Abstract
The incidence of food-borne salmonellosis due to Salmonella enterica serotype Weltevreden is reported to be on the increase in Malaysia. The pulsed-field gel electrophoresis (PFGE) subtyping method was used to assess the extent of genetic diversity and clonality of Salmonella serotype Weltevreden strains from humans and the environment. PFGE of XbaI-digested chromosomal DNA from 95 strains of Salmonella serotype Weltevreden gave 39 distinct profiles with a wide range of Dice coefficients (0.27 to 1.00), indicating that PFGE is very discriminative and that multiple clones of Salmonella serotype Weltevreden exist among clinical and environmental isolates. Strains of one dominant pulsotype (pulsotype X1/X2) appeared to be endemic in this region, as they were consistently recovered from humans with salmonellosis between 1996 and 2001 and from raw vegetables. In addition, the sharing of similar PFGE profiles among isolates from humans, vegetables, and beef provides indirect evidence of the possible transmission of salmonellosis from contaminated raw vegetables and meat to humans. Furthermore, the recurrence of PFGE profile X21 among isolates found in samples of vegetables from one wet market indicated the persistence of this clone. The environment in the wet markets may represent a major source of cross-contamination of vegetables with Salmonella serotype Weltevreden. Antibiotic sensitivity tests showed that the clinical isolates of Salmonella serotype Weltevreden remained drug sensitive but that the vegetable isolates were resistant to at least two antibiotics. To the best of our knowledge, this is the first study to compare clinical and environmental isolates of Salmonella serotype Weltevreden in Malaysia.
Acute gastroenteritis caused by Salmonella enterica continues to be a worldwide public health problem. The Centers for Disease Control and Prevention has estimated that food-borne Salmonella infections are responsible for 1.3 million illnesses annually worldwide, resulting in 16,000 hospitalizations and 600 deaths. In Malaysia, of 8,640 cases of food poisoning reported by the Ministry of Health for the year 1999, 811 (9.4%) were due to Salmonella infections. The two predominant agents associated with food-borne nontyphoidal salmonellosis were S. enterica serotype Enteritidis and S. enterica serotype Typhimurium (10, 23). S. enterica serotype Typhimurium was the serotype most often isolated in a survey of animals conducted over a 10-year period from 1966 to 1975 (11). However, during the next decade, Salmonella serotype Weltevreden became the most commonly isolated serotype, having been isolated from cattle, beef, mutton, duck, prawn, dog, monkey, and rat (12). A report on the prevalence of nontyphoidal Salmonella by the Institute of Medical Research of Malaysia during the period from 1989 to 1994 indicated that Salmonella serotype Weltevreden was the most common serotype isolated (31% [2,163 of 6,937] of the Salmonella isolates recovered) (23). In addition to the importance of raw and undercooked meat, poultry, eggs, and dairy products as potential vehicles of human salmonellosis, there are increasing reports of outbreaks associated with fresh fruits and vegetables. An outbreak of 492 cases of S. enterica serotype Bovismorbificans infections that occurred in Finland and Sweden in 1994 was due to fresh alfalfa sprouts germinated from seeds imported from Australia (18). More recently identified vehicles include tomatoes, cantaloupes, watermelons, fresh unpasteurized orange juice, and mangoes (8). Ait Melloul et al. (2) demonstrated that contamination of vegetables with Salmonella is common in areas where raw, untreated wastewater is used for irrigation.
The emergence of antimicrobial-resistant Salmonella strains is of great concern worldwide (19). Furthermore, the widespread use of antimicrobials in humans and animals is often implicated in the emergence of multidrug-resistant strains of S. enterica. A link between the agricultural use of antibiotics and drug-resistant strains in human infections has been suggested (17). A recent study by Chee-Sanford et al. (7) demonstrated that the prevalence of tet genes in the environment is the result of agriculture and that groundwater could be a potential source of antibiotic resistance in the food chain. Generally, the clinical Salmonella serotypes prevalent in Malaysia remain sensitive to the commonly prescribed antibiotics. A survey of human salmonellosis carried out by the Salmonella Reference Centre at the Institute for Medical Research from 1989 to 1994 showed that the proportion of drug-resistant S. enterica serotype Enteritidis strains increased slightly from 8.5% (5 of 59) to 11.6% (31 of 267) (23). Analysis of the Salmonella serotypes isolated from children in the University Hospital, Kuala Lumpur, Malaysia, between 1994 and 1996 also showed that the strains remained highly sensitive to commonly prescribed antibiotics (14). Antibiotic therapy is usually not recommended for routine treatment of salmonellosis; however, appropriate antimicrobial therapy can be lifesaving for patients with invasive disease (5). Hence, Salmonella isolates from both humans and the environment or animals should be monitored for antimicrobial resistance.
