Abstract
Multidrug-resistant Pseudomonas aeruginosa nosocomial infections are increasingly recognized worldwide. The existence of metallo-β-lactamase- and extended-spectrum β-lactamase-producing isolates exhibiting resistance to most β-lactam antimicrobial agents greatly complicates the clinical management of patients infected with such isolates. Since 1998, P. aeruginosa isolates resistant to all commercially available antimicrobial agents have been detected at a university-affiliated public hospital in Rio de Janeiro, Brazil. The present study was designed to characterize the antimicrobial resistance profiles and the genetic diversity of the P. aeruginosa strains isolated at this hospital and four private hospitals in Rio de Janeiro. Between April 1999 and March 2000, 200 consecutive isolates were obtained and analyzed for antimicrobial resistance. The genetic diversity of a selected number of them was evaluated by pulsed-field gel electrophoresis and PCR with the ERIC-2 primer. A predominant genotype, designated genotype A, was identified among isolates from four of the five hospitals evaluated. Eighty-four ceftazidime-resistant isolates were evaluated for metallo-β-lactamase production, which was detected in 20 (91%) of 22 genotype A isolates and 11 (18%) of 62 isolates belonging to other genotypes (P < 0.05). Two metallo-β-lactamase-producing genotype A isolates also produced an extended-spectrum β-lactamase. The occurrence of multidrug-resistant P. aeruginosa strains belonging to a unique genotype in different hospitals in Rio de Janeiro underscores the importance of the contribution of a single clone to the increase in the incidence of multidrug-resistant P. aeruginosa nosocomial infections.
Pseudomonas aeruginosa is a leading cause of nosocomial infections. In the United States, P. aeruginosa ranked second among all nosocomial pathogens related to pneumonia in intensive care units reported to the National Nosocomial Infection Surveillance System in the last decade (17). Among 70,067 isolates obtained from patients admitted to hospitals in five different geographic areas and evaluated by the SENTRY antimicrobial resistance surveillance program, the prevalence rates of P. aeruginosa infections were higher in the Latin American and the Asia-Pacific regions (11.4% of total isolates in each region) than in Europe (9.3%), the United States (8.7%), and Canada (8.6%) (8). The reasons for such differences are unclear and may be related to the use of suboptimal infection control practices in some of these regions. Infections due to this microorganism are becoming difficult to treat because of limited choices of effective antimicrobial agents (10).
In addition to the constitutive low level of susceptibility of this organism to antimicrobial agents, new mechanisms of resistance have been identified in P. aeruginosa, including the production of β-lactamases belonging to each of the Ambler classes (5). The class A clavulanic acid-inhibited extended-spectrum β-lactamases (ESBLs) described in P. aeruginosa include TEM- and SHV-related ESBLs as well as PER-1 and VEB-1, which hydrolyze ceftazidime and cefepime (18). The class B enzymes, called metallo-β-lactamases (M-βlas), are IMP- and VIM-related enzymes (14, 26) and have the broadest hydrolysis profiles; they hydrolyze all β-lactam agents except monobactams. Class D enzymes include a group of ESBLs comprising OXA-18, OXA-2, and OXA-10 derivatives as well as OXA-24 and ARI-1, which hydrolyze carbapenems (4).
None of these enzymes has yet been described in P. aeruginosa isolates from Brazil. Because multidrug resistance among hospital pathogens is such a serious problem in some parts of Brazil, polymyxin, a highly toxic antimicrobial agent used until the early 1980s to treat infections caused by gram-negative rods, is again being used to treat infections caused by strains of P. aeruginosa susceptible only to this compound (13). Such multidrug-resistant isolates have been detected since 1998 in a public university teaching hospital in the city of Rio de Janeiro. The present study was designed to assess the magnitude of antimicrobial resistance and β-lactamase production in P. aeruginosa isolates obtained at this hospital and determine the genetic diversity among the isolates. We also examined P. aeruginosa isolates obtained at four private hospitals in the state of Rio de Janeiro to assess the possible interhospital spread of multidrug-resistant strains.
