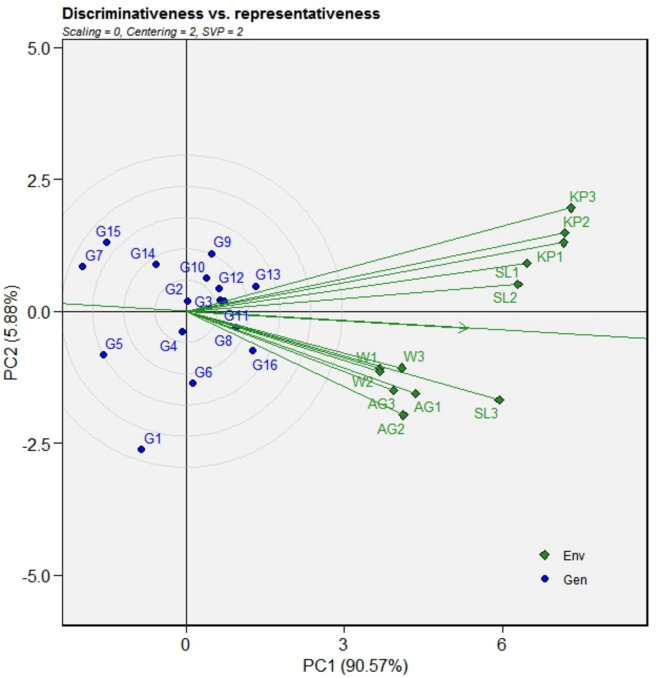
The association among experimental sites is illustrated by the environmental vector aspect of GGE biplot, where every domain is connected by a line representing its environment vector. The cosine of the angle between two environmental vectors indicates the association between them. A firm relationship was observed between Kupwara and Shalimar and between Wadura and Anantnag across all the three years. It has been observed that Shalimar in third year (SL-3) also shows close association with Wadura and Anantnag across all years. These locations displayed acute angles, signifying agreement in their genotypic response to CLS intensity.
In the GGE biplot approach, three variables are crucial for evaluating test locations: “discrimination power”, “representativeness”, and the “desirability index”45. The “discriminating ability” specified by extent of the environmental vector that corresponds to standard deviation within experimental location. Throughout these years, Kupwara and Shalimar exhibit the longest environment vectors and Wadura had the shortest projection (Fig. 3). Consequently, Shalimar and Kupwara were recognized as “discriminating locations” due to their ability to differentiate between genotypes.
Fig. 3.
The perspective of delimitation versus representativeness of experimental sites was compared using a GGE biplot of 16 mungbean genotypes evaluated at four experimental sites. The data were not transformed (transform = 0), and centered using mean values of the environments (centering = 2). The biplot was created using “row metric preserving”. The locations are: For year 1 (2019): W1, AG1, KP1, SL1; Year 2 (2020) W2, AG2, KP2, SL2; Year 3 (2021) W3, AG3, KP3, SL3.
The “representativeness” of experimental sites is driven by acute angle formed by the environment vectors and the “AEC abscissa.” Over these years, Shalimar and Wadura showed the acute angles with the “AEC abscissa,” identifying them as “most representative” experimental sites. The “desirability index” of experimental sites combines both its “discriminative” ability and “representativeness.” Consequently, Shalimar, with the maximum desirability index, pointed out as an “ideal” experimental site for CLS screening. Furthermore, Wadura could be designated as “supplementary” experimental site.
