Abstract
Although the virulences and host ranges differ among members of the Mycobacterium tuberculosis complex (TBC; M. tuberculosis, M. africanum, M. canettii, M. microti, M. bovis, and M. bovis BCG), commercially available molecular assays cannot differentiate these organisms because of the genetic identities of their 16S rRNA gene sequences. Comparative genomic analyses with the complete DNA sequence of M. tuberculosis H37Rv has provided information on regions of difference (RD 1 to RD 16) deleted in members of the TBC other than M. tuberculosis. To determine whether deletion analysis could accurately differentiate members of TBC, we used PCR to assess the presence or absence of specific regions of the genome in 88 well-characterized isolates of M. tuberculosis, M. africanum, M. microti, M. bovis, and M. bovis BCG. The identifications obtained by use of the specific deletion profiles correlated 100% with the original identifications for all TBC members except M. africanum, but further characterization resulted in profiles specific for all members. Although six RD regions were used in the analyses with the original 88 isolates, it was found that the use of RD 1, RD 9, and RD 10 was sufficient for initial screenings, followed by the use of RD 3, RD 5, and RD 11 if the results for any of the first three regions were negative. When 605 sequential clinical isolates were screened, 578 (96%) were identified as M. tuberculosis, 6 (1%) were identified as M. africanum, 8 (1%) were identified as M. bovis, and 13 (2%) were identified as M. bovis BCG. Since PCR-based assays can be implemented in most clinical mycobacteriology laboratories, this approach provides a rapid and simple means for the differentiation of members of TBC, especially M. bovis and M. tuberculosis, when it is important to distinguish between zoonotic sources (i.e., cattle and unpasteurized dairy products) and human sources of tuberculosis disease.
The Mycobacterium tuberculosis complex (TBC) (4, 34) comprises the closely related organisms M. tuberculosis, M. africanum, M. bovis, the M. bovis BCG vaccine strain, and two rarely seen members, M. microti and M. canettii (35). Differentiation of the members of the TBC is necessary for the treatment of individual patients and for epidemiological purposes, especially in areas of the world where tuberculosis has reached epidemic proportions or wherever the transmission of M. bovis between animals or animal products and humans is a problem. In addition, it can be important to rapidly identify isolates of M. bovis BCG recovered from immunocompromised patients.
Although no clear-cut means of differentiation of the members of the TBC was found in the past by using numerical classification (34), a few conventional methods have been useful. Those methods include assays for the ability to metabolize glycerol or pyruvate in Loewenstein-Jensen medium, oxygen preference (aerophilic versus microaerophilic), niacin accumulation, nitrate reductase activity, colony morphology, and resistance to two compounds, thiophen-2-carboxylic acid hydrazide (TCH) and pyrazinamide (PZA) (12, 19, 38). Partially due to the slow growth of the TBC, interpretation of the results of these assays can be highly subjective, especially interpretation of differences in colony morphology (19), which can be due to the loss of virulence or to mutations associated with drug resistance. An alternative approach is the use of high-performance liquid chromatography; however, only the profile for M. bovis BCG differs from those for the other members of the complex (10).
Testing for resistance to TCH has been reported to be the only single test that assigned isolates to any specific member of the TBC; classical M. tuberculosis isolates are resistant to TCH, irrespective of their resistance to isoniazid (8). Alternatively, the Asian strain of M. tuberculosis and all other members of the TBC are TCH susceptible (39, 40). However, cross-resistance to TCH has been seen in isolates of M. bovis expressing resistance to high levels of isoniazid (0.4 μg/ml) (L. M. Parsons, unpublished observations, 2001). Multidrug-resistant TBC isolates can be resistant to PZA, but resistance only to PZA (monodrug resistance) is found almost exclusively in M. bovis and M. bovis BCG (12, 38; M. Salfinger, L. B. Reller, and F. M. Kafader, Abstr. 90th Annu. Meet. Am. Soc. Microbiol. 1990, abstr. U-55, p. 150, 1990). Because M. africanum has a phenotype intermediate between those of M. tuberculosis and M. bovis and is susceptible to PZA, this member of the TBC was originally called M. bovis, Afro-Asian variety (23). However, difficulties in the precise definition of M. africanum were further complicated when variants associated with different geographic regions were described (variant I was an M. bovis-like organism that was nitrate negative and that was from West Africa; variant II was an M. tuberculosis-like organism that was nitrate positive and that was from East Africa) (7). A more recent study from West Africa suggested that similar numbers of the two variants are present in the population in Guinea-Bissau (14).
In DNA-DNA hybridization assays, the four members of the TBC tested were found to share 85 to 100% DNA-DNA relatedness (15). Subsequently, completely conserved DNA sequences were reported for the 16S rRNA gene (rDNA) and 16S-23S rDNA spacers (20). Furthermore, no significant nucleotide sequence variations either in 26 structural genes or in 24 genes coding for proteins that are targets of the host immune system were found among a diverse group of isolates of M. tuberculosis (24, 31).
