Abstract
The rapid detection and identification of Candida species in clinical laboratories are extremely important for the management of patients with hematogenous candidiasis. The presently available culture and biochemical methods for detection and species identification of Candida are time-consuming and lack the required sensitivity and specificity. In this study, we have established a seminested PCR (snPCR) using universal and species-specific primers for detection of Candida species in serum specimens. The universal outer primers amplified the 3′ end of 5.8S ribosomal DNA (rDNA) and the 5′ end of 28S rDNA, including the internally transcribed spacer 2 (ITS2), generating 350- to 410-bp fragments from the four commonly encountered Candida species, viz., C. albicans, C. tropicalis, C. glabrata, and C. parapsilosis. The species-specific primers, complementary to unique sequences within the ITS2 of each test species, amplified species-specific DNA in the reamplification step of the snPCR. The sensitivity of Candida detection by snPCR in spiked serum specimens was close to 1 organism/ml. Evaluation of snPCR for specific identification of Candida species with 76 clinical Candida isolates showed 99% concordant results with the Vitek and/or ID32C yeast identification system. Further evaluation of snPCR for detection of Candida species in sera from culture-proven (n = 12), suspected (n = 16), and superficially colonized (n = 10) patients and healthy subjects (n = 12) showed that snPCR results were consistently negative with sera from healthy individuals and colonized patients. In culture-proven candidemia patients, the snPCR results were in full agreement with blood culture results with respect to both positivity and species identity. In addition, snPCR detected candidemia due to two Candida species in five patients, compared to three by blood culture. In the category of suspected candidemia with negative blood cultures for Candida, nine patients (56%) were positive by snPCR; two of them had dual infection with C. albicans and either C. tropicalis or C. glabrata. In conclusion, the snPCR developed in this study is specific and more sensitive than culture for the detection of Candida species in serum specimens. Moreover, the improved detection of cases of candidemia caused by more than one Candida species is an additional advantage.
Nosocomial candidiasis is a major fungal infection occurring mostly in patients undergoing prolonged hospitalization due to a variety of underlying conditions (26). Bloodstream infections due to Candida are now regarded as the fourth most frequent cause of septicemia, with a mortality rate of about 50% (29). Diagnosis of candidemia or hematogenous candidiasis has been problematic due to the low positivity of blood cultures. Even in patients with autopsy-proven systemic candidiasis, the rate of recovery from blood cultures ranged between 40 and 60% (27). Although various laboratory tests based on detection of Candida-specific antibodies, antigens, or metabolites have been developed, they all suffer from lack of specificity and/or sensitivity, besides being time-consuming (36). Moreover, these tests fail to clearly discriminate between the infecting Candida species, information that is crucial for initiating specific antifungal therapy since several non-C. albicans Candida species are known to be inherently less susceptible to commonly used antifungal drugs (14, 28).
In order to overcome the limitations of conventional diagnostic tests, DNA-based methods have been developed for the detection of Candida species and offer a potentially more sensitive means of diagnosing systemic candidiasis (4, 30). The use of PCR-based tests to detect Candida DNA in body fluids has produced encouraging results (5, 13, 15, 16, 23). However, detection of Candida species by PCR lacks sensitivity when the test is performed with blood or serum specimens (5, 10). DNA amplification with universal fungal primers followed by detection using species-specific probes greatly improved the sensitivity of Candida detection (7, 10, 28, 31, 35), but probing methods involved the use of radioactivity and/or laborious and time-consuming additional steps.
In this study, we have used the species-specific primers described previously (10) for species-specific detection of PCR-amplified ribosomal DNAs (rDNAs) of four commonly encountered Candida species, i.e., C. albicans, C. parapsilosis, C. tropicalis, and C. glabrata, by Southern hybridization and enzyme-linked immunosorbent assay, to develop a seminested PCR (snPCR). By employing universal fungal primers, a portion of the 5.8S and 28S rDNAs including the intervening internally transcribed spacer (ITS2) was amplified. The amplicon so obtained was reamplified in an snPCR using species-specific primers complementary to unique sequences within the ITS2 together with a generic fungal primer. The snPCR thus developed was evaluated for its specificity and sensitivity by using clinical Candida isolates and sera of patients with suspected and proven candidemia in comparison with conventional diagnostic and identification methods.
MATERIALS AND METHODS
Reference organisms.
The reference strains used in the study were C. albicans ATCC 76615, C. parapsilosis ATCC 10233, C. tropicalis ATCC 750, C. glabrata ATCC 15545, C. dubliniensis type strain CD 36, a C. krusei clinical isolate, and a C. lusitaniae clinical isolate. In addition, bacterial strains of Escherichia coli (ATCC 25922), Staphylococcus aureus (ATCC 25923), Salmonella enterica serovar Typhimurium (clinical isolate), and Legionella pneumophila (ATCC 33152) were also included in the study. All strains, including clinical isolates, were stored at −20°C in sterile distilled water.
Clinical Candida isolates.
To test the specificity of snPCR, 76 clinical Candida isolates identified to species level with the Vitek commercial yeast identification system (bioMerieux, Marcy l'Etoile, France) were evaluated by snPCR. The sources of these Candida isolates are presented in Table 1.
TABLE 1.