MATERIALS AND METHODS
Bacterial isolates and identification procedures.
From April 1999 to March 2000, 200 consecutive P. aeruginosa isolates were recovered from 115 patients admitted to the Hospital Universitário Clementino Fraga Filho (HUCFF), a 490-bed university hospital, and 85 patients admitted to four other small private hospitals (which together have a total of 174 beds). Only one isolate per patient was included in the study. Three of these hospitals are also located in Rio de Janeiro City and one is located in Niterói, a city located across the bay from Rio de Janeiro. The isolates were obtained from different clinical specimens, including urine specimens (27%), respiratory tract secretions (25.5%), wounds (16%), blood specimens (14%), catheter tips (7.5%), and specimens from patients with various deep-seated infections (10%). Bacterial isolates were identified with the GNI VITEK system card (BioMérieux Vitek Inc., Hazelwood, Mo.) and by conventional biochemical tests: cytochrome oxidase reaction, pigment production, glucose oxidation, arginine dihydrolase activity, and growth at 42°C (12). They were stored as suspensions in a 10% (wt/vol) skim milk solution containing 10% (vol/vol) glycerol at −20°C until additional tests were performed, as described below.
Antimicrobial susceptibility testing and evaluation of β-lactamase production.
The susceptibilities of the bacterial isolates to 11 antimicrobial agents (amikacin, aztreonam, ceftazidime [CAZ], cefepime, ciprofloxacin, gentamicin, imipenem, meropenem, piperacillin-tazobactam, polymyxin B, and ticarcillin-clavulanate) were determined by the disk diffusion method in accordance with NCCLS guidelines (16). Interpretation of zone diameters obtained for polymyxin followed a protocol suggested by Gales et al. (9). The study period was separated into three parts: period 1, April 1999 to July 1999; period 2, August 1999 to November 1999; and period 3, January 2000 to March 2000. No isolates were collected during December 1999. CAZ-resistant isolates were evaluated for M-βla production by a disk approximation test with disks containing CAZ and 2-mercaptopropionic acid, as described by Arakawa et al. (1). Briefly, a suspension of each isolate was spread onto a Mueller-Hinton agar plate and two disks, each of which contained 30 μg of CAZ (Difco Laboratories, Detroit, Mich.), were placed on the agar surface. The distance between the two CAZ disks was kept at about 5 cm, and a blank filter disk was placed near one of the CAZ disks (center-to-center distance, 2 cm). Three microliters of a 1.2-g/ml solution of 2-mercaptopropionic acid (Sigma Chemical Co., St. Louis, Mo.) was added to the blank filter disk, and the cultures were incubated at 37°C overnight. The observation of an increase in the inhibition zone or the appearance of an inhibition zone around the CAZ disk was interpreted as a positive test result. All isolates showing resistance to CAZ and/or cefepime (51 isolates) were evaluated for ESBL production by the double diffusion test (11) with disks containing CAZ, cefepime and piperacillin-tazobactam, and amoxicillin-clavulanate as inhibitors; the disks were placed 2.0 cm apart (center to center) (24).
Genotyping.