Unfortunately, the high degree of sequence conservation among the members of the TBC has resulted in difficulties for the clinical mycobacteriology laboratory since commercial DNA probe and amplification assays based on 16S rDNA sequences, identical for all TBC members, cannot be used to differentiate members of the complex. Further complications have arisen following the addition to the complex of three recently described members that are positive by commercial hybridization assays: M. canettii (35), M. tuberculosis subsp. caprae (1), and the unnamed seal bacillus (41).
Despite the difficulties described above, advances in molecular methods and the accumulating knowledge of the M. tuberculosis genome have resulted in methods designed to rapidly identify the members of the TBC. These methods are based on differences in various alleles and repetitive regions, mutations associated with drug resistance, and transposition of mobile elements (Table 1). While these variations in molecular characteristics have enabled scientific distinctions to be made between the different members of the complex, the complexities of the methods have hindered development of a single, direct assay for rapid identification.
TABLE 1.
Genetic differences among members of the TBC
| Component evaluated | Difference | Reference(s) |
|---|---|---|
| Variable alleles | ||
| oxyR nucleotide 285 | A in M. bovis, G in other members of the TBC | 30 |
| pncA nucleotide 169 | G in M. bovis and M. bovis BCG, C in other members of the TBC | 28 |
| katG codon 463 | CTG (Leu) in group 1, CGG (Arg) in group 2, CGG (Arg) in group 3a | 31 |
| gyrA codon 95 | ACC (Thr) in group 1, ACC (Thr) in group 2, AGC (Ser) in group 3a | |
| gyrB | Sequence differences among members of the TBC | 17, 25 |
| Variable sequences for spacers between direct repeatsb | ||
| Spacers 33 to 36 (derived from BCG) | M. tuberculosis does not hybridize to the spacers | 16 |
| Spacers 39 to 43 (derived from M. tuberculosis) | M. bovis and BCG do not hybridize to the spacers | 16 |
| Spacers 37 and 38 | M. microti has a very short direct repeat region; many strains only hybridize to spacers 37 and 38 | 36 |
| Spacers 8, 9 and 39 | M. africanum does not hybridize to the spacers | 37 |
Group 1 contains M. tuberculosis, M. africanum, M. microti, and M. bovis; groups 2 and 3 contain only M. tuberculosis.
As determined by spoligotyping.
Recently, comparative genomics with the complete DNA sequence of M. tuberculosis H37Rv has resulted in the demonstration of 16 regions of the genome (regions of difference [RD]) deleted in M. bovis and M. bovis BCG; subsequent studies found that some of these regions are also deleted in other members of the TBC (2, 3, 4, 11, 22). On the basis of these data, the aim of the present study was to investigate whether molecular amplification methods for determination of the presence or absence of specific regions of the genome could be used by clinical laboratories as rapid and simple assays for the precise identification of individual members of the TBC. This approach was validated with 88 well-characterized TBC isolates and was then used to identify 605 members of the TBC recovered from clinical specimens from March 2000 through June 2001.
MATERIALS AND METHODS
Bacterial isolates.
In the first phase of the study, 88 members of the TBC were obtained from specimens submitted to the following laboratories: the Clinical Mycobacteriology Laboratory, Wadsworth Center, Albany, N.Y.; the Armauer Hansen Institute, German Leprosy Relief Association, Würzburg, Germany; the National Institute of Public Health and the Environment, Bilthoven, The Netherlands; and the Florida Department of Health, Jacksonville. In addition, strains of M. africanum and M. microti were purchased from the American Type Culture Collection. Each laboratory that submitted isolates confirmed that the isolates belonged to the TBC by commercial nucleic acid amplification or hybridization methods, with the final identification based on conventional phenotypic methods (12, 19, 38). Once the study protocol was established with the 88 previously identified isolates, the additional 605 isolates belonging to the TBC were obtained from specimens submitted to the Wadsworth Center from March 2000 through June 2001.
PCR deletion analyses.
The genomes of the isolates were analyzed by PCR for the presence or the absence of six regions (RD 1, RD 3, RD 5, RD 9, RD 10, and RD 11) originally described as being deleted in the genomes of BCG isolates relative to the sequence of M. tuberculosis H37Rv (2, 3, 11, 22). A multiprimer PCR assay with three primers was used to detect RD 1, RD 9, and RD 10. In these assays, two primers complementary to sequences flanking the deleted region amplified a small product from strains from which the region was deleted. A third primer complementary to internal sequences and one of the flanking primers amplified a product of a different size when the region was present since the two flanking primers are too far apart to efficiently amplify the entire region for those strains. The primer sequences and PCR product sizes are listed in Table 2, with the sequence of the primer for RD 1 (9,455 bp) taken from that used in the previously published RD 1 multiprimer assay (32). The sequences for the primers for RD 9 (2,030 bp) and RD 10 (1,903 bp) were determined for this study by retrieving the sequences of the deleted regions (Sequence Retrieval Service, Institut Pasteur [www.srs.pasteur.fr]; accession numbers, Y181604 and AJ131209 for RD 9 and RD 10, respectively) and the flanking sequences for these two regions published by Gordon and coworkers (11). Primers were selected by importing the sequences into the Primer 3 program at www.genome.wi.mit.edu. For these three assays, the 50-μl reaction mixture contained each of the two flanking primers at a concentration of 10 mM, 50 mM internal primer, 50 mM KCl, 10 mM Tris-HCl (pH 8.3), 1.5 mM MgCl2, 0.01% gelatin, each deoxynucleoside triphosphate at a concentration of 200 μM, 1.25 U of Taq DNA polymerase, and 10 μl of heat-killed (1 h at 80°C) bacterial cells. After denaturation at 95°C for 5 min, the reaction mixtures were incubated for 40 cycles at 94°C for 30 s and 1 min at 65°C, followed by 10 min at 72°C, in a GeneAmp 9700 (Perkin-Elmer Biosystems, Foster City, Calif.). Fifteen-microliter samples were run at 125 V for 1 h on a 2% agarose gel, and the band size was estimated by comparison to a 1-kb-plus DNA ladder (Gibco BRL, Life Technologies, Gaithersburg, Md.).