Molecular subtyping of Salmonella isolates is an invaluable epidemiological tool that can be used to track the source of infection and to determine the epidemiological link between isolates from human and environmental sources. Pulsed-field gel electrophoresis (PFGE) provides information that can be used to evaluate epidemiological associations with a high degree of confidence. PFGE has been shown to be very discriminative compared to serotyping, ribotyping, or other restriction fragment length polymorphism methods (13, 16, 22). Hence, the objective of this study was to determine the extent of genetic variation and clonality among the clinical and environmental isolates of Salmonella serotype Weltevreden from different parts of Malaysia by using PFGE. In view of the increasing incidence of gastroenteritis due to Salmonella serotype Weltevreden in Malaysia, it is timely to establish the molecular typing data for this important Salmonella serovar from various sources. The molecular typing data will be useful in allowing investigators to recognize and identify new infectious strains from outbreaks or sporadic cases of gastroenteritis.
MATERIALS AND METHODS
Bacterial strains.
A total of 95 isolates of Salmonella serotype Weltevreden from humans (n = 40), uncooked vegetables (n = 40), well water (n = 8), beef (n = 3), raw meat (n = 1), and poultry products (chicken liver, n = 2; spicy chicken, n = 1) were analyzed. The 40 clinical isolates (isolates SW1 to SW18, SW20, SW22, SW23, and SW30 to SW49) were from the feces of patients with sporadic cases of gastroenteritis admitted to various public hospitals in different parts of Malaysia from 1996 to 2001. The 40 vegetables isolates (isolates SWE1 to SWE11, and SWE13 to SWE41) were recently sampled (from 1999 to 2001) from four different types of local vegetables, that is, kangkong (Ipomoea aquatica), selom (Oenanthe stolonifera), pegaga (Centella asiatica), and kesum (Polygonum minus). These vegetables are commonly eaten raw as ulam (salad) or moderately cooked. These vegetables were purchased retail from three different wet markets in the vicinity of Kuala Lumpur, namely, the Puchong, Kajang, and Sungai Besi wet markets. The vegetable samples were washed for 15 min in 500 ml of sterile buffered peptone water (Oxoid, Basingstoke, United Kingdom), and thereafter, the solution was examined for the presence of Salmonella. The isolation and biochemical identification of the organisms were carried out by standard laboratory methods. Serotyping was done according to the Kauffmann-White scheme by the slide agglutination method both at the Salmonella Reference Centre, Institute for Medical Research, Kuala Lumpur, and at the Veterinary Research Institute, Perak, Malaysia.
Antibiotic disk diffusion testing.
The isolates were tested for their susceptibilities to ampicillin (10 μg), chloramphenicol (30 μg), co-trimoxazole (25 μg), nalidixic acid (30 μg), kanamycin (30 μg), streptomycin (10 μg), sulfamethoxazole (25 μg), and tetracycline (30 μg) by using the Kirby-Bauer disk diffusion method (6). Only the vegetable isolates were tested for their susceptibilities to additional antimicrobial agents, that is, erythromycin (15 μg), nitrofurantoin (10 μg), cephalothin (30 μg), neomycin (10 μg), gentamicin (10 μg), norfloxacin (10 μg), and amoxicillin (25 μg).
DNA preparation and PFGE.