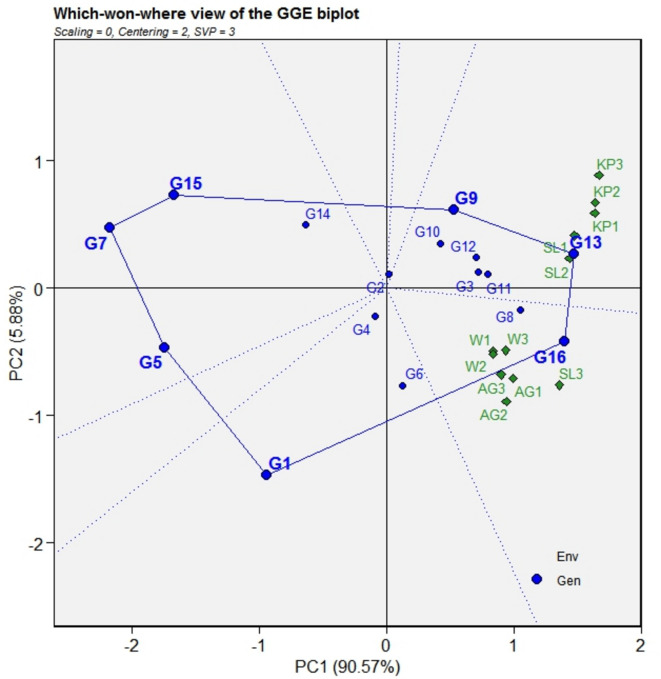
Identification of mega environments and “which won where”
The diagram represented as a polygon showing “which won where” is formed by intercalating the genotypes in a limited extent from the origin. Vertical lines, the so-called “equality lines,” are then subsequently drawn from the origin to both sides of polygon to divide it into sectors33. The relationship between genotypes and environments is assessed using “symmetric scaling,” which involves singular value partitioning that focuses on both genotype and environment. This graph helps to pinpoint major environments and genotypes with specialized resilience. Typically, genotypes located at polygon vertices revealed the most significant genotype-by-environment association, indicating either the top performers or the poorest performers for the specific environment in that area46,47. In this study, the polygon is dissected into 4 distinct sectors representing 4 “mega environments (ME),” which confirms the existence of crossover interactions (Fig. 4). It is noteworthy that the Shalimar and Kupwara test sites did not interact with each other for years and formed a single “mega environment.” Wadura and Anantnag showed consistent genotypic responses to CLS severity over these years. Based on the genotype positions at the vertices of every “mega environment,” MEI included Kupwara and Shalimar (SL-1, SL-2) along with (9), (13) emerging as the top-performing genotype. In MEII encompassing Wadura, Anantnag and Shalimar (SL-3) during the third year along with WMB-1 (16) were identified as the winning genotypes for each mega environment. MEIII includes WMB-4 (1) and SM-27 (5). Similarly, in MEIV SK-39 (7), SM-89 (15) exhibited the best performance.
Fig. 4.
“Which-won-where” perspective of GGE biplot used to analyze 16 mungbean genotypes across 4 experimental sites. Data was not transformed (transform = 0), and were centered by means of the environments (centering = 2). The biplot was based on “row metric preserving”. Locations are: For year 1 (2019): W1, AG1, KP1, SL1; year 2 (2020) W2, AG2, KP2, SL2; year 3 (2021) W3, AG3, KP3, SL3.
Discussion
CLS represents a significant fungal threat to mungbean crops, with no highly resistant sources identified to date. The task of identifying CLS-resistant mungbean genotypes is further complicated by pathogenic variability and a broad host range. Identifying genotypes with low CLS intensity and consistent performance across different environments is essential for incorporating these resistant sources into future breeding programs and management strategies. However, the existence of genotype-by-environment association complicates multi-environment testing against the disease. The intricate dynamics of host-pathogen interactions further add to the complexity of selecting durable resistant sources across various conditions and times. Evaluating numerous genotypes across multiple locations poses a significant challenge. In addition to detecting genotypes with enduring resistance, identifying suitable testing locations is essential for conducting cost-effective multi-environment trials39,48. The GGE biplot methodology offers a promising solution to these challenges by facilitating the assessment of genotypes and to pinpoint optimal experimental locations, which can then be tagged into distinct “mega environments,” regardless of their agroecological zones49.
In this study, 16 mungbean genotypes were assessed across four distinct sites over three successive years, revealing a noteworthy genotype × environment association concerning CLS intensity. ANOVA indicated that the environment had the most substantial impact on CLS severity, followed by genotype × environment (G x E) association. The average performance of the genotypes evaluated in various sites revealed disparate genotypic performances and the presence of crossover interaction. Previous research has shown that variations in environmental variables at experimental sites significantly influence CLS severity. Additionally, differences in genotypic variability and pathogen virulence may affect disease development. The significant presence of G x E association underscores the necessity of multilocation evaluation of genotypes against CLS prior to finalizing genotype rankings41,50.