TABLE 2.
Primer sequences for deletion analyses
| Region and primer | Sequence |
|---|---|
| RD 1 (region present, 150 bp; region absent, 200 bp) | |
| ET1 | 5′-AAG-CGG-TTG-CCG-CCG-ACC-GAC-C-3′ |
| ET2 | 5′-CTG-GCT-ATA-TTC-CTG-GGC-CCG-G-3′ |
| ET3 | 5′-GAG-GCG-ATC-TGG-CGG-TTT-GGG-G-3′ |
| RD 3 (region present, 500 bp; region absent, no product) | |
| RD3intF | 5′-TTA-TCT-TGG-CGT-TGA-CGA-TG-3′ |
| RD3intR | 5′-CAT-ATA-AGG-GTG-CCC-GCT-AC-3′ |
| RD 5 (region present, 152 bp; region absent, no product) | |
| mtp40F | 5′-CTG-GTC-GAA-TTC-GGT-GGA-GT-3′ |
| mtp40R | 5′-ATG-GTC-TCC-GAC-ACG-TTC-GAC-3′ |
| RD 9 (region present, 306 bp; region absent, 206 bp) | |
| RD9 FF | 5′-GTG-TAG-GTC-AGC-CCC-ATC-C-3′ |
| RD9 Int | 5′-CAA-TGT-TTG-TTG-CGC-TGC-3′ |
| RD9 FR | 5′-GCT-ACC-CTC-GAC-CAA-GTG-TT-3′ |
| RD 10 (region present, 308 bp; region absent, 202 bp) | |
| RD10 FF | 5′-CTG-CAA-CCA-TCC-GGT-ACA-C-3′ |
| RD10 Int | 5′-GAA-GTC-GTA-ACT-CAC-CGG-GA-5′ |
| RD10 FR | 5′-AAG-CGC-TAC-ATC-GCC-AAG-3′ |
| RD 11 (region present, 454 bp; region absent, no product) | |
| RD11intF | 5′-CGG-CAG-CTA-GAC-GAC-CTC-3′ |
| RD11intR | 5′-AAC-GTG-CTG-CGA-TAG-GTT-TT-3′ |
Because RD 5 is flanked by a repetitive region and RD 3 and RD 11 contain potentially mobile bacteriophages, only internal primers were used for the detection of these regions. The assay for RD 5 amplified a 152-bp product from mtp40 (plcA), as described previously (33). The primers for RD 3 and RD 11 were obtained for this study by using the Primer 3 program from the integrase genes from lysogenic bacteriophages φRv1 and φRv2, respectively. The primers, listed in Table 2, were used to amplify 500- and 454-bp products when RD 3 (φRv1) and RD 11 (φRv2) were present. For these three assays, the same reaction components described above were used, but with different cycling temperatures and times. For RD 5, the samples were denatured at 95°C for 5 min and were then incubated at 94°C for 30 s, 64°C for 30 s, and 30 s at 72°C for 40 cycles, followed by 10 min at 72°C. For RD 3 and RD 11, following denaturation at 95°C for 5 min, the samples were incubated at 95°C for 1 min, 55°C for 1 min, and 1 min at 72°C for 40 cycles, followed by 10 min at 72°C. The products were analyzed as described above.
Phenotypic characterization.
Drug resistance assays were performed by standard radiometric procedures with the BACTEC instrument and included assays for resistance to 100 μg of PZA per ml and 1 μg of TCH per ml (13, 27, 29). The PZA assay was completed at the time when the growth index (GI) for the control vial was equal to or greater than 200. If the GI for the PZA-containing vial was greater than 10% of that for the control vial at that time, the isolate was considered resistant to PZA. Resistance to TCH was noted when a steady increase in the GI for the TCH-containing vial was greater than the increase seen for a control vial with no drug. In previous studies, 5 μg of TCH per ml was used in assays with solid medium, whereas 1 to 2 μg of TCH per ml was used in the broth-based radiometric test (8, 29).