Evaluation of chromosomal polymorphisms was performed by pulsed-field gel electrophoresis (PFGE) of isolates by a previously reported method (22), with modifications, and by PCR with primer ERIC-2 (19). For PFGE, the bacteria were grown overnight at 37°C on Trypticase soy agar plates. Cells were suspended in Pett IV buffer, and the turbidity of the suspension was adjusted to match that of a 6 McFarland density standard. A 0.5-ml aliquot of this suspension was mixed with an equal volume of 2.0% low-melting-temperature agarose, and the mixture was distributed into plug molds. The DNA in the plugs was digested with SpeI (Boehringer Mannheim Biochemicals, Indianapolis, Ind.) at 37°C according to the instructions of the manufacturer. The restriction fragments were separated by PFGE in 1.2% agarose gels, and electrophoresis was carried out at 6 V/cm for 24 h with pulse times ranging from 5 to 35 s and at a temperature of 11°C in a CHEF DR III system (Bio-Rad Laboratories, Richmond, Calif.). Agarose gels were stained with ethidium bromide, visualized by UV transillumination, and photographed. The banding patterns were interpreted by visual inspection, based on the criteria of relatedness proposed by Tenover et al. (23), and with the Molecular Analyst Fingerprinting Plus software package (version 1.12) of the Image Analysis System (Bio-Rad) by using the Dice index and the unweighted pair group method with arithmetic averages. For analysis by PCR with primer ERIC-2 (19), chromosomal DNA was obtained from bacterial suspensions grown overnight in Luria broth with shaking, suspended in 100 ml of sterile water, and boiled for 10 min. PCRs were performed in total volumes of 25 μl containing 50 mM KCl, 10 mM Tris HCl (pH 8.3), 5 mM MgCl2, each deoxynucleotide triphosphate at a concentration of 0.25 mM, 2.5 U of Taq polymerase, and 1.25 mM primer (primer ERIC-2 [5′-AAG TAA GTG ACT GGG GTG AGC G-3′]). Amplification conditions were 94°C for 2 min of initial denaturation; 40 cycles in which each cycle consisted of denaturation at 94°C for 30 s, annealing at 50°C for 1 min, and extension at 72°C for 4 min; and a final extension at 72°C for 1 min. The agarose gels were stained with ethidium bromide, visualized by UV transillumination, and photographed. Isolates showing more than one band difference by PCR with primer ERIC-2 were considered to belong to separate genotypes.
RESULTS
Antimicrobial susceptibility testing and β-lactamase production.
All isolates were susceptible to polymyxin. The rates of resistance to each of the other antimicrobial agents tested were usually lower among strains isolated at HUCFF than among those isolated from the private hospitals, as shown in Table 1. Coresistance to 8, 9, or 10 antimicrobial agents was detected in 37 (32%) of the 115 isolates obtained at HUCFF and 34 (40%) of the 85 isolates from the private hospitals (P = 0.3). All CAZ-resistant isolates detected at the five hospitals (84 of the 200 isolates included in the study) were assessed for M-βla production. A clear positive result was observed for 31 isolates (37% of the 84 CAZ-resistant isolates). It was revealed that three isolates produced an ESBL, and two of them also produced an M-βla. A positive result was obtained by using either clavulanic acid or tazobactam as the inhibitor.
TABLE 1.
Distribution of antimicrobial resistance rates among P. aeruginosa isolates isolated in Rio de Janeiro State, Brazil, from April 1999 to March 2000
| Antimicrobial agent | No. (%) of resistant isolates
|
Range of % resistance at four private hospitals | |
|---|---|---|---|
| HUCFF (na = 115) | Private hospitals (n = 85) | ||
| Amikacin | 41 (35.6) | 38 (44.7) | 26.1-80.0 |
| Aztreonam | 50 (43.5) | 49 (57.6) | 50.0-73.3 |
| CAZ | 42 (36.5) | 42 (49.4) | 26.1-73.3 |
| Cefepimeb | 47 (41.0) | 53 (62.3) | 55.5-93.3 |
| Ciprofloxacinb | 49 (43.0) | 58 (68.2) | 44.4-95.0 |
| Gentamicin | 55 (48.0) | 52 (61.2) | 44.4-85.0 |
| Imipenem | 44 (38.3) | 36 (42.3) | 30.4-66.6 |
| Meropenem | 35 (30.4) | 36 (42.3) | 30.0-80.0 |
| Piperacillin-tazobactamb | 42 (36.5) | 44 (51.8) | 30.4-80.0 |
| Ticarcillin-clavulanateb | 61 (53.0) | 61 (71.8) | 55.5-93.3 |
n, total number of isolates
A significant difference (P < 0.05). in rates of resistance to the antimicrobial was detected between isolates from HUCFF and those from private hospitals considered together.