Candida isolates used
| Source of isolation | No. of isolates | No. of isolates of the following Candida species as identified by Vitek:
|
|||
|---|---|---|---|---|---|
| C. albicans | C. parapsilosis | C. tropicalis | C. glabrata | ||
| Blood culture | 33 | 26 | 4 | 3 | 0 |
| Mouth swab | 9a | 2 | 1 | 3 | 2 |
| Urine | 8 | 1 | 2 | 2 | 3 |
| ET aspirate | 6 | 0 | 2 | 3 | 1 |
| Skin swab | 13 | 2 | 5 | 1 | 5 |
| Rectal swab | 7 | 1 | 2 | 1 | 3 |
One isolate was unidentified.
Subjects.
Thirty-eight patients admitted to the intensive care unit and other wards of Mubarak Al-Kabeer Hospital, Jabriya, Kuwait, were included in the study. For the purpose of these investigations, they were broadly divided into three groups. (i) Colonized patients (n = 10) were those yielding Candida species from one or more anatomic sites with no clinical suspicion of Candida infection. (ii) Patients with suspected candidiasis (n = 16) were those with three or more of the following conditions: having an extended period of hospitalization (>2 weeks), yielding Candida from one or more anatomic sites, having an inadequate response to broad-spectrum antibiotics, and having Candida antigen titers of ≥1:2 and blood cultures negative for Candida species. (iii) Candidemic patients (n = 12) were clinically suspected patients with blood cultures yielding Candida species on one or more occasions. In addition, 12 apparently healthy individuals with no complaints of oral or vaginal Candida infection were included as controls.
Culture.
Clinical specimens, such as sputum, bronchoalveolar lavage (BAL), endotracheal aspirate (ET), and urine specimens, were processed for the isolation of Candida species according to standard procedures (21). All cultures were made on Sabouraud glucose agar (SGA) (glucose [15 g], peptone [10 g], and agar [158 g] in 1 liter of distilled water, pH 6.8) at 30°C. Blood cultures were processed either with the BACTEC 9240 system (Becton Dickinson, Paramus, N.J.) or with a lysis centrifugation system (Isostat; Wampole Laboratories, Cranbury, N.J.) and the growth so obtained was subcultured on Sabouraud glucose agar plates (SGA) for further identification.
Identification.
All of the yeast isolates were examined by wet mount and tested for germ tube formation. The germ tube-positive isolates were provisionally identified as C. albicans, and their identities were further confirmed with the Vitek and ID32C yeast identification systems (bioMerieux). Likewise, all non-C. albicans Candida species were identified by both systems.
Serum samples.
Fifty serum samples, 38 from patients and 12 from healthy volunteers, were analyzed by the snPCR for the detection of Candida DNA. Blood samples were obtained in plain tubes, and sera were separated by centrifugation and stored frozen at −20°C until used.
Antigen detection.
Cand-Tec (Ramco Inc., Houston, Tex.), a latex agglutination test based on rabbit antibodies to a heat-labile antigen, was used for the detection of Candida antigen in the sera. The test was performed according to the manufacturer's instructions.
Extraction of Candida DNA from cultures and sera.
DNA was extracted from broth cultures by the method of Lee (18) with an additional step of DNA purification by extraction in phenol-chloroform (24:1). DNA from serum was extracted by the method of Sandhu et al. (31). To remove PCR inhibitors, the samples were heated with Chelex-100 (Sigma) (30 mg/ml) before precipitation of the DNA.
PCR primers.
A 22-bp forward primer, CTSF (5′-TCGCATCGATGAAGAACGCAGC-3′), and a 25-bp reverse primer, CTSR (5′-TCTTTTCCTCCGCTTATTGATATGC-3′), capable of amplifying the 3′ end of 5.8S rDNA and the 5′ end of 28S rDNA, including the intervening spacer region, were synthesized by Genemed Synthesis, Inc., San Francisco, Calif. Species-specific oligonucleotide primers for snPCR were derived from the ITS2 regions of C. albicans (CADET, 5′-ATTGCTTGCGGCGGTAACGTCC-3′), C. parapsilosis (CPDET, 5′-ACAAACTCCAAAACTTCTTCCA-3′), C. tropicalis (CTDET, 5′-AACGCTTATTTTGCTAGTGGCC-3′), and C. glabrata (CGDET, 5′-TAGGTTTTACCAACTCGGTGTT-3′) (10).
DNA amplification and detection.
Amplification of target DNA was carried out in thin-walled 0.2-ml PCR tubes in a total volume of 50 μl containing 1× AmpliTaq PCR buffer I, 1 U of AmpliTaq DNA polymerase, 10 pmol each of CTSF and CTSR primers, 1 μl of DNA extracted from culture or 5 μl of DNA extracted from serum, and 0.1 mM each deoxynucleoside triphosphate. After amplification in the first step, 1 μl of the product was further amplified using the initial reverse primer (CTS1R) and a species-specific forward primer in four separate tubes corresponding to each of the Candida species to be detected. For snPCR, the reaction mixture consisted of 1× AmpliTaq PCR buffer I; 1 U of AmpliTaq DNA polymerase; 5 pmol of CTSR together with 5 pmol of CADET, CPDET, CGDET, or CTDET; 1 μl of the first PCR product; and 0.1 mM each deoxynucleoside triphosphate. All reagents except primers were obtained from Perkin-Elmer Corp., Norwalk, Conn. PCR cycling was carried out in a Perkin-Elmer cycler (GeneAmp PCR system 2400) under the following conditions: denaturation at 94°C for 1 min, annealing at 60°C for 30 s, and extension at 72°C for 1 min. An initial denaturation step at 94°C for 3 min and a final extension step at 72°C for 10 min were also included. Optimum amplification was determined to be obtained with 30 cycles of the first PCR followed by 20 cycles of the snPCR for DNA extracted from broth cultures and with 35 cycles followed by 25 cycles for DNA extracted from sera. To avoid the risk of contamination of PCR samples, the precautions and guidelines advocated by Kwok and Higuchi (17) were followed. The area where the PCR mixtures were prepared was physically separated from the laboratory where DNA extraction was performed. Amplicon carryover was prevented by using aerosol-guarded pipette tips. Appropriate negative controls were included in each test run, including controls omitting the DNA template during PCR assays.