An oxygen preference assay (12, 21) was performed by inoculating 0.2 ml of an actively growing bacterial suspension into 10 ml of Middlebrook 7H9 broth containing 0.2% agar (in a screw-cap tube). The location of the growth in this semisolid medium was evaluated after 3 weeks of incubation at 37°C. A preference for aerophilic conditions was suggested by growth on the surface or no more than 5 mm below the surface of the medium. A preference for microaerophilic conditions was suggested by a band of growth approximately 10 to 15 mm below the surface. The percentage of agar used in the oxygen preference assay was increased to 0.2% from the 0.1% described previously (12) because the results could not be clearly visualized when the lower percentage was used. The use of 0.2% agar resulted in a clear distinction between aerophilic and microaerophilic growth. Additional assays included tests for niacin accumulation and nitrate reductase, performed by standard methods with organisms from a 2- to 3-week-old growth on Loewenstein-Jensen medium (19).
RESULTS
Results of deletion analyses with 88 previously characterized isolates.
At the time when this study began, 16 regions had been found to be deleted in M. bovis BCG relative to the sequence of M. tuberculosis H37Rv (2, 3, 11, 22). It was shown that some of these regions were also absent from other members of the TBC, thus suggesting that the presence or absence of these regions could be useful in differentiation of members of the complex. For this study, six regions from various locations in the chromosome were selected for use as possible identification tools. The regions were RD 1, absent only in M. bovis BCG; RD 5, present in most strains of M. tuberculosis, M. africanum, and M. microti but absent in M. bovis and M. bovis BCG; RD 3 and RD 11, lysogenic bacteriophages found to be useful in differentiating M. bovis and M. microti; and RD 9 and RD 10, absent in isolates of M. africanum (2, 3, 4, 11, 22).
Typical reactions demonstrating the presence or absence of the six RD regions are shown in Fig. 1, with the results for the 88 previously characterized strains listed in Table 3. The 27 strains of M. tuberculosis were all positive (100%) for RD 1, RD 9, RD 10, and RD 11; most (96%) were positive for RD 5; and there were variable results for RD 3 (26%). The 25 strains previously identified as M. africanum were more genetically diverse, with 100% positivity seen only for RD 1. However, 6 of the 25 strains were positive for all six regions, and further analysis of representative strains (see below) resulted in a phenotype consistent with that of M. tuberculosis. When the results for the 19 other M. africanum strains were evaluated alone, a distinct profile was seen (Table 3): RD 1 was always present and RD 9 was always absent, with variable results obtained for the other regions. Although only five strains of M. microti were analyzed, a distinct profile was seen for these strains: all were positive for RD 1 and RD 11, variable for RD 5 (60%), and negative for RD 3, RD 9, and RD 10. The 14 M. bovis strains also contained RD 1, were often positive for RD 3 (71%), and were negative for RD 5, RD 9, RD 10, and RD 11. Finally, all six regions were absent in the 17 BCG strains tested.
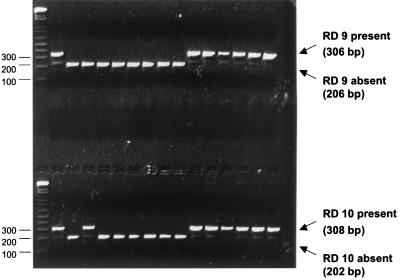
FIG. 1.
Results of PCR for presence of RD 9 and RD 10. The larger PCR products indicate the presence of RD 9 (upper lanes) or RD 10 (lower lanes), while the smaller products indicate that the region has been deleted. The size standards (in base pairs) are labeled on the left side of the same 2% agarose gel prepared with a double comb.
TABLE 3.
Percentages of the 88 members of TBC that contain the six RD regions and selected phenotypes of the strains
| RD region or phenotype | % of isolates
|
|||||
|---|---|---|---|---|---|---|
| M. tuberculosis (27)a | M. africanumb (25) | M. africanumc (19) | M. microti (5) | M. bovis (14) | BCG (17) | |
| RD 1d | 100 | 100 | 100 | 100 | 100 | 0 |
| RD 3 | 26 | 32 | 11 | 0 | 71 | 0 |
| RD 5 | 96 | 96 | 95 | 60 | 0 | 0 |
| RD 9 | 100 | 24 | 0 | 0 | 0 | 0 |
| RD 10 | 100 | 56 | 42 | 0 | 0 | 0 |
| RD 11 | 100 | 56 | 42 | 100 | 0 | 0 |
| Monodrug resistance to PZA | 0 | 0 | 0 | 0 | 100 | 100 |
| Resistance to TCH | 100 | 4 | 0 | 0 | 0 | 0 |
| Preference for O2e | A | A or M | M | A | M | A |
| Niacin positive | 93 | 90 | 88 | NDf | 0 | 0 |
| Nitrate positive | 86 | 0 | 0 | ND | 0 | 25 |
The values in parentheses are the numbers of isolates.
Six of the 25 isolates originally identified as M. africanum were positive for all six regions; however, two of the six were found to phenotypically resemble M. tuberculosis upon retesting.
The six isolates resembling M. tuberculosis phenotypically were omitted from this group.
The boldface indicates the three regions selected for initial screening.
A, aerophilic growth preferred; M, microaerophilic growth preferred.