Intact, chromosomal DNA was prepared in agarose by the modified protocol described previously (21). Briefly, cells were scraped from Luria-Bertani agar plates and directly transferred to 1 ml of SB buffer (10 mM Tris-HCl [pH 7.5], 1 M NaCl). The cells were washed twice by centrifugation at 8,000 rpm (6,000 × g; Sigma Laborzentrifugen 2K15) at 4°C for 5 min and resuspended in SB buffer, mixed with an equal volume of 1.5% low-melting-point agarose, and then allowed to set immediately in a mold for 10 min at 4°C. The solidified agarose-cell plugs were incubated with 2 ml of prelysis buffer (10 mM Tris-HCl [pH 7.5], 100 mM NaCl, 0.5% 20 cetyl ether, 0.2% deoxycholate, 0.5% Sarkosyl, 1 mg of RNase per ml, 1 mg of lysozyme per ml) for 2 h at 37°C. The buffer was then discarded, and fresh lysis buffer (0.5 M EDTA [pH 8.0], 1% Sarkosyl) containing 1 mg of proteinase K per ml was added, followed by further incubation at 55°C for 12 h. The lysis buffer was then discarded and the DNA plugs were washed thoroughly three times with TE buffer (1 mM Tris-HCl [pH 8.0], 1 mM EDTA [pH 8.0]) at room temperature with gentle agitation. To save money, only one restriction endonuclease, XbaI, was used to restrict the chromosomal DNA. A slice of DNA plug (2 by 3 by 3 mm) was digested with 10 U of Xbal at 37°C for 2 h. PFGE of XbaI-digested chromosomal DNA was carried out on a CHEF-DR II system for 26 h at 6 V/cm with a ramped pulsed time of 2 to 40 s. A bacteriophage lambda DNA concatemer pulsed-field gel marker was used as a DNA size standard. Fingerprinting profiles were examined visually as well as by use of GelCompar software (version II; Applied Maths, Kortrijk, Belgium). The profiles were scored for the presence and absence of bands, and strains that differed by one band were assigned different pulsed-field profiles (PFPs). The extent of variability was determined by use of the Dice coefficient (F), which is given by the formula 2nxy/(nx + ny), where nx is the total number of DNA fragments present in isolate x, ny is the total number of DNA fragments present in isolate y, and nxy is the total number of DNA fragments present in both isolates x and y. An F value of 1 indicates that the two isolates are indistinguishable, and an F value of 0 indicates that the isolates are dissimilar. Clustering was based on the unweighted pair group average method (UPGMA) and was performed with GelCompar software (version II; Applied Maths).
RESULTS
Thirty-eight of the 40 clinical isolates were sensitive to all antibiotics tested. Two very recent isolates, isolates SW41 and SW47 (July to August, 2001), were each resistant to a single antibiotic (tetracycline and streptomycin, respectively). In contrast, all the 40 vegetable isolates were resistant to both erythromycin and sulfamethoxazole; and some of the isolates were resistant to tetracycline (n = 4), streptomycin (n = 4), and chloramphenicol (n = 1). One vegetable isolate (isolate SWE14) was resistant to five antibiotics (erythromycin, tetracycline, sulfamethoxazole, streptomycin, and chloramphenicol). Two beef isolates were resistant to streptomycin, while one isolate from well water was resistant to both co-trimoxazole and ampicillin.
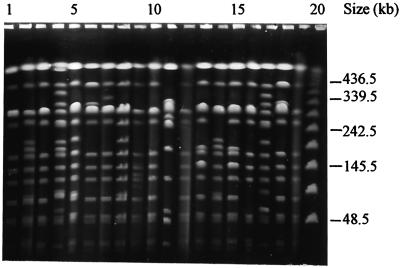
Ninety-five isolates of XbaI-digested Salmonella serotype Weltevreden gave 39 distinct PFPs with 10 to 18 resolvable fragments (F = 0.41 to 1.0). The 40 clinical isolates gave 20 PFPs with two predominant profiles, X1 (11 of 40 isolates) and X2 (8 of 40 isolates), which differed by only one DNA fragment (F = 0.96) (Fig. 1). Both these profiles were consistently observed among isolates collected from 1996 to 2001. These two PFPs were also present among isolates from different hospitals in Kuala Lumpur, Ipoh, Penang, and Alor Setar and from the environment. Both clinical isolates from Kuala Pilah Hospital were identical and had a distinct profile (PFP X15) (Fig. 1, lanes 6 and 18, respectively). Seventeen of the 40 clinical isolates had unique PFPs (PFPs X3 to X20), indicating that PFGE analysis is useful in discriminating Salmonella serotype Weltevreden isolates (Fig. 1; Table 1).