Genotyping.
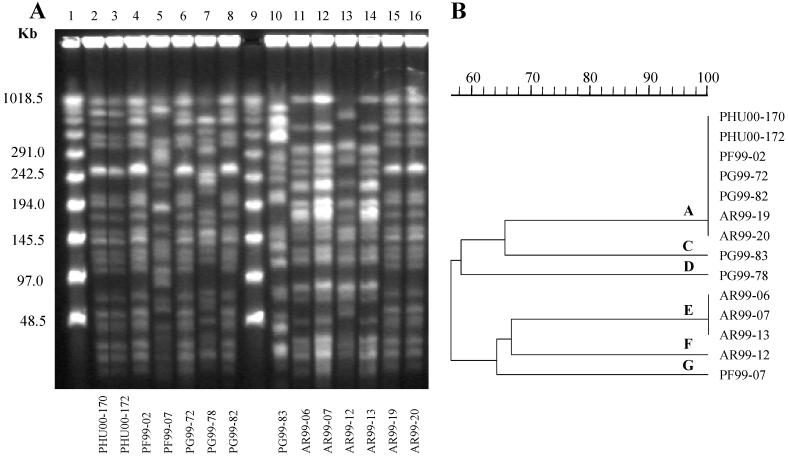
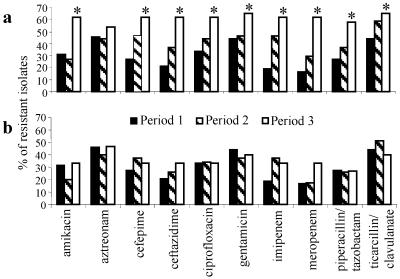
A total of 88 (44%) isolates were typed, including 60 of 71 isolates with combined resistance to 8, 9, or 10 of the 11 antimicrobial agents tested, 6 isolates randomly selected from among 55 isolates that were susceptible to all agents tested, and 22 isolates randomly selected from among 74 isolates with combined resistance to 1 to 7 agents. Forty-two distinct PFGE patterns were observed among these isolates. Twenty-two (25%) of 88 isolates tested had a single predominant PFGE genotype, named genotype A (Table 2). For 6 of the 17 patients at HUCFF infected with genotype A strains, the clinical specimen was obtained after 48 h of admission to the intensive care unit. The other 11 patients were admitted to different units at HUCFF. Surprisingly, isolates of genotype A were also detected among isolates from three of the four private hospitals, all of them in Rio de Janeiro City. Among the isolates tested, 17 (27%) of 63 isolates from HUCFF were genotype A. Although a few band differences were noted among some of the genotype A isolates, representative isolates from all four hospitals showed identical PFGE profiles (Fig. 1). Genotype A isolates were not detected among isolates from the hospital in the city of Niterói, located about 30 km from the hospitals in Rio de Janeiro. All genotype A isolates had combined resistance to 8, 9, or 10 antimicrobial agents, and 20 of the 22 isolates produced M-βla. The occurrence of genotype A isolates at HUCFF during the study period was associated with a significant increase in the overall rates of resistance to the antimicrobial agents tested with the exception of aztreonam, as shown in Fig. 2a. In examining the increase in P. aeruginosa resistance rates over the three periods at this hospital, much of the increase itself may be explained by the emergence of genotype A strains in August 1999 and the subsequent dissemination of the genotype A strains; when genotype A strains are excluded from the analysis, the background resistance rates over the same period did not show a significant change (Fig. 2b). Fourteen of the 22 isolates found to be genotype A by PFGE were also examined by PCR with primer ERIC-2. All 14 isolates had the same pattern by PCR with primer ERIC-2.
TABLE 2.