ND, not done.
Selected phenotypic assays.
Because there was some overlap in the deletion profiles, assays for the detection of resistance to TCH and PZA and oxygen preference were added to confirm the identifications for the isolates. Although the Asian strain of M. tuberculosis can be TCH susceptible, previous classification schemes have listed only classical M. tuberculosis isolates as TCH resistant; likewise, monodrug resistance to PZA has been listed only for M. bovis and M. bovis BCG, and a preference for microaerophilic conditions differentiates M. africanum and M. bovis from the other members of the complex (Fig. 2) (12, 19, 38).
FIG. 2.
Oxygen preference test (Middlebrook 7H9 broth with 0.2% agar). Four different members of the M. tuberculosis complex are shown, with the oxygen preference typical for each member indicated. Tube 1, M. tuberculosis; tube 2, M. africanum; tube 3, M. bovis; tube 4, M. bovis BCG.
These three assays were used to test viable representatives of the original 88 isolates. The following results were consistent for each member of the TBC, as shown in Table 3: M. tuberculosis, TCH resistant, no monodrug resistance to PZA, preference for aerophilic conditions; M. microti, TCH susceptible, no monodrug resistance to PZA, preference for aerophilic conditions; M. bovis, TCH susceptible, monodrug resistance to PZA, preference for microaerophilic conditions; and M. bovis BCG, TCH susceptible, monodrug resistance to PZA, preference for aerophilic conditions.
For the group of isolates that were previously identified as M. africanum, monodrug resistance to PZA was not seen; however, variable results were obtained in the assays for resistance to TCH and oxygen preference. The 19 isolates that were negative for RD 9 were consistently susceptible to TCH and preferred microaerophilic conditions. However, two viable representatives of the six isolates that were positive for all RD regions were aerophilic, with one isolate being resistant to TCH and the other being susceptible. These isolates were found to be positive for niacin and negative for nitrate. The original identification was based solely on the phenotype; however, some phenotypic assays are variable for both M. tuberculosis and M. africanum. In some instances, strains of M. tuberculosis can be susceptible to TCH or have a negative reaction for nitrate reductase, phenotypes that are usually listed for M. africanum. Thus, we propose that a more precise differentiation of the members of the TBC should include an analysis for the presence or absence of these RD regions, and for an isolate to be identified as M. africanum, RD 9 should be absent.
Use of deletion analyses for screening of 605 clinical isolates.
For the screening of the 605 clinical isolates obtained from a diverse patient population in New York State including New York City, the following strategy was used. After detection of growth of a pure culture of TBC in a BACTEC 12B vial, PZA and TCH were inoculated as part of the routine susceptibility testing. In addition, a portion of the bacterial suspension was heat killed and PCR assays for RD 1, RD 9, and RD 10 were performed. If any of the regions were absent, PCR assays for RD 3, RD 5, and RD 11 were performed. If the organism was susceptible to TCH, an oxygen preference assay was inoculated. Some TCH-resistant isolates were also tested for oxygen preference.
Table 4 summarizes the results of these assays. Of the 605 isolates, 578 (96%) were identified as M. tuberculosis, 6 (1%) were identified as M. africanum, 8 (1%) were identified as M. bovis, and 13 (2%) were identified as M. bovis BCG.
TABLE 4.
Results from screening of 605 clinical isolates belonging to TBC
| Profilea | Identification | No. (%) of isolates
|
Oxygen preferenceb | ||
|---|---|---|---|---|---|
| Total | TCH resistant | Resistant only to PZA | |||
| A | M. tuberculosis | 578 | 517 (89) | 0 | Aerophilic (87/90 [97]) |
| B | M. africanum | 6 | 0 | 0 | Microaerophilic (6/6 [100]) |
| C | M. bovis | 8 | 0 | 8 (100) | Microaerophilic (8/8 [100]) |
| D | M. bovis BCG | 13 | 0 | 13 (100) | Aerophilic (13/13 [100]) |
A, positive for RD 1, RD 9, RD 10 (tests for RD 3, RD 5, and RD 11 were not done); B, positive for RD 1, negative for RD 9, and variable for RD 3, RD 5, RD 10, and RD 11; C, positive for RD 1, variable for RD 3, and negative for RD 5, RD 9, RD 10, and RD 11; D, negative for all six regions.
The values in parentheses indicate the number of isolates with the indicated oxygen reference/total number of isolates tested (percent).
Of the 578 isolates identified as M. tuberculosis on the basis of the presence of RD 1, RD 9, and RD 10, 89% were the “classical” human strain resistant to TCH, 11% were the TCH-susceptible Asian strain, and none were drug resistant only to PZA. There were slight differences in the percentages of niacin- and nitrate-positive isolates in the two groups. Among the isolates of the classical strain tested, 99% (438 of 441) were niacin positive and 97% (426 of 441) were nitrate positive, while among the isolates of the Asian strain tested, 100% (52 of 52) were niacin positive and 88% (46 of 52) were nitrate positive. The oxygen preferences of the two groups also differed slightly, with 100% (38 of 38) of the isolates of the classical strain and 94% (49 of 52) of the isolates of the Asian strain preferring aerophilic conditions. These results demonstrate that the assay is capable of rapidly identifying M. tuberculosis isolates, even those with an atypical phenotype.