Among the locations examined, Kupwara (KP-2) and Shalimar (SL-2) exhibited relatively lower CLS severity, showing resistant to moderately resistant responses. Conversely, Anantnag (AG-1, AG-2 & AG-3) and Wadura (W-1, W-2) displayed highly susceptible to susceptible responses. Hence, Anantnag and Wadura were identified as “hot spots” for screening mungbean averse to CLS. The heightened CLS severity during the kharif season in Anantnag reaffirmed its aggressiveness. Anantnag experienced a greater number of rainy days in these years, contributing to increased disease intensity. The congruous susceptibility of the check “WMB-1” across years and locations confirms the presence of enough infection throughout screening, facilitating specific genotypic selection. In multi-location evaluations, screening genotypes specific to a mega environment with non-crossover association is crucial. So selecting an exemplary genotype, both its average performance and high stability should be taken into account51,53. In GGE biplot view, the “AEC abscissa” denotes elevated average performance, representing genotype benefaction (G), while the “AEC ordinate” indicates genotype stability and depicts the contribution of genotype to genotype × environment interaction51,52,54,61. Among the evaluated genotypes, SK-89 (7) was located very near to the “AEC abscissa,” exhibiting minimal prognostication on the “AEC ordinate” with high stability, thus being contemplated an “ideal” genotype and the categorization of other genotypes is based on their biplot interspaced from the ideal genotype. Consequently, SM-21 (4) was identified as a “desirable” genotype with nearly reconcilable performance and lower CLS intensity. Moreover, SK-89 (15) and WMB-9 (14) should be considerably utilized in mungbean resistance breeding programs against CLS.
The distinctive feature of the GGE biplot lies in its capacity to eliminate unnecessary testing locations while maintaining trial heritability and genetic gain during selection in a very profitable manner53,55. In this investigation, a strong association was observed between Kupwara and Shalimar, suggesting consistent genotypic responses to CLS severity. Given Kupwara minimal CLS severity, it could be excluded from further mungbean genotype screening. In GGE biplot analysis, an “ideal” experimental location should be designated depending upon its potential to distinguish genotypes, represent the “mega environment,” and possess a high “desirability index"45,51,55,56. A “mega environment” refers to a cluster of identical sites that consistently elicit the best genotypic responses throughout the year57,60. Experimental sites with longer vector lengths are recommended as perspicacious, while those with acute angles relative to the “AEC abscissa” are deemed representative58. The desirability index of a genotype, derived from both its discriminatory power and the representativeness of the environment, is crucial for selecting experimental locations to optimize resource allocation63. Anantnag, identified as “hot spot” for CLS, was designated as the “ideal” experimental site, with Wadura designated as “supplementary” experimental location for future mungbean genotype screening against CLS. These two sites could aid in identifying genotypes with broader adaptability. Hinge on desirability index, Shalimar and Kupwara were designated as the ineffectual sites for CLS screening, with finite ability to identify superior genotypes.
In this study, all the experimental sites were categorized into four different “mega environments,” each associated with triumphant genotypes, highlighting the existence of a crossover genotype-by-environment association and the importance of breeding for specific adaptability. A “mega environment” is characterized as a collection of similar locations that exhibit homogenous genotypic responses and consistently allocate principal set of genotypes throughout the year64. The spotting of mega environments effectively addresses the evaluation of genotypes and experimental sites, guiding the deployment of specific genotypes within each location59. Partitioning experimental sites into different sub-regions depending upon their repeatability holds significant relevance for mungbean resistance breeding programs. Therefore, this study provides insights into the optimal mega environments for evaluating mungbean genotypes against CLS. The most promising genotypes, SK-89 (15) and WMB-9 (14), were identified, which could contribute to the development of resistance against this disease in mungbean.
Materials and methods
Initial testing
In 2019–2020, a comprehensive evaluation was conducted on 110 mungbean genotypes, including released varieties, to assess their resistance to CLS (Table S1). After a preliminary screening, a group of 16 mungbean genotypes that showed resistance to CLS were selected for further evaluation in multiplication and multiyear trials.
Screening under in-vitro conditions
Cercospora canescens was cultured on potato dextrose agar (PDA) in a BOD incubator. A spore suspension (1 × 10⁶ conidia mL⁻¹) was prepared by flooding the plates with sterile distilled water, scraping the surface, filtering through sterile cheesecloth, and adjusting the concentration using a hemocytometer. Thirty-day-old mungbean plants were thoroughly sprayed with the spore suspension using a glass atomizer, and wetness was maintained for 72 h by covering the plants with clear polyethylene bags. A control plant remained uninoculated under identical conditions. After 72 h, the covers were removed, and the plants were transferred to a greenhouse for three weeks for revalidation. Symptoms were observed at regular intervals, and a disease rating scale (1–9), adapted from Iqbal63 was employed for disease evaluation. To confirm its identity through Koch’s postulates, the pathogen was re-isolated from symptomatic leaves and compared the new cultures with the original (Fig. 5).