To detect amplified DNA fragments, agarose gel electrophoresis was performed as described previously (16). The gels were exposed to UV light and photographed. The sizes of amplified DNA fragments were identified by comparison with molecular size marker DNA (100-bp DNA ladder).
RESULTS
Standardization of snPCR.
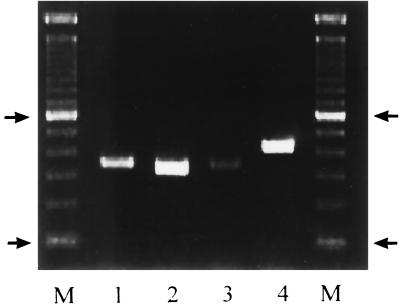
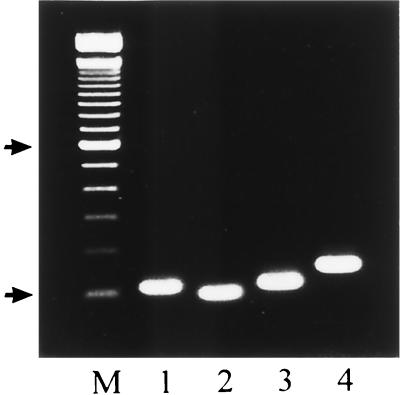
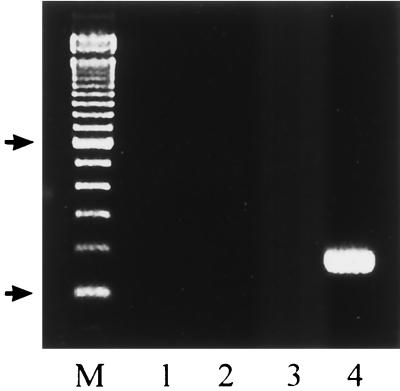
The PCR amplification of rDNAs from the four Candida species, viz., C. albicans, C. parapsilosis, C. tropicalis, and C. glabrata, using universal fungal primers (CTSF and CTSR) resulted in amplification of a single DNA fragment of the expected size (Fig. 1) (2, 10). Similar results were obtained when genomic DNAs prepared from C. krusei, C. lusitaniae, and C. dubliniensis were used as templates (data not shown). Reamplification of the product of the first PCR with CTSR and the species-specific primers corresponding to the ITS2 sequences from C. albicans, C. parapsilosis, C. tropicalis, and C. glabrata resulted in specific amplification of single DNA products of the expected sizes (Fig. 2). For example, CTSR and CGDET amplified a ∼140-bp product in snPCR only when the first PCR was performed with template DNA from C. glabrata and not when it was performed with DNAs from C. albicans, C. parapsilosis, and C. tropicalis (Fig. 3), as well as C. krusei, C. lusitaniae, and C. dubliniensis (data not shown). Similar results were obtained with other primer combinations for specific detection of C. albicans, C. parapsilosis, and C. tropicalis (data not shown). No amplification was detected with genomic DNAs from E. coli, S. aureus, L. pneumophila, S. enterica serovar Typhimurium, and a human cell line in the first as well as the second step of snPCR (data not shown). The results of these experiments established the species specificity of the snPCR.
FIG. 1.
PCR amplification of genomic DNAs of C. albicans (lane1), C. parapsilosis (lane 2), C. tropicalis (lane 3), and C. glabrata (lane 4) with universal fungal primers. Lanes M, 100-bp molecular size marker. Arrows indicate positions of 100 and 600 bp in ascending order.
FIG. 2.
Lanes 1 to 4, snPCR amplification of DNAs from C. albicans, C. parapsilosis, C. tropicalis, and C. glabrata, respectively, using primer CTSR with primers CADET, CPDET, CTDET, and CGDET, respectively. Lane M, 100-bp molecular size marker. Arrows indicate positions of 100 and 600 bp in ascending order.
FIG. 3.
Lanes 1 to 4, snPCR amplification using primers CTSR and CGDET and DNAs from C. albicans, C. parapsilosis, C. tropicalis, and C. glabrata, respectively. Lane M, 100-bp molecular size marker. Arrows indicate positions of 100 and 600 bp in ascending order.
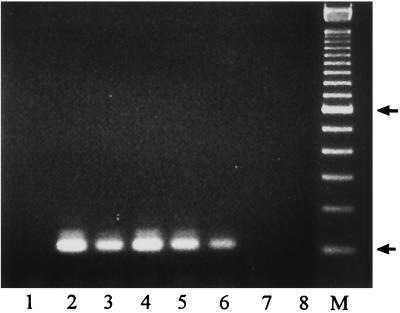
To determine the sensitivity of snPCR for detection of Candida in clinical specimens, experiments were performed with total DNA isolated from serum specimens from a healthy individual spiked with different concentrations of C. albicans DNA. The results showed that snPCR was positive in specimens spiked with 4 fg of C. albicans DNA isolated from 200-μl spiked specimens (Fig. 4). The sensitivity of snPCR was therefore 20 fg of DNA/ml, which is equivalent to one C. albicans genome per milliliter of serum.