Of the six isolates identified as M. africanum on the basis of the presence of RD 1, the absence of RD 9, and variable results for RD 3, RD 5, RD 10, and RD 11, 100% (six of six isolates) were susceptible to TCH and PZA and preferred microaerophilic conditions. In addition, 83% (five of six) were niacin positive and 33% (two of six) were nitrate positive.
As was the case for the M. bovis and M. bovis BCG isolates among the original 88 isolates tested (Table 3), M. bovis and M. bovis BCG were the easiest to differentiate from the other members of the TBC. The 8 isolates of M. bovis (which were positive for RD 1 and variable for RD 3, with RD 5, RD 9, RD 10, and RD 11 being absent) and the 13 isolates of M. bovis BCG (from which all six regions were absent) were all susceptible to TCH and resistant only to PZA. These variants differed in their oxygen preferences, with M. bovis preferring microaerophilic conditions and M. bovis BCG preferring aerophilic conditions.
DISCUSSION
The housekeeping genes and genes encoding immunogenic proteins are highly conserved among members of the TBC (24, 31), such that commercially available nucleic acid probes and amplification assays cannot differentiate the different organisms in the complex. Comparative genomics of the members of the TBC by use of subtractive hybridization (22), bacterial artificial chromosome arrays (3, 11), or DNA microarrays (2) identified 16 regions ranging in size from 2 to 12.7 kb that were present in M. tuberculosis H37Rv and absent in most BCG derivatives and also in other members of the TBC. These results suggested that deletion of genomic regions has been important in generating genetic diversity within the complex (4, 18, 26). Deletions often arise from recombination between insertion sequence (IS) elements (5, 9), and the M. tuberculosis genome contains greater than 40 ISs and mobile genetic elements that could mediate deletions (6). Furthermore, the presence of variable regions in members of the TBC suggested that this approach could be used as a tool for differentiation of members of the TBC. On the basis of the results of these studies, we sought to validate the use of deletion analysis as a solution to the problem faced by clinical mycobacteriology laboratories for differentiation of the members of the TBC.
The junction sequences flanking the variable regions used in this study suggest that at least two different mechanisms are responsible for the deletions. First, RD 3, RD 5, and RD 11 all contain mobile genetic elements (prophage φRv1, insertion element IS6110, and prophage φRv2, respectively). The distribution of these regions in the members of the TBC can be variable. In contrast, the junction regions bordering RD 1, RD 9, and RD 10 do not contain repetitive sequences; these deletions occur in coding regions and result in the truncation of genes (11). The exact mechanism for this type of deletion remains obscure, but DNA polymerase slippage errors may be responsible.
Because of the conservation of junction sequences flanking RD 1, RD 9, and RD 10, multiprimer PCR assays were used to detect these regions. Three primers were included in each assay; two primers were specific for the sequences that flanked the region, and the third was specific for an internal sequence close to one of the flanking primers. Thus, the size of the PCR product was used to determine the presence or the absence of these regions. In contrast, the junction sequences flanking variable regions RD 3, RD 5, and RD 11 are not highly conserved because these regions contain mobile genetic elements whose distributions can differ. Thus, only two primers complementary to internal regions were used for these assays.
First, 88 well-characterized members of the TBC were tested for the presence or the absence of six regions located in various parts of the chromosome. Specific deletion profiles were found for each of the members of the TBC (Table 3), with only M. tuberculosis strains containing regions RD 1, RD 9, and RD 10. Significantly, a recent study on the evolution of the members of the TBC also found that the sequences of M. tuberculosis strains are highly conserved with respect to RD 1, RD 9, and RD 10 and that these three regions can be used to differentiate M. tuberculosis strains from the other members of the TBC (4).
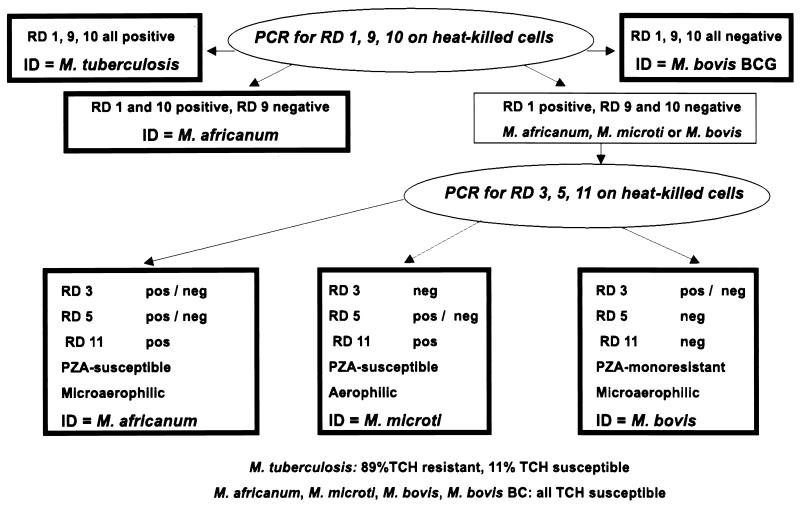
On the basis of the results presented here, a new approach (Fig. 3) was used to rapidly differentiate 605 members of the TBC isolated from patient specimens (Table 4). This approach used prescreening of the isolates by three PCR assays (for RD 1, RD 9, and RD 10). When all three regions were present, the isolate was identified as M. tuberculosis. When all three regions were absent, the isolate was identified as M. bovis BCG. Isolates that lacked only RD 9 were identified as M. africanum, a result also reported in a recent study (4). Those lacking both RD 9 and RD 10 were tested for the presence of RD 3, RD 5, and RD 11; and the results were used to identify M. bovis and M. africanum (and potentially M. microti). This approach provided rapid identifications of members of the TBC that were later confirmed by slower conventional assays, such as TCH and PZA susceptibility tests and assays for oxygen preference, niacin accumulation, and nitrate reductase activity.