FIG. 1.
Representative PFPs of clinical isolates of Salmonella serotype Weltevreden in Malaysia. Lanes 1 to 20, PFPs X1, X11, X1, X12, X13, X15, X16, X13, X17, X1, X18, X1, X2, X11, X2, X19, X20, X15, X1, and bacteriophage lambda DNA concatemer standard marker, respectively. Numbers on the right indicate the positions of molecular mass markers.
TABLE 1.
Sources and PFGE profiles of Salmonella serotype Weltevreden isolates from Malaysia
| Isolate source | Location (mo/yr of isolation) | No. of isolates tested | PFP(s) (no. of isolates) |
|---|---|---|---|
| Humans (stools isolates) | Universiti Hospital Kuala Lumpur (1/96-9/99) | 22 | X1 (7), X2 (6), X3 (1), X4 (1), X5 (1), X6 (1), X7 (1), X8 (1), X9 (1), X10 (1), X14 (1) |
| General Hospital KL (5/01-7/01) | 3 | X1, X12, X17 | |
| Hospital Klang (5/01-6/01) | 2 | X11, X12 | |
| Hospital Alor Setar (6/01-8/01) | 3 | X1, X2, X16 | |
| Hospital Ipoh (7/01-9/01) | 3 | X1 (1), X2 (1), X11 (1) | |
| Hospital Kuala Pilah (7/01-9/01) | 2 | X15 (2) | |
| Hospital K. Terengganu (7/01) | 1 | X13 | |
| Hospital Pulau Pinang (7/01) | 1 | X1 | |
| Hospital Selayang (7/01) | 1 | X18 | |
| Hospital Tumpat (8/01) | 1 | X19 | |
| Hospital Sungai Petani (8/01) | 1 | X20 | |
| Vegetables | Kajang (8/00-10/00) | 8 | X1 (2), X21 (4), X27 (2) |
| Selom | Puchong (12/99-10/20) | 12 | X14 (5), X23 (4), X24 (2), X39 (1) |
| Sungai Besi (10/00) | 2 | X1, X2 | |
| Kangkong | Kajang (8/00-10/00) | 6 | X1 (1), X21 (4), X25 (1) |
| Sungai Besi (1/00) | 1 | X25 | |
| Kesum | Kajang (8/00) | 7 | X21 (6), X20 (1) |
| Puchong (1/00) | 1 | X25 | |
| Pegaga | Kajang (8/00) | 2 | X21 (2) |
| Sungai Besi (10/00) | 1 | X22 | |
| Well water | Kelantan (1/01-11/01) | 8 | X30, X31, X32, X33, X34, X35, X36, X38 |
| Beef | Klang (2/98) | 2 | X26, X37 |
| Kelantan (2/98) | 1 | X1 | |
| Spicy chicken | Kelantan (9/00) | 1 | X2 |
| Chicken liver | Poultry farm (10/01) | 2 | X28 |
| Raw meat | Johor Baru (11/01) | 1 | X29 |
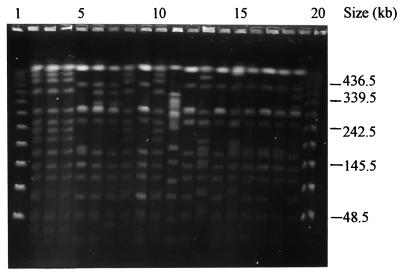
The 40 isolates from four different kinds of vegetables had 10 different PFPs, with F values ranging from 0.19 to 1.0 (Fig. 2). Eight of these profiles (PFPs X21 to X27 and X39) were unique for the vegetables isolates. Some of the vegetable isolates had PFPs indistinguishable from those of the clinical isolates. For example, four vegetable isolates from a wet market had PFPs X1 and X14 (Fig. 2, lanes 9 and 6, respectively), while the three isolates from the Sungai Besi wet market had PFPs X1, X2, and X2, respectively. Another five isolates from selom from the Puchong wet market had the same profile (PFP X14) as a clinical isolate (isolate SW35); both sets of isolates were recovered in 1999 (Fig. 2, lanes 7 and 19, respectively). Sixteen isolates from four different types of vegetables (selom, kangkong, kesum, and pegaga) from the same locality (the Kajang wet market) had a similar, unique profile (PFP X21), indicating a possible point source of contamination within the wet market. Another observation was that isolates with PFP X25 were present in all three wet markets from which isolates were obtained (Table 1).