Distribution of P. aeruginosa isolates belonging to genotype A among five hospitals in Rio de Janeiro
| Resistance phenotype | Genotype | No. of isolates
|
||||
|---|---|---|---|---|---|---|
| HUCFF | Private hospitals
|
|||||
| Hospital I | Hospital II | Hospital III | Hospital IV | |||
| Coresistance to 8-10 agents (60/71)a | A | 17 | 1 | 2 | 2 | 0 |
| Non-A | 18 | 1 | 2 | 4 | 13 | |
| Coresistance to 1-7 agents (22/74) | A | 0 | 0 | 0 | 0 | 0 |
| Non-A | 22 | 0 | 0 | 0 | 0 | |
| Susceptibility to all tested agents (6/55) | A | 0 | 0 | 0 | 0 | 0 |
| Non-A | 6 | 0 | 0 | 0 | 0 | |
| All isolates typed | 63 | 2 | 4 | 6 | 13 | |
The values in parentheses are the number of isolates typed/total number of isolates showing the resistance phenotype.
FIG. 1.
(A) PFGE profiles of SpeI-digested chromosomal DNA of P. aeruginosa isolates recovered at four hospitals in Rio de Janeiro. Lanes 1 and 9, molecular size markers; lanes 2 and 3, genotype A isolates from HUCFF; lane 4, genotype A isolate from hospital III; lanes 6 and 8, genotype A isolates from hospital I; lanes 15 and 16, genotype A isolates from hospital II; lane 5, genotype G isolate from hospital III; lane 7, genotype D isolate from hospital I; lane 10, genotype C isolate from hospital I; lanes 11, 12, and 14, genotype E isolates from hospital II; lane 13, genotype F isolate from hospital II. (B) Dendrogram resulting from computer-assisted analysis of the profiles shown in panel A.
FIG. 2.
(a) Rates of resistance of 115 P. aeruginosa isolates obtained at HUCFF to 10 antimicrobial agents. (b) Same as panel a, but without genotype A isolates. ∗, P < 0.05 compared to study period 1.
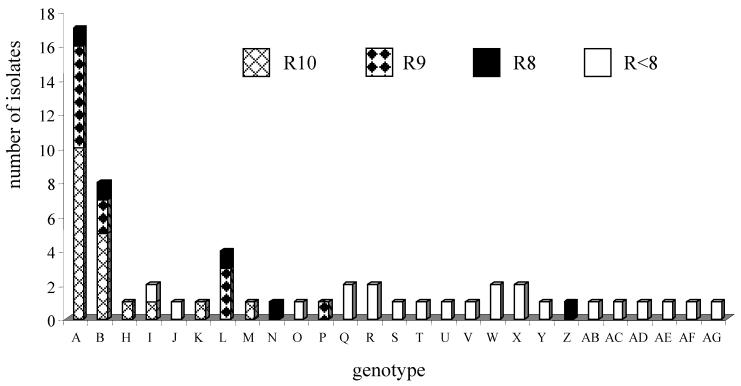
Among other isolates showing resistance to more than seven antimicrobial agents, a cluster of a second genotype (genotype B) was observed by PFGE, but only among the isolates at HUCFF, as shown in Fig. 3. However, genotype B isolates were equally distributed among all isolates evaluated during the study period. No other clusters of isolates were detected by PFGE.
FIG. 3.
Distribution of genotypes, as determined by PFGE, among P. aeruginosa isolates recovered at HUCFF by antimicrobial resistance patterns. R10, isolates coresistant to 10 antimicrobial agents; R9, isolates coresistant to 9 antimicrobial agents; R8, isolates coresistant to 8 antimicrobial agents; R<8, isolates coresistant to less than 8 antimicrobial agents.