FIG. 3.
Flow chart for identification of TBC isolates by deletion analysis.
This study has opened new perspectives for the rapid identification of individual strains of the TBC in the clinical laboratory. Although more than 90% of the isolates encountered in this study were identified as M. tuberculosis, the results suggest that it would be advantageous to use this approach in areas of the world where unusual members of the TBC are more common. The application of the pathway presented here might become even more powerful by including an assay for the M. tuberculosis-specific deletion (TbD1) that was identified only recently (4). Finally, we propose that this PCR-based approach, which is simple to perform, can be incorporated into the laboratory routine by many clinical mycobacteriology laboratories. While assays requiring a multistep hybridization technique such as spoligotyping or DNA sequencing may not be within the scope of a clinical diagnostic laboratory, many laboratories use amplification procedures. In our experience, this approach has provided a means for the rapid and clear differentiation of members of TBC and determination of the prevalence of each of the members within the population in New York State. Moreover, most of these isolates are clinically significant, and patient treatment is dependent on their correct and timely identification, suggesting that use of this method is likely to enhance primary care and public health services.
Acknowledgments
We acknowledge the technical contributions of Tiffany Bratts, Jennifer Greggo, Svetlana Popova, Jacquelin Hotaling, Dianne O'Donnell, Alfred Waring, and Andreas Kilmartin and thank Ron Limberger and Judit Mester for critical review of the manuscript.
Ákos Somoskövi was supported by grants 1D43TW00915 and 2D43TW00233 from the Fogarty International Center, National Institutes of Health, Bethesda, Md.
REFERENCES
- 1.Aranaz, A., E. Liebana, E. Gomez-Mampaso, J. C. Galan, D. Cousins, A. Ortega, J. Blazquez, F. Baquero, A. Mateos, G. Suarez, and L. Dominguez. 1999. Mycobacterium tuberculosis subsp. caprae subsp. nov.: a taxonomic study of a new member of the Mycobacterium tuberculosis complex isolated from goats in Spain. Int. J. Syst. Bacteriol. 49:1263-1273. [DOI] [PubMed] [Google Scholar]
- 2.Behr, M. A., M. A. Wilson, W. P. Gill, H. Salamon, G. K. Schoolnik, S. Rane, and P. M. Small. 1999. Comparative genomics of BCG vaccines by whole-genome DNA microarray. Science 284:1520-1523. [DOI] [PubMed] [Google Scholar]
- 3.Brosch, R., S. V. Gordon, A. Billault, T. Garnier, K. Eiglmeier, C. Soravito, B. G. Barrell, and S. T. Cole. 1998. Use of a Mycobacterium tuberculosis H37Rv bacterial artificial chromosome library for genome mapping, sequencing, and comparative genomics. Infect. Immun. 66:2221-2229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brosch, R., S. V. Gordon, M. Marmiesse, P. Brodin, C. Buchrieser, K. Eiglmeier, T. Garnier, C. Gutierrez, G. Hewinson, K. Kremer, L. M. Parsons, A. S. Pym, S. Samper, D. van Soolingen, and S. T. Cole. 2002. A new evolutionary scenario for the Mycobacterium tuberculosis complex. Proc. Natl. Acad. Sci. USA 99:3684-3689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brosch, R., W. J. Philipp, E. Stavropoulos, M. J. Colston, S. T. Cole, and S. V. Gordon. 1999. Genomic analysis reveals variation between Mycobacterium tuberculosis H37Rv and the attenuated M. tuberculosis H37Ra strain. Infect. Immun. 67:5768-5774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cole, S. T., R. Brosch, J. Parkhill, T. Garnier, C. Churcher, D. Harris, S. V. Gordon, K. Eiglmeier, S. Gas, C. E. Barry III, F. Tekaia, K. Badcock, D. Basham, D. Brown, T. Chillingworth, R. Connor, R. Davies, K. Devlin, T. Feltwell, S. Gentles, N. Hamlin, S. Holroyd, T. Hornsby, K. Jagels, B. G. Barrell, et al. 1998. Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature 393:537-544. [DOI] [PubMed] [Google Scholar]
- 7.Collins, C. H., and M. D. Yates. 1984. Mycobacterium africanum and the ‘African' tubercle bacilli. Med. Lab. Sci. 41:410-413. [PubMed] [Google Scholar]
- 8.Collins, T., and P. N. Levett. 1989. Radiometric studies on the use of selective inhibitors in the identification of Mycobacterium spp. J. Med. Microbiol. 30:175-181. [DOI] [PubMed] [Google Scholar]