FIG. 2.
Representative PFPs from environmental and clinical isolates of Salmonella serotype Weltevreden. Lanes 1 and 20, bacteriophage lambda DNA concatemer standard marker; lanes 2 to 19, PFPs X21 (vegetable), X21 (vegetable), X21 (vegetable), X2 (vegetable), X14 (vegetable), X14 (vegetable), X21 (vegetable), X1 (vegetable), X21 (vegetable), X22 (vegetable), X1 (beef), X4 (clinical), X1 (clinical), X6 (clinical), X3 (clinical), X2 (chicken meat), X10 (clinical), and X14 (clinical), respectively. Numbers on the right indicate the positions of molecular mass markers.
Isolates from well water had diverse PFPs, each showing a unique patterns (profiles X30 to X38), with F values ranging from 0.36 to 0.92 (Table 1; Fig. 3). Isolates from beef and spicy chicken also had the predominant profiles, PFPs X1 and X2, respectively. The isolates from chicken liver and raw meat had unique profiles, PFPs X28 and X29, respectively.
FIG. 3.
Dendrogram showing the results of cluster analysis of the PFGE patterns of XbaI-digested DNA from 95 strains of Salmonella serotype Weltevreden generated with GelCompar software by the UPGMA method, based on the matrix of F values. The different PFPs and the sources and number of isolates are indicated.
Cluster analysis of all the Salmonella serotype Weltevreden strains based on the matrix of F values (at 80% similarity) generated four clusters, with each cluster comprising some subclusters (Fig. 3). Cluster I consisted of 15 PFPs from 47 clinical and environmental isolates. Cluster II comprised eight PFPs from 4 clinical isolates and 9 environmental isolates. Cluster III consisted of only three PFPs from 18 environmental isolates. Cluster IV consisted of five PFPs from nine isolates. The sharing of similar genotypes between the isolates from clinical and environmental samples provides an indication of the possible transmission of Salmonella serotype Weltevreden from vegetables, well water, and cooked food to humans.
DISCUSSION
Many interrelated factors including increased urbanization, inadequate supplies of clean water, inadequate sanitary measures, the development of a large-scale food industry, and a lack of adequate food hygiene and food safety measures have been implicated in the increased incidence of food poisoning. The availability of detailed and accurate data related to the molecular epidemiology of nontyphoidal Salmonella spp. is crucial for effective surveillance and prevention. Genomic macrorestriction fragment length analysis by PFGE is now a widely used molecular tool for the subtyping of Salmonella spp. Only XbaI was used in this study, as it is more discriminative and cheaper and has been shown to be a useful restriction enzyme for Salmonella spp. (22). In our previous study, PFGE analysis of 20 clinical isolates of Salmonella serotype Weltevreden from a single hospital showed that the strains were very closely related and clustered in one group (based on 85% similarity) (9). We have now extended the study to include more clinical isolates from wider geographical areas in Malaysia as well as isolates from different environmental sources, namely, well water, vegetables, beef, raw meat, and chicken products.
Overall, PFGE was shown to be very useful in delineating the genetic variability of the strains and was an invaluable epidemiological tool, as the DNA fingerprints generated were stable and reproducible and all the strains were typeable. In addition, the methodology is now relatively simple and only about 2 days is required to prepare chromosomal DNA for PFGE analysis.