Fig. 5.
Symptoms of Cercospora Leaf Spot on infected mungbean plants. (a) Infected mungbean leaf. (b) Cercospora canescens culture (c) Conidiophore of Cercospora canescens. (d) Multi-septate spore of Cercospora canescens.
Multi environment testing
An evaluation across multiple environments was conducted with 16 potential mungbean genotypes, including susceptible check (WMB-1), to evaluate their susceptibility to CLS across four diverse locations over three consecutive seasons (2019–21), under natural epiphytotic conditions. These locations encompassed FoA Wadura Sopore, SKUAST-K Shalimar, KVK Kupwara, and MRCFC Khudwani. FoA Wadura Sopore, and MRCFC, Khudwani were specifically chosen for continued testing over three years due to the persistent prevalence of CLS, designating them as hotspots for the disease. Comprehensive data regarding the plant materials used in this research (Table 2). The different genotypes were grown in accordance with standard agricultural practices in a randomized block design (RCBD) with three replicates and appropriate plant spacing. Inoculum pressure was induced by planting a row of susceptible genotype (WMB-1) after every third row of test genotypes, surrounding the test block. Additionally meteorological data of all experimental sites are given (Fig. 6).
Table 2.
Plant material used in the study.
| S. no. | Genotype | Code | S. No. | Genotype | Code |
|---|---|---|---|---|---|
| 1 | WMB-4 | G1 | 9 | SK-77 | G9 |
| 2 | SM-6 | G2 | 10 | WMB-78 | G10 |
| 3 | SM-10 | G3 | 11 | SK-87 | G11 |
| 4 | SM-21 | G4 | 12 | SM-92 | G12 |
| 5 | SM-27 | G5 | 13 | SM-107 | G13 |
| 6 | WMB-34 | G6 | 14 | WMB-9 | G14 |
| 7 | SK-39 | G7 | 15 | SM-89 | G15 |
| 8 | SM-66 | G8 | 16 | WMB-1 | G16 |
Screening and disease documenting
CLS intensity was critically evaluated once the susceptible check exhibited 50% disease severity, with each genotype additionally subjected to infection following Bakr62. Typically appearing post-flowering in mungbean plants, leaves from 10 randomly selected plants were used to access CLS severity during the pod filling stage (60 days). A disease rating scale (1–9), adapted from Iqbal63 was employed for disease evaluation. This scale ranged from 1 denoting no observable disease symptoms to 9 indicating over 70.1% foliage affected. Intermediate categories were defined, such as 2 for 0.1–10% foliage affected, 3 for 10.1–20% foliage affected, and so forth, up to 8 for 60.1–70% foliage affected. Disease intensity data from all experimental sites were combined each year for every genotype to conduct GGE biplot study.
Construction of GGE biplot
Yan et al.51 extended the use of the GGE biplot to score genotypes and identify mega-environments within multi-environment trial (MET) data; by prioritizing the genotype main effect (G) and genotype-environment interaction (GEI) by eliminating trivial environmental main effects (E). The GGE biplot is constructed by plotting the values of genotypes and environments on the first principal component (PC1) against their values on the second principal component (PC2), obtained by singular value decomposition (SVD) of environment-centered data using the given equation.

Where:
Yij is the mean severity of ith genotype (i = 1,….,i) in the jth environment (j = 1,…,j). µ is the grand mean. N is the number of principal components retained in the model.  is environment deviations from grand mean. λn is the eigenvalue associated with principal component analysis axis. γ in is the genotype PC score for axis. δ
jn is the environment PC score for axis. εij is the residual error term.
is environment deviations from grand mean. λn is the eigenvalue associated with principal component analysis axis. γ in is the genotype PC score for axis. δ
jn is the environment PC score for axis. εij is the residual error term.