FIG. 4.
Lanes 2 to 8, snPCR using DNA extracted from serum spiked with 40 and 4 pg and 400, 40, 4, 0.4, and 0.04 fg of C. albicans rDNA, respectively. Lane 1, negative control with water in place of template DNA. Lane M, 100-bp molecular size marker. Arrows indicate positions of 100 and 600 bp in ascending order.
Species identification of Candida isolates by snPCR.
The specificity of the snPCR was further investigated in species identification with 76 clinical Candida isolates. These isolates were first identified to species level with the Vitek and ID32C identification systems (Tables 1 and 2). The results of snPCR in species identification were concordant with those of the Vitek and ID32C systems for 68 of 76 (89%) and 71 of 76 (93%) isolates, respectively. The identification results for the 13 isolates showing discrepancy between snPCR and one or both biochemical tests are listed in Table 3. The results further showed that compared to snPCR, the discordant results in the identification of C. albicans were more marked between Vitek and ID32C (Table 2). It may be noted that the species identification of only one isolate was different by snPCR from that of both the Vitek and ID32C (Tables 3 and 4). Thus, the overall concordance of the snPCR compared to the two biochemical tests together in the identification of Candida species can be taken to be 75 of 76 or ∼99%.
TABLE 2.
Identification of clinical Candida isolates by commercial assimilation methods and agreement with snPCR
| Candida species | No. of isolates identified by:
|
% Agreement with PCRa
|
|||
|---|---|---|---|---|---|
| PCR | Vitek | ID32C | Vitek | ID32C | |
| C. albicans | 27 | 32 | 23 | 84 | 85 |
| C. parapsilosis | 20 | 17 | 20 | 85 | 100 |
| C. tropicalis | 14 | 13 | 13 | 93 | 93 |
| C. glabrata | 15 | 13 | 15 | 87 | 100 |
Percent agreement = (number positive by PCR/number positive by Vitek-ID32C) × 100. However, if the number of isolates positive by Vitek or ID32C exceeded the number of PCR-positive isolates, percent agreement = number positive by Vitek or ID32C/number of PCR-positive isolates × 100.
TABLE 3.
Candida isolates identified differently by snPCR and by the Vitek and ID32C methods
| Isolate | Identification by:
|
Germ tube formation | ||
|---|---|---|---|---|
| PCR | ID32Ca | Vitekb | ||
| 431/98c | C. tropicalis | C. parapsilosis | C. parapsilosis | − |
| 82/97 (626) | C. albicans | C. pelliculosa | C. albicans | + |
| 82/97 (660) | C. albicans | C. pelliculosa | C. albicans | + |
| 602/99 | C. albicans | Unidentified | C. albicans | + |
| 548/98 | C. tropicalis | C. tropicalis | C. albicans | − |
| 829/98 | C. parapsilosis | C. parapsilosis | C. albicans | − |
| 576/99 | C. albicans | Unidentified | C. albicans | + |
| 706/2k | C. parapsilosis | C. famata | C. parapsilosis | − |
| P1R(3) | C. parapsilosis | C. parapsilosis | C. albicans | − |
| P1M(5) | C. parapsilosis | C. parapsilosis | C. albicans | − |
| P1G(3) | C. parapsilosis | C. parapsilosis | C. albicans | − |
| P26U(7)B | C. glabrata | C. glabrata | C. tropicalis | − |
| P34M(1)B | C. glabrata | C. glabrata | Unidentified | − |
Read at 48 h.
Read at 24 and 48 h.
The identification was reconfirmed as C. tropicalis by PCR with species-specific primers amplifying the gene encoding P450 lanosterol-α-demethylase (16).
TABLE 4.
Agreement between snPCR and biochemical test results
| Candida species | No. of clinical isolates identified by:
|
% Agreement | |
|---|---|---|---|
| snPCR | Vitek and ID32C | ||
| C. albicans | 27 | 27 | 100 |
| C. parapsilosis | 19 | 20a | 95 |
| C. tropicalis | 15a | 14 | 93 |
| C. glabrata | 15 | 15 | 100 |
One isolate was identified as C. tropicalis by snPCR and as C. parapsilosis by both biochemical tests.
Comparison of blood culture versus snPCR.
The comparative results of blood culture and the snPCR for the detection of Candida species are presented in Table 5. None of the blood or serum samples from 12 healthy volunteers were positive for Candida by culture or snPCR. Similarly, culture and snPCR from sera were negative in all 10 patients colonized with Candida species (Table 5). Among 16 clinically suspected cases of candidemia or hematogenous candidiasis, 9 (∼56%) yielded positive results by snPCR, while blood cultures remained negative for all of them. Six of the positive snPCR results were due to C. albicans, one was due to C. parapsilosis, and two were due to dual infection, i.e., C. albicans with C. tropicalis and C. tropicalis with C. glabrata (Table 5). All of the snPCR-positive patients had the corresponding Candida species isolated from one or more specimens other than blood (Table 5). Ten of the 16 patients demonstrated Candida antigen titers ranging from 1:2 to 1:16. Of the nine snPCR-positive patients, one demonstrated antigen titers of 1:16, two had antigen titers of 1:8, one each had antigen titers of 1:4 and 1:2, and four were negative (Table 5).
TABLE 5.