The present study showed that many PFGE subtypes of Salmonella serotype Weltevreden are present in humans as well as in agricultural produce and well water. The DNA profiles of most of the Salmonella serotype Weltevreden isolates from various hospitals varied greatly and thus indicated that the isolates belong to different clones. Comparative analysis of clinical isolates obtained 6 years apart, in 1996 and 2001, also indicated that these genotypes (PFPs X1 and X2) of Salmonella serotype Weltevreden are stable and persist over a considerable period of time.
Salmonella serotype Weltevreden isolates recovered from the Kajang wet market at different times were indistinguishable, indicating the persistence of this clone in the environment. PFP X21 was found repeatedly in samples of vegetables during August 2000 in the Kajang wet market. This supports the notion that contamination was caused by the same type strain of Salmonella serotype Weltevreden and may indicate cross-contamination between products and the environment.
The emergence of multiresistant strains of Salmonella species is of great concern to clinicians worldwide. The resistance of Salmonella species to chloramphenicol, ampicillin, and trimethoprim-sulfamethoxazole was reported in Southeast Asia (3, 4). Clinical isolates of Salmonella spp. isolated in Malaysia between 1989 and 1996 were, however, highly sensitive (14, 23). In the present study, the majority of the recent isolates from humans and well water (85 and 87%, respectively) remained sensitive to all the antibiotics tested. On the other hand, all the vegetable isolates were resistant to at least two antibiotics (erythromycin and sulfamethoxazole), and about 11% were resistant to multiple antimicrobials. Many researchers (1, 20) have reported the presence of antimicrobial-resistant Salmonella strains in raw vegetables. The presence of resistant Salmonella strains in all the vegetables sampled in the present study is a cause for concern. This has obvious public health implications since multidrug resistance limits the possible therapeutic treatments.
Salmonellosis in humans is usually due to the consumption of contaminated food and/or water. The effluents from infected animals and humans are important sources of contamination of the environment and the food chain. Salmonella serotype Typhimurium DT104 appeared to spread from farm to farm via raw water (2). Vegetables are not known to harbor Salmonella, and the presence of Salmonella in vegetables could be due to contamination with sewage-tainted water or handling by humans. A number of studies have demonstrated a close relationship between Salmonella isolates from irrigation water and Salmonella isolates from irrigated vegetables (2, 20). Consumption of these contaminated crops could result in the transmission of pathogenic microorganisms to consumers. Ait Melloul and Hassani (1) noted that the rate of salmonellosis among children living in an area where crops were irrigated with raw wastewater was high compared to the rate among children from another area that did not irrigate crops with sewage water. In addition, in the present study, since all the isolates from the four different types of vegetables from one particular locality (a wet market) had the same PFP (PFP X21), it is unclear whether the vegetables were contaminated at the source or whether the isolates spread locally from stall to stall. Contamination of vegetables with human sewage or animal manure may occur at any point in the food chain, either on the farm or at the point of sale. In Malaysia, water from irrigation ditches, ponds, and swamps is used to cultivate the types of vegetables studied. Often, liquid waste from slaughterhouses and processing plants is illegally released in these bodies of water. Madden (15) reported that contaminated irrigation water, improper handling of food by humans, contaminated containers, and animal-waste fertilizers can be sources of microbial contamination of vegetables. Unlike the other three vegetables, pegaga is a topsoil creeper, and soil can be a source of contamination if animal waste is used as a fertilizer for crops of this vegetable. Similar clones of Salmonella serotype Weltevreden (PFPs X1 and X2) were associated with humans, vegetables, spicy chicken, and beef; and this probably indicates cross-contamination. Food hygiene is therefore of utmost importance in the prevention of salmonellosis, as it is impossible to eradicate Salmonella primarily because it survives as a zoonotic agent in the environment.
Although the PFGE patterns obtained were helpful in identifying similar strains from different hospitals and environmental sources, some of these isolates had different antibiotic resistance patterns. It is possible that differences in susceptibility may be due to point mutations or minor genetic changes that were insufficient to alter the PFGE patterns; in these cases only a large alteration in the DNA or a mutation occurring within the recognition site for the restriction enzyme used may alter the PFGE patterns.