The MLT dataset concerning the response of mungbean genotypes to CLS severity underwent analysis without scaling (scaling = 0), with the objective of constructing an environment-centered (Centering = 2) G × E table47. The genotype assessment depends upon genotype- based single value portioning (SVP = 1), while the environment-based single value portioning (SVP = 2) was used for the environmental evaluation33. The optimal genotype was determined based on mean performance and stability in different environments. The evaluation of genotypes was executed and visualized using “average environmental coordination” (AEC) perspective of the GGE biplot, which facilitates the comparison of mean disease score and stability in environments within a mega-environment33,49. The evaluation of experimental site suitability was demonstrated by utilizing “discriminatory power and representativeness perspective” of the GGE biplot. Discriminatory power was insistent on the magnitude of the environmental vector, while representativeness was indicated by the acute angle formed with the “Average Environment Coordinate/Axis” (AEC/AEA)19. Additionally, effectiveness of genotypes in various experimental conditions and the categorization of test conditions into distinct “mega-environments” were established using GGE biplots “which won where” approach57.
Data analysis
The impact of environments, genotypes and their interactions on CLS were assessed using analyses of variance (ANOVA) with mixed model analyses and GGE biplots; using the “METAN” package in R software version 4.3.3.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgements
We acknowledge Princess Nourah bint Abdulrahman University Researchers Supporting Project number (PNURSP2025R402), Princess Nourah bint Abdulrahman University, Riyadh, Saudi Arabia. We also appreciate the plant breeders and pathologists of SKUAST-K at experimental sites for their support in screening the CLS disease.
Author contributions
Mohammad Irfan devises and drafted the study. Mohammad Irfan, Uzma Rashid, Mohd Ashraf Bhat and Farooq Ahmed Bhat administer field tests and collected data. Mohammad Irfan, Uzma Rashid and Khairiah Mubarak Alwutayd interpret the results. Mohammad Irfan, Uzma Rashid, Mohd Ashraf Bhat, Farooq Ahmed Bhat and Khairiah Mubarak Alwutayd provided censorious inputs during the course of study and edited the manuscript. All the authors made an equal contribution to the article and have given their approval for the submitted version.
Data availability
The data that support the findings of this study are available from the corresponding author upon reasonable request.
Competing interests
The authors declare no competing interests.
Ethical statement
Statement specifying permission, we acquired permission to study mungbean issued by the Department of Genetics and Plant breeding, SKUAST –Kashmir, India.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Mohammad Irfan, Email: irather046@gmail.com.
Uzma Rashid, Email: uzmawani.1994@gmail.com.
References
- 1.Dahiya, P. K. et al. Technological and nutritional potential. Crit. Rev. Food Sci. Nutr.55(5), 670–688 (2015). [DOI] [PubMed] [Google Scholar]
- 2.Deraz, S. F. & Khalil, A. A. Strategies to improve protein quality and reduce antinutritional factors in Mungbean. Food2(1), 25–38 (2008). [Google Scholar]
- 3.Ahmed, H. G. M. D. et al. Enriching the content of proteins and essential amino acids in legumes. In Legumes Biofortification 417–447 (Springer International Publishing, 2023).
- 4.Nair, R. M. et al. Biotic and abiotic constraints in Mungbean production—progress in genetic improvement. Front. Plant Sci.10, 1340 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hanumantha Rao, B., Nair, R. M. & Nayyar, H. Salinity and high temperature tolerance in Mungbean [Vigna radiata (L.) Wilczek] from a physiological perspective. Front. Plant Sci.7, 186318 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Babar, M., Ijaz, S., Haq, I. U. & Khan, M. S. Development of molecular marker linked with Cercospora leaf spot (CLS) disease resistance in Vigna radiata, cloning, and expression for evaluating antifungal activity against Cercospora canescens 1289–1300 (2023).
- 7.Mogali, S. C. & Hegde, G. M. Recent advances in mungbean breeding: a perspective.In Accelerated Plant Breeding, Volume 3: Food Legumes 235–282 (2020).
- 8.Sunani, S. K., Rout, A. K., Kumar, R. & Choudhary, D. K. Major diseases of green gram, black gram and their integrated management strategies. SATSA Mukhapatra-Annu. Tech. Issue28, 257–271 (2024). [Google Scholar]
- 9.Reddy, P. P. Emerging Crop Pest Problems: Redefining Management Strategies (Scientific, 2018).
- 10.Kimber, R. B. E. & Paull, J. G. Identification and genetics of resistance to Cercospora leaf spot (Cercospora zonata) in faba bean (Vicia faba). Euphytica177(3), 419–429 (2011). [Google Scholar]
- 11.Biradar, S. A. et al. Prospects and challenges in linseed (Linum usitatissimum L.) production: a review. J. Oilseeds Res.33(1), 1–13 (2016). [Google Scholar]
- 12.Sahoo, L. Induced resistance mechanism in plant and its importance in agriculture. Available at SSRN 4764708 (2024).