Comparative results of blood cultures and snPCR
| Candida status | Subject no. | Age/sexa | Underlying conditionb | Source or site of Candida isolation | Candida spp. isolated | Antigen titerc | Blood culture resultsc | snPCR resultsc |
|---|---|---|---|---|---|---|---|---|
| Colonized | 1 | 65/F | Allergy | Oral | C. albicans | − | − | − |
| 2 | 78/M | CVA | Urine | C. tropicalis | − | − | − | |
| 3 | 88/M | CVA | Urine | C. tropicalis | − | − | − | |
| 4 | 6 mo/F | Chest infection | ET | C. albicans | 1:2 | − | − | |
| 5 | 70/F | Chest infection | Urine | C. glabrata | − | − | − | |
| 6 | 30/F | UTI | Urine | C. albicans | − | − | − | |
| 7 | 35/F | Vaginitis | Vaginal swab | C. albicans | 1:2 | − | − | |
| 8 | 42/F | Vaginitis | Vaginal swab | C. albicans | − | − | − | |
| 9 | 29/F | Vaginitis | Vaginal swab | C. albicans. C. glabrata | − | − | − | |
| 10 | 38/F | Vaginitis | Vaginal swab | C. albicans | − | − | − | |
| Suspected candidiasis | 11 | 35/M | RTA, head trauma | ET | C. albicans | − | − | C. albicans |
| 12 | 17/M | Head trauma, epilepsy | Urine | C. albicans | − | − | − | |
| 13 | 66/M | Chest infection, chronic renal failure | Urine; BAL | C. tropicalis, C. tropicalis, C. glabrata | 1:4 | − | − | |
| 14 | 44/F | Bronchial asthma | Sputum | C. albicans | 1:8 | − | − | |
| 15 | 36/M | RTR | Sputum | C. albicans | 1:4 | − | C. albicans | |
| 16 | 45/F | RTR, chest infection | Urine; ET | C. parapsilosis, C. krusei | 1:4 | − | − | |
| 17 | 45/M | RTR, chest infection | Urine; pus swab | C. tropicalis, C. glabrata, C. albicans | 1:8 | − | C. albicans | |
| 18 | 52/M | RTR, pneumonia | Urine | C. albicans | 1:8 | − | − | |
| 19 | 80/M | Septicemia, fever | Skin swab | C. albicans | − | − | C. albicans, C. tropicalis | |
| 20 | 85/F | Fever of unidentified origin | Skin swab | C. parapsilosis | − | − | C. parapsilosis | |
| 21 | 52/F | Chest infection | Urine, ET | C. albicans, C. albicans | 1:8 | − | C. albicans | |
| 22 | 45/M | RTR, pneumonia | BAL | C. albicans | − | − | − | |
| 23 | 45/M | RTR | Urine | C. albicans | 1:4 | − | − | |
| 24 | 41/M | Burns | Skin swab | C. tropicalis | 1:2 | − | C. tropicalis, C. glabrata | |
| 25 | 72/M | Cholecystectomy | ET | C. albicans | 1:16 | − | C. albicans | |
| 26 | 70/F | Pulmonary edema | Urine | C. albicans | − | − | C. albicans | |
| Candidemic | 27 | 48/M | DM, HTN, CVA | Urine | C. albicans | 1:8 | C. albicans | C. albicans |
| 28 | 77/F | Bronchial asthma, pulmonary embolism | Urine, ET | C. glabrata, C. glabrata | 1:2 | C. parapsilosis | C. parapsilosis, C. glabrata | |
| 29 | 7/M | Chest infection, fever | − | − | 1:8 | C. albicans, C. parapsilosis | C. albicans, C. parapsilosis | |
| 30 | 70/F | DM, HTN, LVF | Urine | C. glabrata | 1:2 | C. glabrata | C. glabrata | |
| 31 | 64/F | HTN, SAH | Urine | C. albicans | 1:2 | C. tropicalis, C. albicans | C. tropicalis, C. albicans | |
| 32 | 1 mo/F | Premature baby | − | − | − | C. parapsilosis | C. parapsilosis | |
| 33 | 50/M | Septicemia, fever | Urine | C. albicans | 1:2 | C. albicans | C. albicans | |
| 34 | 29/F | Chronic renal failure | − | − | − | C. albicans (Vitek), C. parapsilosis (ID32C) | C. parapsilosis | |
| 35 | 67/F | Tracheostomy, DM | − | − | − | C. albicans | C. albicans | |
| 36 | 60/F | Brain abscess | − | − | − | C. albicans | C. albicans, C. tropicalis | |
| 37 | 3 mo/F | Lung infection | − | − | − | C. albicans | C. albicans | |
| 38 | 74/F | LVF, CVA, rectal ulcer | Urine | C. tropicalis | 1:4 | C. albicans, C. tropicalis | C. albicans, C. tropicalis |
Ages are in years unless otherwise indicated. F, female; M, male.
RTA, road traffic accident; DM, diabetes mellitus; UTI, urinary tract infection; RTR, renal transplant; LVF, left ventricular failure; CVA, cardiovascular accident; SAH, subarachinoid hemorrhage; HTN, hypertension.
−, negative.