The multiple-antibiotic-resistance pattern of the environmental strain with PFP X1 suggests that other selective pressures may be responsible for the continued carriage of the resistance genes in these strains. However, it is possible that the isolate with PFP X1 may have acquired the genes for resistance to multiple drugs from other enteric bacteria. At this stage of our study, we cannot speculate whether the antimicrobial resistance genes in the multidrug-resistant isolates were carried on plasmids or were chromosomally mediated.
A noteworthy finding in this study was that two lineages (PFPs X1 and X2) could be considered endemic in the study area because organisms implicated in human salmonellosis between 1996 and 2000 as well as organisms isolated from raw vegetables had these PFPs, suggesting that raw vegetables may be one of the reservoirs of such organisms.
In addition, a new lineage (PFP X14) seems to have emerged. Isolates of this lineage were recovered from raw vegetables in 1999 and were found to cause salmonellosis. These bacterial clones establish themselves in the environment, multiply, and act as infectious agents. Other clones with PFP X21 also persisted in a wet market. Thus, the use of effective cleaning and disinfecting practices is necessary.
Precise discrimination is needed to identify the specific settings and routes of transmission so that food, animal, and environmental sources of Salmonella isolates can be investigated promptly to maximize the likelihood of recovering potentially causative organisms. Outbreaks in a small geographical region may involve nationally distributed products and would require wider investigations of the potential links of Salmonella serotype Weltevreden strains that are causing infections.
The presence of Salmonella isolates from various hospitals with several PFPs probably reflects the microbiological variation of the source of infection. This study highlighted the importance of continuous surveillance in detecting the introduction of new clones into a hospital or the environment. This observation suggests an epidemiological linkage between the strains of Salmonella serotype Weltevreden isolated from the stools of patients in hospitals and those isolated from raw vegetables in the environment.
If cross-transmission occurs efficiently, vegetables may play an important role in the epidemiology of Salmonella serotype Weltevreden, a fact that should be considered when control programs are planned.
Acknowledgments
The work described here is supported by IRPA grant 06-02-03-0750 from the Ministry of Science, Technology and Environment of Malaysia.
REFERENCES
- 1.Ait Melloul, A., and L. Hassani. 1999. Antibiotic resistance of Salmonella strains isolated from children living in the waste-waters spreading field of Marrakesh city (Morocco). World J. Microbiol. Biotoechnol. 15:91-96. [Google Scholar]
- 2.Ait Melloul, A., L. Hassani, and L. Rafouk. 2001. Salmonella contamination of vegetables irrigated with untreated wastewater. World J. Microbiobiol. Biotechnol. 17:207-209. [Google Scholar]
- 3.Akhtiar, M. A., K. A. Karamt, A. Z. Malik, A. Hashmi, and Q. M. Khan. 1989. Efficacy of ofloxacin in typhoid fever, particularly in drug resistant cases. Rev. Infect. Dis. 2(Suppl.):S11-S93. [Google Scholar]
- 4.Anand, A. C., V. K. Kataeria, W. Singh, and S. K. Chatterjee. 1990. Epidemic multiresistant enteric fever in Eastern India. Lancet 315:352.. [DOI] [PubMed] [Google Scholar]
- 5.Aserkof, B., and J. V. Bennett. 1969. Effect of antibiotic therapy in acute salmonellosis on the fecal excretion of salmonellae. N. Engl. J. Med. 281:634-640. [DOI] [PubMed] [Google Scholar]
- 6.Bauer, A. W., M. W. Kirby, J. C. Sherris, and M. Turck. 1966. Antibiotic susceptibility testing by a standardized single disc method. Am. J. Clin. Pathol. 45:493-496. [PubMed] [Google Scholar]
- 7.Chee-Sanford, J. C., R. I. Aminov, I. J. Krapac, N. Garrigues-Jeanjean, and R. I. Mackie. 2001. Occurrence and diversity of tetracycline resistance genes in lagoons and groundwater underlying two swine production facilities. Appl. Environ. Microbiol. 67:1494-1502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.D'Aoust, J. Y. 2000. Salmonella, p. 1233-1299. In B. M. Lund, A. C. Parker-Baird, and G. W. Gould (ed.), The microbiological safety and quality of food. Aspen, Gaithersburg, Md.