DISCUSSION
In the present study, high levels of resistance to all antimicrobial agents commercially available in Brazil were detected among P. aeruginosa isolates obtained from five hospitals in Rio de Janeiro State. Interestingly, the overall resistance rates were lower at HUCFF, a university teaching hospital, than at the four private hospitals. HUCFF has an infection control committee that strongly restricts antimicrobial use, whereas at the private hospitals, the recommendations of infection control staff may not be as strictly adhered to as they are at HUCFF. These resistance rates in Rio de Janeiro are higher than those reported by the SENTRY surveillance program for five different regions of the world except for the rate of resistance to aztreonam in Latin America (8). In addition, for the first time we provide evidence of the presence of ESBL- and M-βla-producing P. aeruginosa clinical isolates in Brazil, including two isolates that produce both kinds of β-lactamases. This combination of β-lactamases in a single strain was recently described for the first time in a P. aeruginosa strain from Italy, which produced the PER-1 ESBL and the VIM-2 M-βla (7). Such an accumulation of resistance determinants in one strain imposes a tremendous limitation on the therapeutic choices available for the treatment of infections caused by gram-negative species. Studies are under way to characterize the specific types of ESBLs and M-βlas produced by P. aeruginosa isolates from Rio de Janeiro.
Multidrug-resistant isolates were clustered into two major genotypes, and isolates of one of these genotypes were identified in at least four different hospitals in the city. To our knowledge, no other studies have shown that a single P. aeruginosa strain unrelated to cystic fibrosis could be detected nearly simultaneously in distinct institutions. This observation could result from the interhospital spread of P. aeruginosa genotype A, interhospital transfers of infected patients, health care staff working at different hospitals, or contaminated products commonly used by hospitals. We do not have information to support any of these possibilities at this time. Alternatively, isolates of genotype A might have emerged independently from a common ancestral strain of P. aeruginosa at these hospitals. The interhospital clonal spread of strains belonging to other bacterial species has been documented by PFGE analyses in the United States (6, 15, 21). Clonal spread of drug-resistant P. aeruginosa isolates may be more frequent than recognized, and this study highlights the importance of using molecular techniques for the typing of strains to evaluate the patterns of distribution of nosocomial drug-resistant pathogens.
At all five hospitals, patients infected with multidrug-resistant organisms are placed under contact precautions. However, a number of factors might have contributed to the dissemination of genotype A isolates at four of these hospitals, including the use of suboptimal aseptic techniques and infection control practices by health care workers, inadequate cleaning and disinfection of the environment and medical equipment, and understaffing.
Multidrug-resistant P. aeruginosa isolates are usually reported to be responsible for outbreaks of nosocomial infections, mainly in intensive care units (3, 25). In such studies, isolates of a single genotype or a few predominant outbreak-related genotypes are detected among isolates obtained from distinct hospital units or a few units in one institution, as described in two hospitals in São Paulo, Brazil (2, 20). The detection of a strain with a unique genotype among different hospitals in the present study indicates that, in addition to inadequate nosocomial infection control practices, certain clones of drug-resistant P. aeruginosa may be better adapted to spread among susceptible hosts in nosocomial settings. Importantly, isolates with this unique genotype comprised nearly half of the multiresistant P. aeruginosa isolates detected at HUCFF, thus accounting for much of the increase in drug-resistant P. aeruginosa infections in this single teaching hospital during the study period. After the results of this study were disclosed, a number of inadequacies in infection control practices were corrected at HUCFF, and these practices might contain the transmission of these organisms. Additional studies are under way to evaluate the effects of these changes.
Acknowledgments
This study was supported by Conselho Nacional de Desenvolvimento Científico e Tecnológico, Coordenação de Aperfeiçoamento de Pessoal de Nível Superior, Fundação de Amparo à Pesquisa do Estado do Rio de Janeiro, and the Ministério Da Ciência e Tecnologia (MCT/PRONEX) in Brazil and by the Fogarty International Program in Research and Training in Emerging Infectious Diseases (grant TW00905) of the National Institutes of Health in the United States.