All of the 12 patients with culture-proven candidemia yielded positive results by snPCR for the corresponding Candida species, with 100% concordance (Table 5). The infecting species identified by blood culture included C. albicans in nine patients, C. parapsilosis and C. tropicalis in two patients each and C. glabrata in one patient. The results of snPCR revealed that five of the patients were infected with more than one Candida species, as against three detected by blood culture. Only 7 of the 12 patients had demonstrable antigen titers: 1:8 in 2 patients, 1:4 in 1 patient, and 1:2 in 4 patients (Table 5). In case 34, Vitek identified the Candida isolate as C. albicans whereas the snPCR with serum identified the species as C. parapsilosis. Further identification of the blood culture isolate by ID32C and snPCR (using genomic DNA purified from the culture) established the species to be C. parapsilosis (Table 5).
DISCUSSION
In the present study, snPCR assays targeting species-specific sequences in the rDNA have been established for the specific detection of four clinically important Candida species by using reference strains. These snPCRs were evaluated for Candida species identification with clinical isolates and for direct detection as well as specific identification of Candida species in serum samples from patients. The target for snPCR amplification was rDNA. Although PCR assays with several other target sequences have been reported in the literature (22, 30), the use of rDNA for sensitive detection of Candida was considered to be most suitable because it is present in multiple copies (50 to 100 copies) per Candida genome (30), and PCR assays with multiple-copy targets are usually more sensitive than those with single-copy targets (22, 30). Moreover, between the highly conserved rDNA subunits are the internally transcribed spacers, which contain sequences unique to each Candida species, and thus the use of primers corresponding to these regions facilitates species identification. Although several PCR assays based on amplification of the rDNA have been recently reported, species identification usually involved further manipulation of the amplified products, i.e., restriction enzyme digestion and analysis (24, 37), use of radioactive and enzyme-labeled probes (2, 10, 15, 20, 23, 32, 33, 35), and DNA sequencing (25). Many of these procedures required prolonged hybridization times and the use of hazardous radioactive materials. Moreover, species identification by the use of a biotinylated probe and detection by enzyme immunoassay showed lower specificity for C. glabrata (10). In contrast, the snPCR established in this study has an average processing time of 9 to 10 h, does not require the use of hybridization probes and radioactive substances, and is specific for the detection of all four Candida species tested. More recently, multiplex PCR assays, targeting rDNA and spacer regions, to detect Candida species have been established (3, 11). However, for a positive result, multiplex PCRs required a minimum of 20 cells, compared to 0.2 cell for snPCR. Thus, the snPCR appears to be at least 100 times more sensitive than the multiplex PCRs. Also, the multiplex PCR assays have not yet been evaluated for their potential use in the diagnosis of Candida infection directly with clinical specimens.
The specificity of the snPCR was established with 99% accuracy, as it correctly identified 75 of the 76 Candida isolates previously identified to species level by conventional biochemical methods. However, a solitary blood culture isolate identified as C. parapsilosis by Vitek and ID32C was shown to be C. tropicalis by snPCR. Although the precise reason for this discrepancy remains unclear, it could be related to the inadequacy of the presently available commercial yeast identification systems (9). While several investigators have used PCR to identify Candida species, their studies have been mostly limited to the use of reference strains and clinical isolates (12, 16, 28). In such studies, there has been no comparison of the identification accuracy between PCR and one or more biochemical methods. This is despite the fact that several studies have shown that commercial yeast identification systems misidentify some percentage of clinical yeast isolates (6, 8, 9, 19, 34). The same is also apparent in the present investigation (Tables 2 and 3).
The snPCR assay was subsequently applied for Candida detection and species identification directly in clinical specimens. Consistent with the results of other investigators (15, 35), our results also showed that all of the sera from healthy volunteers were PCR negative. In addition, sera of patients colonized with Candida spp., with no suggestive clinical indications for systemic candidiasis, also yielded uniformly negative results. This observation suggests that patients with superficial or mucosal colonization (oral thrush and vaginitis, etc.) may not give rise to detectable levels of Candida DNA in the serum, thereby reducing the possibility of false positivity. However, 9 (56%) of the 16 blood culture-negative patients clinically suspected of having invasive candidiasis yielded positive snPCR results. In two of these patients, snPCR was positive for two Candida species (Table 5). Interestingly, all of the snPCR-positive patients had the respective Candida species isolated from one or more anatomic sites. In addition to supporting the diagnosis of candidiasis in the nine suspected cases, snPCR provided specific information about the incriminating Candida species. Considering the detection limit of 0.2 Candida genome and the possibility of detecting nonviable cells, the increased sensitivity of the snPCR compared to blood cultures in patients with suspected candidiasis is understandable. This is also consistent with several other studies where PCR has been found to be more sensitive than conventional culture methods in the diagnosis of Candida infections (5, 16, 35).
When applied to serum samples from 12 blood culture-proven cases of candidemia, the snPCR showed 100% concordant results and provided evidence of dual infections in 5 patients (41%). The therapeutic implications of this finding are quite apparent, since Candida species may have different antifungal susceptibility profiles. It also underscores the fact that the occurrence of candidemia due to more than one species may be more frequent than what is reported in the literature (1). In a study on the epidemiology of hematogenous candidiasis (1), 20 (4%) of the 491 episodes investigated had ≥2 species isolated in blood cultures. Besides C. albicans, 18 of these patients had coinfection with C. parapsilosis, C. tropicalis, and C. glabrata, while the remaining two had C. tropicalis isolated with C. parapsilosis or C. glabrata. Consistent with these observations, among the five PCR-positive candidemic patients identified in this study, three had concomitant infection with C. albicans and C. tropicalis, one had concomitant infection with C. parapsilosis and C. glabrata, and one had concomitant infection with C. albicans and C. parapsilosis. Since different Candida species have similar colonial morphological appearances on routine culture media, such as SGA, cases of candidemia caused by more than one species could be missed, especially if one of the coinfecting species yields only a few colonies. Probably, like polymicrobial (bacterial) septicemias, Candida infection involving more than one species may also have its origin from the gastrointestinal tract. Considering the various susceptibilities of Candida species to antifungal agents (more so with azoles), the problem of candidemia due to multiple species may be encountered with greater frequency in the future. A further point to note is that there appeared to be no correlation between Candida antigen titers and culture or snPCR positivity. Although our results are consistent with a previous study using a murine model of systemic candidiasis (16), the kit used for antigen detection in that study is known to have low sensitivity.