- 9.Goh, Y. L., S. D. Puthucheary, and K. L. Thong. 2000. Application of ribosomal RNA gene restriction patterns analysis and pulsed field gel electrophoresis in distinguishing Salmonella weltevreden isolates in Malaysia. Southeast Asian J. Trop Med. Public Health 31:697-701. [PubMed] [Google Scholar]
- 10.Jegathesan, M. 1984. Salmonella serotypes isolated from man in Malaysia over the 10-year period 1973-1982. J. Hyg. Camb. 92:395-399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Joseph, P. G., M. Anwar, and M. Jegathesan. 1978. Animal salmonellosis in peninsular Malaysia. II. Annual and zoological distribution of Salmonella serotypes over a 10-year period 1966-75. Am. J. Trop. Med. Hyg. 27:562-566. [PubMed] [Google Scholar]
- 12.Joseph, P. G., S. P. Sivanandan, H. T. Yee, S. Saroja, Z. Mahmud, and M. Jegathesan. 1986. Animal Salmonella surveillance in peninsular Malaysia, 1976-1980. Trop. Biomed. 3:79-84. [Google Scholar]
- 13.Lee, L. A., N. D. Puhr, E. K. Maloney, N. H. Bean, and R.V. Tauxe. 1994. Increase in antimicrobial-resistant Salmonella infections in the United States, 1989-1990. J. Infect. Dis. 170:128-134. [DOI] [PubMed] [Google Scholar]
- 14.Lee, W. S., S. D. Puthucheary, and C. C. M. Boey. 1998. Nontyphoid Salmonella gastroenteritis J. Pediatr. Child Health 34:387-390. [DOI] [PubMed] [Google Scholar]
- 15.Madden, J. M. 1992. Microbial pathogens in fresh produce—the regulatory perspective. J. Food Prot. 55:821-823. [DOI] [PubMed] [Google Scholar]
- 16.Maslow, J. N., M. E. Mulligan, and R. D. Arbeit. 1993. Molecular epidemiology: application of contemporary techniques to the typing of microorganisms. Clin. Infect. Dis. 17:153-164. [DOI] [PubMed] [Google Scholar]
- 17.Molbak, K., D. L. Baggesen, F. M. Aarestrup, J. M. Ebbesen, J. Engberg, K. Frydendahl, Gerner-Smidt, A. M. Petersen, and H. C. Wegener. 1999. An outbreak of multidrug-resistant quinolone-resistant Salmonella enterica serotype Typhimurium DT104. N. Engl. J. Med. 341:1420-1425. [DOI] [PubMed] [Google Scholar]
- 18.Ponka, A., Y. Anderson, A. Siitonen, B. de Jong, M. Jahkola, and O. Haikapa. 1995. Salmonella in alfalfa sprouts. Lancet 345:462-463. [DOI] [PubMed] [Google Scholar]
- 19.Rowe, B., L. R. Ward, and E. J.Threfall. 1990. Spread of multiresistant Salmonella typhi. Lancet 337:1065.. [PubMed] [Google Scholar]
- 20.Ruiz, G. V. B. 1987. A comparative study of strains of Salmonella isolated from irrigation waters, vegetables and human infections. Epidemiol. Infect. 98:271-274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Thong, K. L., and T. Pang. 1996. A rapid, simplified method for preparation of chromosomal DNA from pathogenic bacteria for use in pulsed-field gel electrophoresis. Asia Pacific J. Mol. Biol. Biotechnol. 4:59-62. [Google Scholar]
- 22.Thong, K. L., Y. M. Cheong, S. D. Puthucheary, C. L. Koh, and T. Pang. 1994. Epidemiological analysis of sporadic Salmonella typhi isolates and those from outbreaks by pulsed-field gel electrophoresis. J. Clin. Microbiol. 32:1135-1141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yasin, R. Y., C. T. Cheah, and M. Jegathesan. 1995. Human salmonellosis in Malaysia for the period 1989-July 1994. Southeast Asian J. Trop. Med. Public Health 26:457-460.