- 13.Spanner, R. E. The molecular basis of demethylation inhibitor fungicide resistance in Cercospora beticola and the role of seed inoculum in cercospora leaf spot disease of sugar beet (Doctoral dissertation, North Dakota State University) (2020).
- 14.Sahu, P. K., Dhole, V. J. & Mondal, S. Molecular breeding for resistance to economically important diseases of pulses. In Disease Resistance in Crop Plants: Molecular, Genetic and Genomic Perspectives 157–198 (2019).
- 15.Boopathi, N. M. & Boopathi, N. M. Recent advances in MAS in major crops. In Genetic Mapping and Marker Assisted Selection: Basics, Practice and Benefits 245–280 (2013).
- 16.Bhatnagar-Mathur, P., Sunkara, S., Bhatnagar-Panwar, M., Waliyar, F. & Sharma, K. K. Biotechnological advances for combating Aspergillus flavus and aflatoxin contamination in crops. Plant Sci.234, 119–132 (2015). [DOI] [PubMed] [Google Scholar]
- 17.Merrick, L. F., Burke, A. B., Chen, X. & Carter, A. H. Breeding with major and minor genes: genomic selection for quantitative disease resistance. Front. Plant Sci.12, 713667 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Das, A. et al. Delineating Genotype× environment interactions towards durable resistance in Mungbean against Cercospora leaf spot (Cercospora canescens) using GGE biplot. Plant. Breed.139(3), 639–650 (2020). [Google Scholar]
- 19.Tamang, S., Saha, P., Bhattacharya, S. & Das, A. Unveiling genotype X environment interactions towards identification of stable sources of resistance in chickpea—collar rot pathosystem exploiting GGE biplot technique. Australas Plant Pathol.2022, 1–12 (2022).
- 20.Maurya, A. K., Navathe, S., Mohapatra, C. & Chand, R. Antioxidants elevates the resistance to Cercospora canescens in interspecific cross of Vigna radiata (Kopergaon) × Vigna mungo (Pant urd 31). Indian Phytopathol.71, 519–528 (2018). [Google Scholar]
- 21.Das, A. et al. Deciphering genotype-by-environment interaction for targeting test environments and rust resistant genotypes in field pea (Pisum sativum L). Front. Plant Sci.10, 825 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chattopadhyay, N., Mandal, R., Roy, A., Bhattacharya, P. M. & Chowdhury, A. K. Assessment of wheat genotypes based on genotype-by-environment interaction for durable resistance to spot blotch disease in hot spot. Cereal Res. Commun.50(1), 95–102 (2022). [Google Scholar]
- 23.Mukuze, C. Genetic Diversity and Genotype× Environment Interaction of Advanced Elite Soybean Genotypes in Uganda (Doctoral dissertation, Makerere University) (2019).
- 24.Singh, B. et al. Delineation of Genotype-by-Environment interactions for identification and validation of resistant genotypes in Mungbean to root-knot nematode (Meloidogyne incognita) using GGE biplot. Sci. Rep.10(1), 4108 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Parihar, A. K., Kushwaha, K. P. & Bhattacharya, S. Retraction notice to ‘use of HA-GGE biplot for interpretation of genotype ¾ environment interaction when assessing sources of durable resistance against powdery mildew in Mungbean. Crop Pasture Sci.71, 794 (2020). [Google Scholar]
- 26.Malosetti, M., Ribaut, J. M. & van Eeuwijk, F. A. The statistical analysis of multi-environment data: modeling genotype-by-environment interaction and its genetic basis. Front. Physiol.4, 37433 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kang, M. S. Genotype-environment interaction and stability analyses: an update. Quant. Genet. Genom. Plant Breed.. 9, 140–161 (2020).