In conclusion, the snPCR that was established and evaluated in this study is a specific and sensitive method for the diagnosis of candidemia or hematogenous candidiasis caused by the four most commonly encountered Candida species, i.e., C. albicans, C. parapsilosis, C. tropicalis, and C. glabrata. Besides being rapid, the snPCR has the added advantage of identifying patients infected with more than one Candida species. This information can facilitate the selection of appropriate therapeutic agents.
Acknowledgments
We are thankful to T. D. Chugh for his encouragement and to Rachel Chandy and Da'ad Farhat for excellent technical assistance.
This work was supported by Kuwait University grant no. MPI 118.
REFERENCES
- 1.Abi-Said, D., E. Anaissie, O. Uzun, I. Raad, H. Pinzcowski, and S. Vartivarian. 1997. The epidemiology of hematogenous candidiasis caused by different Candida species. Clin. Infect. Dis. 24:1122-1128. [DOI] [PubMed] [Google Scholar]
- 2.Botelho, A. R., and R. J. Planta. 1994. Specific identification of Candida albicans by hybridization with oligonucleotides derived from ribosomal DNA internal spacers. Yeast 10:709-717. [DOI] [PubMed] [Google Scholar]
- 3.Chang, H. C., S. N. Leaw, A. H. Huang, T. L. Whu, and T. C. Chang. 2001. Rapid identification of yeasts in positive blood cultures by a multiplex PCR method. J. Clin. Microbiol. 39:3466-3471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cheung, L. L., and J. B. Hudson. 1988. Development of DNA probes for Candida albicans. Diagn. Microbiol. Infect. Dis. 10:171-179. [DOI] [PubMed] [Google Scholar]
- 5.Chryssanthou, E., B. Andersson, B. Petrini, S. Lofdahl, and J. Tollemar. 1994. Detection of Candida albicans DNA in serum by polymerase chain reaction. Scand. J. Infect. Dis. 26:479-485. [DOI] [PubMed] [Google Scholar]
- 6.Crist, A. E., Jr., L. M. Johnson, and P. J. Burke. 1996. Evaluation of the Microbial Identification System for identification of clinically isolated yeasts. J. Clin. Microbiol. 34:2408-2410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Einsele, H., H. Hebart, G. Roller, J. Loffler, I. Rothenhofer, C. A. Muller, R. A. Bowden, J. van Burik, D. Engelhard, L. Kanz, and U. Schumacher. 1997. Detection and identification of fungal pathogens in blood by using molecular probes. J. Clin. Microbiol. 35:1353-1360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Espinel-Ingroff, A., L. Stockman, G. Roberts, D. Pincus, J. Pollack, and J. Marler. 1998. Comparison of RapID yeast plus system with API 20C system for identification of common, new, and emerging yeast pathogens. J. Clin. Microbiol. 36:883-886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fenn, J. P., H. Segal, B. Barland, D. Denton, J. Whisenant, H. Chun, K. Christofferson, L. Hamilton, and K. Carroll. 1994. Comparison of updated Vitek Yeast Biochemical Card and API 20C yeast identification systems. J. Clin. Microbiol. 32:1184-1187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fujita, S., B. A. Lasker, T. J. Lott, E. Reiss, and C. J. Morrison. 1995. Microtitration plate enzyme immunoassay to detect PCR-amplified DNA from Candida species in blood. J. Clin. Microbiol. 33:962-967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fujita, S., Y. Senda, S. Nakaguchi, and T. Hashimoto. 2001. Multiplex PCR using internal transcribed spacer 1 and 2 regions for rapid detection and identification of yeast strains. J. Clin. Microbiol. 39:3617-3622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Guiver, M., K. Levi, and B. A. Oppenheim. 2001. Rapid identification of Candida species by TaqMan PCR. J. Clin. Pathol. 54:362-366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Haynes, K. A., T. J. Westerneng, J. W. Fell, and W. Moens. 1995. Rapid detection and identification of pathogenic fungi by polymerase chain reaction amplification of large subunit ribosomal DNA. Med. Vet. Mycol. 33:319-325. [DOI] [PubMed] [Google Scholar]
- 14.Johnson, E. M., D. W. Warnock, J. Luker, S. R. Porter, and C. Scully. 1995. Emergence of azole drug resistance in Candida species from HIV-infected patients receiving prolonged fluconazole therapy for oral candidosis. J. Antimicrob. Chemother. 35:103-114. [DOI] [PubMed] [Google Scholar]
- 15.Kan, V. L. 1993. Polymerase chain reaction for the diagnosis of candidemia. J. Infect. Dis. 168:779-783. [DOI] [PubMed] [Google Scholar]
- 16.Khan, Z. U., and A. S. Mustafa. 2001. Detection of Candida species by polymerase chain reaction (PCR) in blood samples of experimentally infected mice and patients with suspected candidemia. Microbiol. Res. 156:95-102. [DOI] [PubMed] [Google Scholar]
- 17.Kwok, S., and R. Higuchi. 1989. Avoiding false positives with PCR. Nature (London) 339:237-238. [DOI] [PubMed] [Google Scholar]
- 18.Lee, F. J. S. 1992. Modified protocol for yeast DNA mini-preparation. BioTechniques 5:677.. [PubMed] [Google Scholar]
- 19.Lo, H. J., Y. A. Ho, and M. Ho. 2001. Factors accounting for misidentification of Candida species. J. Microbiol. Immunol. Infect. 34:171-177. [PubMed] [Google Scholar]
- 20.Martin, C., D. Roberts, M. van Der Weide, R. Rossau, G. Jannes, T. Smith, and M. Maher. 2000. Development of a PCR-based line probe assay for identification of fungal pathogens. J. Clin. Microbiol. 38:3735-3737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.McGinnis, M. R. 1994. Mycology, p. 6.1-6.12. In H. D. Isenberg (ed.), Clinical microbiology procedure handbook. American Society for Microbiology, Washington D.C.