REFERENCES
- 1.Arakawa, Y., N. Shibata, K. Shibayama, H. Kurokawa, T. Yagi, H. Fujiwara, and G. Masafumi. 2000. Convenient test for screening metallo-β-lactamase-producing gram-negative bacteria by using thiol compounds. J. Clin. Microbiol. 38:40-43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arruda, E. A. G., I. S. Marinho, M. Boulos, I. S. Sumiko, H. H. F. Caiaffa, C. M. Mendes, C. P. Oplustil, H. Sader, C. E. Levy, and A. S. Levin. 1999. Nosocomial infections caused by multiresistant Pseudomonas aeruginosa. Infect. Control Hosp. Epidemiol. 20:620-623. [DOI] [PubMed] [Google Scholar]
- 3.Bingen, E., S. Bonacorsi, P. Rohrlich, M. Duval, S. Lhopital, N. Brahimi, E. Vilmer, and R. V. Goering. 1996. Molecular epidemiology provides evidence of genotypic heterogeneity of multidrug-resistant Pseudomonas aeruginosa serotype O:12 outbreak isolates from a pediatric hospital. J. Clin. Microbiol. 34:3226-3229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bou, G., A. Oliver, and J. Martinez-Beltran. 2000. OXA-24, a novel class D β-lactamase with carbapenemase activity in an Acinetobacter baumannii clinical strain. Antimicrob. Agents Chemother. 44:1556-1561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bush, K., G. A. Jacoby, and A. A. Medeiros. 1995. A functional classification for β-lactamases and its correlation with molecular structure. Antimicrob. Agents Chemother. 39:1211-1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chow, J. W., A. Kuritza, D. M. Shlaes, M. Green, D. F. Sahm, and M. J. Zervos. 1993. Clonal spread of vancomycin-resistant Enterococcus faecium between patients in three hospitals in two states. J. Clin. Microbiol. 31:1609-1611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Docquier, J.-D., F. Luzzaro, G. Amicosante, A. Toniolo, and G. M. Rossolini. 2001. Multidrug-resistant Pseudomonas aeruginosa producing PER-1 extended-spectrum serine-β-lactamase and VIM-2 metallo-β-lactamase. Emerg. Infect. Dis. 7:910-911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gales, A. C., R. N. Jones, J. Turnidge, R. Rennie, and R. Ramphal. 2001. Characterization of Pseudomonas aeruginosa isolates: occurrence rates, antimicrobial susceptibility patterns, and molecular typing in the global SENTRY antimicrobial surveillance program, 1997-1998. Clin. Infect. Dis. 32:S146-S155. [DOI] [PubMed] [Google Scholar]
- 9.Gales, A. C., A. O. Reis, and R. N. Jones. 2001. Contemporary assessment of antimicrobial susceptibility testing methods for polymyxin B and colistin: review of available interpretative criteria and quality control guidelines. J. Clin. Microbiol. 39:183-190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Harris, A., C. Torres-Viere, L. Vinkataraman, P. de Girolanie, M. Samore, and Y. Carmeli. 1999. Epidemiology and clinical outcomes of patients with multiresistant Pseudomonas aeruginosa. Clin. Infect. Dis. 28:1128-1133. [DOI] [PubMed] [Google Scholar]
- 11.Jarlier, V., M. J. Nicolas, G. Fournier, and A. Phillipon. 1988. Extended broad-spectrum β-lactamases conferring transferable resistance to newer β-lactam agents in Enterobacteriaceae: hospital prevalence and susceptibility patterns. Rev. Infect. Dis. 10:867-878. [DOI] [PubMed] [Google Scholar]
- 12.Kiska, D. L., and P. H. Gilligan. 1999. Pseudomonas, p. 516-526. In P. R. Murray, E. J. Baron, M. A. Pfaller, F. C. Tenover, and R. H. Yolken (ed.), Manual of clinical microbiology, 7th ed. American Society for Microbiology, Washington, D.C.