- 28.Rakshit, S. et al. Analysis of Indian post-rainy sorghum multi-location trial data reveals complexity of genotype X environment interaction. J. Agric. Sci.155(1), 44–59 (2017). [Google Scholar]
- 29.Scavo, A., Mauromicale, G. & Ierna, A. Genotype× environment interactions of potato tuber quality characteristics by AMMI and GGE biplot analysis. Sci. Hort.310(15), 111750 (2023). [Google Scholar]
- 30.Khan, M. M. H., Rafii, M. Y., Ramlee, S. I. & Jusoh, M. Al Mamun, M. AMMI and GGE biplot analysis for yield performance and stability assessment of selected Bambara groundnut (Vigna subterranea L. Verdc.) genotypes under the multi-environmental trials (METs). Sci. Rep.11(1), 22791 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ajayi, A. et al. Genotype X environment interaction and adaptation of Cowpea genotypes across six planting seasons. Front. Life Sci. Relat. Technol.3(1), 7–15 (2022). [Google Scholar]
- 32.Hashim, N. et al. Integrating multivariate and univariate statistical models to investigate genotype–environment interaction of advanced fragrant rice genotypes under rainfed condition. Sustainability13(8), 4555 (2021). [Google Scholar]
- 33.Linus, R. A., Olanrewaju, O. S., Oyatomi, O., Idehen, E. O. & Abberton, M. Assessment of yield stability of Bambara groundnut (Vigna subterranea (L.) Verdc.) using genotype and genotype–environment interaction biplot analysis. Agronomy13(10), 2558 (2023). [Google Scholar]
- 34.Sujitha, R. et al. X insights into yield stability: a comparative analysis of regression, AMMI indices and biplot methods in Pearl millet. Electron. J. Plant. Breed.15(1), 42–52 (2020). [Google Scholar]
- 35.Kuwar, C. B. et al. Unraveling Genotype-by-Environment interaction in maize using cutting edge statistical tools: innovative empirical selection for increased yield stability. Ecol. Genet. Genomics2024, 100249 (2024).
- 36.Greveniotis, V. et al. Genotype-by-Environment interaction analysis for quantity and quality traits in Faba beans using AMMI, GGE models, and stability indices. Plants12(21), 3769 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yang, X. et al. Yield adaptability and stability in field pea genotypes using AMMI, GGE, and GYT biplot analyses. Agriculture13(10), 1962 (2023). [Google Scholar]
- 38.Tollo, J. A., Ojwang, P. P. O., Karimi, R., Mafurah, J. J. & Nzioki, H. S. Genotype-by-environment interaction and stability of resistance in Mungbean landraces against common bacterial blight across semi-arid environments. Euphytica216, 1–16 (2020). [Google Scholar]
- 39.Parihar, A. K. et al. Biplot evaluation of test environments and identification of lentil genotypes with durable resistance to fusarium wilt in India. Crop Pasture Sci.68(11), 1024–1030 (2017). [Google Scholar]
- 40.Parihar, A. K. et al. Targeting test environments and rust-resistant genotypes in lentils (Lens culinaris) by using heritability-adjusted biplot analysis. Crop Pasture Sci.69(11), 1113–1125 (2018). [Google Scholar]
- 41.Alam, A. M., Somta, P., Jompuk, C., Chatwachirawong, P. & Srinives, P. Evaluation of Mungbean genotypes based on yield stability and reaction to Mungbean yellow mosaic virus disease. Plant. Pathol. J.30(3), 261 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bhartiya, A. et al. Evaluation of Indigenous and exotic soybean accessions for yield, resistance to frog-eye leaf spot and yellow mosaic virus diseases. Plant. Genetic Resour.21(6), 513–519 (2023). [Google Scholar]
- 43.Azam, M. G. et al. Genetic analyses of Mungbean [Vigna radiata (L.) Wilczek] breeding traits for selecting superior genotype (s) using multivariate and multi-traits indexing approaches. Plants12(10), 1984 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kaur, L., Singh, P. & Sirari, A. Biplot analysis for locating multiple disease resistance diversity in Mungbean germplasm. Plant. Disease Res.26(1), 55–60 (2011). [Google Scholar]
- 45.Mohammadi, R., Jafarzadeh, J., Poursiahbidi, M. M., Hatamzadeh, H. & Amri, A. Genotype-by-environment interaction and stability analysis for grain yield in durum wheat using GGE biplot and genotypic and environmental covariates. Agricultural Res.12(4), 364–374 (2023). [Google Scholar]