- 22.Mitchell, T. G., R. L. Sandin, B. H. Bowman, W. Meyer, and W. G. Merz. 1994. Molecular mycology: DNA probes and applications of PCR technology. J. Med. Vet. Mycol. 32(Suppl. 1):351-366. [DOI] [PubMed] [Google Scholar]
- 23.Miyakawa, Y., T. Mabuchi, and Y. Fukazawa. 1993. New method for detection of Candida albicans in human blood by polymerase chain reaction. J. Clin. Microbiol. 31:3344-3347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Morace, G., L. Pagano, M. Sanguinetti, B. Posteraro, L. Mele, F. Equitani, G. D'Amore, G. Leone, and G. Fadda. 1999. PCR-restriction enzyme analysis for detection of Candida DNA in blood from febrile patients with hematological malignancies. J. Clin. Microbiol. 37:1871-1875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Niesters, H. G., W. H. Goessens, J. F. Meis, and W. G. Quint. 1993. Rapid, polymerase chain reaction-based identification assays for Candida species. J. Clin. Microbiol. 31:904-910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pfaller, M. A. 1996. Nosocomial candidiasis: emerging species, reservoirs, and modes of transmission. Clin. Infect. Dis. 22(Suppl. 2):S89-S94. [DOI] [PubMed]
- 27.Pizzo, P. A., and T. J. Walsh. 1990. Fungal infections in the pediatric cancer patient. Semin. Oncol. 173(Suppl. 6):6-9. [PubMed] [Google Scholar]
- 28.Posteraro, B., M. Sanguinetti, L. Masucci, L. Romano, G. Morace, and G. Fadda. 2000. Reverse cross blot hybridization assay for rapid detection of PCR-amplified DNA from Candida species, Cryptococcus neoformans, and Saccharomyces cerevisiae in clinical samples. J. Clin. Microbiol. 38:1609-1614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Reiss, E., and C. J. Morrison. 1993. Nonculture methods for diagnosis of disseminated candidiasis. Clin. Microbiol. Rev. 6:311-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Reiss, E., K. Tanaka, G. Bruker, V. Chazalet, D. Coleman, J. P. Debeaupuis, R. Hanazawa, J. P. Latge, J. Lortholary, K. Makimura, C. J. Morrison, S. Y. Murayama, S. Naoe, S. Paris, J. Sarfati, K. Shibuya, D. Sullivan, K. Uchida, and H. Yamaguchi. 1998. Molecular diagnosis and epidemiology of fungal infections. Med. Mycol. 36(Suppl. 1):249-257. [PubMed] [Google Scholar]
- 31.Sandhu, G. S., B. C. Kline, L. Stockman, and G. D. Roberts. 1995. Molecular probes for diagnosis of fungal infections. J. Clin. Microbiol. 33:2913-2919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shin, J. H., F. S. Nolte, and C. J. Morrison. 1997. Rapid identification of Candida species in blood cultures by a clinically useful PCR method. J. Clin. Microbiol. 35:1454-1459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.van Deventer, A. J., W. H. Goessens, A. van Belkum, H. J. van Vliet, E. W. van Etten, and H. A. Verbrugh. 1995. Improved detection of Candida albicans by PCR in blood of neutropenic mice with systemic candidiasis. J. Clin. Microbiol. 33:625-628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Verweij, P. E., I. M. Breuker, A. J. Rijs, and J. F. Meis. 1999. Comparative study of seven commercial yeast identification systems. J. Clin. Pathol. 52:271-273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wahyuningsih, R., H. J. Freisleben, H. G. Sonntag, and P. Schnitzler. 2000. Simple and rapid detection of Candida albicans DNA in serum by PCR for diagnosis of invasive candidiasis. J. Clin. Microbiol. 38:3016-3021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Walsh, T. J., and S. J. Chancock. 1997. Laboratory diagnosis of invasive candidasis: a rationale for complementary use of culture and non-culture based detection systems. Int. J. Infect. Dis. 1(Suppl.):511-519.
- 37.Williams, D. W., M. J. Wilson, M. A. Lewis, and A. J. Potts. 1995. Identification of Candida species by PCR and restriction fragment length polymorphism analysis of intergenic spacer regions of ribosomal DNA. J. Clin. Microbiol. 33:2476-2479. [DOI] [PMC free article] [PubMed] [Google Scholar]