- 13.Levin, A. S., A. A. Barone, J. Penço, M. V. Santos, I. S. Marinho, E. A. G. Arruda, E. I. Manrique, and S. F. Costa. 1999. Intravenous colistin as therapy for nosocomial infections caused by multidrug-resistant Pseudomonas aeruginosa and Acinetobacter baumannii. Clin. Infect. Dis. 28:1008-1111. [DOI] [PubMed] [Google Scholar]
- 14.Livermore, D. M., and N. Woodford. 2000. Carbapenemases: a problem in waiting? Curr. Opin. Microbiol. 3:489-495. [DOI] [PubMed] [Google Scholar]
- 15.Moreno, F., P. Grota, and C. Crisp. 1995. Clinical and molecular epidemiology of vancomycin-resistant Enterococcus faecium during its emergence in a city in southern Texas. Clin. Infect. Dis. 21:1234-1237. [DOI] [PubMed] [Google Scholar]
- 16.National Committee for Clinical Laboratory Standards. 2000. Performance standards for antimicrobial disk susceptibility testing. Document M100-S9. National Committee for Clinical Laboratory Standards, Wayne, Pa.
- 17. National Nosocomial Infections Surveillance System. 1999. National Nosocomial Infections Surveillance (NNIS) System report, data summary from January 1990-May 1999, issued June 1999. Am. J. Infect. Control 26:520-523. [DOI] [PubMed] [Google Scholar]
- 18.Nordmann, P., and M. Guibert. 1998. Extended-spectrum β-lactamases in Pseudomonas aeruginosa. J. Antimicrob. Chemother. 42:128-131. [DOI] [PubMed] [Google Scholar]
- 19.Renders, N., U. Romling, H. Verbrugh, and A. Van Belkum. 1996. Comparative typing of Pseudomonas aeruginosa by random amplification of polymorphic DNA or pulsed-field gel electrophoresis of DNA macrorestriction fragments. J. Clin. Microbiol. 34:3190-3195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sader, H. S., A. C. Pignatari, I. L. Leme, M. N. Burattini, R. Trancresi, R. J. Hollis, and R. N. Jones. 1993. Epidemiologic typing of multiply drug-resistant Pseudomonas aeruginosa isolated from an outbreak in an intensive care unit. Diagn. Microbiol. Infect. Dis. 17:13-18. [DOI] [PubMed] [Google Scholar]
- 21.Sousa, M. A., M. Miragaia, I. S. Sanches, S. Ávila, I. Adamson, S. T. Casagrande, M. C. C. Brandileone, R. Palacio, L. Dell'Acqua, M. Hortal, T. Camou, A. Rossi, M. E. Velazquez-Meza, G. Echaniz-Aviles, F. Solorzano-Santos, I. Heitmann, and H. deLencastre. 2001. Three-year assessment of methicillin resistant Staphylococcus aureus clones in Latin America from 1996 to 1998. J. Clin. Microbiol. 39:2197-2205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Teixeira, L. M., M. G. S. Carvalho, V. L. C. Merquior, A. G. Steigerwalt, D. J. Brenner, and R. R. Facklam. 1997. Phenotypic and genotypic characterization of Vagococcus fluvialis including strains isolated from human sources. J. Clin. Microbiol. 35:2778-2781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tenover, F. C., R. D. Arbeit, R. V. Goering, P. A. Mickelsen, B. E. Murray, D. H. Persing, and B. Swaminathan. 1995. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J. Clin. Microbiol. 33:2233-2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Thomson, K. S. 2001. Controversies about extended-spectrum and AmpC beta-lactamases. Emerg. Infect. Dis. 7:333-336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Widmer, A. F., R. P. Wenzel, A. Trilla, M. J. Bale, R. N. Jones, and B. N. Doebbeling. 1993. Outbreak of Pseudomonas aeruginosa infections in a surgical intensive care unit: probable transmission via hands of a health care worker. Clin. Infect. Dis. 16:372-376. [DOI] [PubMed] [Google Scholar]
- 26.Yan, J.-J., P.-R. Hsueh, K. Wen-Chien, K.-T. Luh, S.-H. Tsai, H.-M. Wu, and J.-J. Wu. 2001. Metallo-β-lactamases in clinical Pseudomonas isolates in Taiwan and identification of VIM-3, a novel variant if the VIM-2 enzyme. Antimicrob. Agents Chemother. 45:2224-2228. [DOI] [PMC free article] [PubMed] [Google Scholar]