- 46.Enyew, M. et al. Genotype by environment interaction, correlation, AMMI, GGE biplot and cluster analysis for grain yield and other agronomic traits in sorghum (Sorghum bicolor L. Moench). Plos One16(10), e0258211 (2021). [DOI] [PMC free article] [PubMed]
- 47.Wagaw, K. et al. Distinguishing of stable genotypes and mega environment for grain yield performance of sorghum [Sorghum bicolor (L.) moench] genotypes using spatial analysis. Am. J. Plant. Sci.12(3), 417–431 (2021). [Google Scholar]
- 48.Munaro, L. B. et al. Exploring long-term variety performance trials to improve environment-specific Genotype× management recommendations: a case-study for winter wheat. Field Crops Res.255, 107848 (2020). [Google Scholar]
- 49.Naik, A. et al. Deciphering Genotype× environment interaction by AMMI and GGE biplot analysis among elite wheat (Triticum aestivum L.) genotypes of Himalayan region. Ekin J. Crop Breed. Genet.8(1), 41–52 (2022). [Google Scholar]
- 50.Alemu, A. et al. Genomic selection in plant breeding: Key factors shaping two decades of progress. Mol. Plant17(4), 552–578 (2024). [DOI] [PubMed]
- 51.Yan, W., Kang, M. S., Ma, B., Woods, S. & Cornelius, P. L. GGE biplot vs. AMMI analysis of genotype by environment data. Crop Sci.47, 643–653 (2007). [Google Scholar]
- 52.Basnet, B. Deciphering genetic variability and phenotype expression, assessing drought stress tolerance and multi-trait stability index of (Vigna radiata) genotypes in Chitwan, Nepal. Cogent Food Agric.10(1), 2417843 (2024). [Google Scholar]
- 53.Yan, W. & Tinker, N. A. Biplot analysis of multi-environment trial data: principles and applications. Can. J. Plant Sci.86(3), 623–645 (2006). [Google Scholar]
- 54.Kunwar, C. B. et al. Multi-model approach for optimizing cold-wave resilient maize selection: unveiling genotype-by-environment interaction and predicting yield stability. CABI Agric. Biosci.5(1), 63 (2024). [Google Scholar]
- 55.Badu-Apraku, B. et al. Application of the GGE biplot as a statistical tool in the breeding and testing of early and extra-early maturing maize in sub-Saharan Africa. Crop Breed. Genet. Genomics2(3), e200012 (2020).
- 56.Basnet, B., Upreti, U. & Thapaliya, K. P. Genotypic variations in post fertility traits and yield components of Mungbean (Vigna radiata (L.) R. Wilczek) germplasms in Chitwan, Nepal. Heliyon10(20), e39226 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Mamo, W., Enyew, M., Mekonnen, T., Tesfaye, K. & Feyissa, T. Phenotypic variability, heritability and GGE biplot analysis for agronomic traits in Ethiopian sorghum [Sorghum bicolor (L.) Moench] genotypes. Ecol. Genet. Genomics. 27, 100170 (2023). [Google Scholar]
- 58.Demelash, H. Genotype by environment interaction, AMMI, GGE biplot, and mega environment analysis of elite Sorghum bicolor (L.) Moench genotypes in humid lowland areas of Ethiopia. Heliyon10, 5 (2024). [DOI] [PMC free article] [PubMed]
- 59.Van Eeuwijk, F. A., Bustos-Korts, D. V. & Malosetti, M. What should students in plant breeding know about the statistical aspects of Genotype× environment interactions? Crop Sci.56(5), 2119–2140 (2016). [Google Scholar]
- 60.Kebede, G., Worku, W., Feyissa, F. & Jifar, H. Genotype by environment interaction and stability analysis for selection of superior fodder yield performing oat (Avena sativa L.) genotypes using GGE biplot in Ethiopia. Ecol. Genet. Genomics. 28, 100192 (2023). [Google Scholar]
- 61.Pour-Aboughadareh, A. et al. Deciphering genotype-by-environment interaction in barley genotypes using different adaptability and stability methods. J. Crop Sci. Biotechnol.26(5), 547–562 (2023). [Google Scholar]
- 62.Bakr, M. A. Management of important disease of major pulses: advances in pulse research in Bangladesh. In Proceedings of the Second National Workshop on Pulse 6–8 June 1998. ICRISAT, India 121–125 (1991).
- 63.Iqbal, S. M., Ikram-ul-Haq, I. U. H., Ahmad Bakhsh, A. B., Ghafoor, A. & Haqqani, A. M. A. G. Screening of chickpea genotypes for resistance against Fusarium wilt. 1–5 (2005).
- 64.Kidie, Y., Tesso, B. & Amsalu, B. AMMI and GGE biplot analysis of genotype by environment interaction and yield stability in early maturing Cowpea [Vigna unguiculata (L) Walp] landraces in Ethiopia. Plant-Env. Interact.3(1), 1–9 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.