- 9.Fang, Z., C. Doig, D. T. Kenna, N. Smittipat, P. Palittapongarnpim, B. Watt, and K. J. Forbes. 1999. IS6110-mediated deletions of wild-type chromosomes of Mycobacterium tuberculosis. J. Bacteriol. 181:1014-1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Floyd, M. M., V. A. Silcox, W. D. Jones, Jr., W. R. Butler, and J. O. Kilburn. 1992. Separation of Mycobacterium bovis BCG from Mycobacterium tuberculosis and Mycobacterium bovis by using high-performance liquid chromatography of mycolic acids. J. Clin. Microbiol. 30:1327-1330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gordon, S. V., R. Brosch, A. Billault, T. Garnier, K. Eiglmeier, and S. T. Cole. 1999. Identification of variable regions in the genomes of tubercle bacilli using bacterial artificial chromosome arrays. Mol. Microbiol. 32:643-655. [DOI] [PubMed] [Google Scholar]
- 12.Grange, J. M., M. D. Yates, and I. N. de Kantor. 1996. Guidelines for speciation within the Mycobacterium tuberculosis complex, p. 1-18. Emerging and other communicable diseases, surveillance and control. Report WHO/EMC/ZOO/96.4. World Health Organization, Geneva, Switzerland.
- 13.Gross, W. M., and J. E. Hawkins. 1985. Radiometric selective inhibition tests for differentiation of Mycobacterium tuberculosis, Mycobacterium bovis, and other mycobacteria. J. Clin. Microbiol. 21:565-568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hoffner, S. E., S. B. Svenson, R. Norberg, F. Dias, S. Ghebremichael, and G. Kallenius. 1993. Biochemical heterogeneity of Mycobacterium tuberculosis complex isolates in Guinea-Bissau. J. Clin. Microbiol. 31:2215-2217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Imaeda, T. 1985. Deoxyribonucleic acid relatedness among selected strains of Mycobacterium tuberculosis, Mycobacterium bovis, Mycobacterium bovis BCG, Mycobacterium microti, and Mycobacterium africanum. Int. J. Syst. Bacteriol. 35:147-150. [Google Scholar]
- 16.Kamerbeek, J., L. Schouls, A. Kolk, M. van Agterveld, D. van Soolingen, S. Kuijper, A. Bunschoten, H. Molhuizen, R. Shaw, M. Goyal, and J. van Embden. 1997. Simultaneous detection and strain differentiation of Mycobacterium tuberculosis for diagnosis and epidemiology. J. Clin. Microbiol. 35:907-914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kasai, H., T. Ezaki, and S. Harayama. 2000. Differentiation of phylogenetically related slowly growing mycobacteria by their gyrB sequences. J. Clin. Microbiol. 38:301-308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kato-Maeda, M., P. J. Bifani, B. N. Kreiswirth, and P. M. Small. 2001. The nature and consequence of genetic variability within Mycobacterium tuberculosis. J. Clin. Investig. 107:533-537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kent, P. T., and G. P. Kubica. 1985. Public health mycobacteriology: a guide for the level III laboratory. Centers for Disease Control, Public Health Service, U.S. Department of Health and Human Services, Atlanta, Ga.
- 20.Kirschner, P., B. Springer, U. Vogel, A. Meier, A. Wrede, M. Kiekenbeck, F. C. Bange, and E. C. Bottger. 1993. Genotypic identification of mycobacteria by nucleic acid sequence determination: report of a 2-year experience in a clinical laboratory. J. Clin. Microbiol. 31:2882-2889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lebek, G. 1958. Ueber den Nachweis des unterschiedlichen Sauerstoffoptimums des humanen and bovinen Mycobacterium tuberculosis. Zentbl. Bakteriol. Parasitenkd. Infektkrankh. Hyg. Abt. 1 Orig. 173:581-587. [PubMed] [Google Scholar]
- 22.Mahairas, G. G., P. J. Sabo, M. J. Hickey, D. C. Singh, and C. K. Stover. 1996. Molecular analysis of genetic differences between Mycobacterium bovis BCG and virulent M. bovis. J. Bacteriol. 178:1274-1282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Marks, J. 1976. A system for the examination of tubercle bacilli and other mycobacteria. Tubercle 57:207-225. [DOI] [PubMed] [Google Scholar]
- 24.Musser, J. M., A. Amin, and S. Ramaswamy. 2000. Negligible genetic diversity of Mycobacterium tuberculosis host immune system protein targets: evidence of limited selective pressure. Genetics 155:7-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Niemann, S., D. Harmsen, S. Rusch-Gerdes, and E. Richter. 2000. Differentiation of clinical Mycobacterium tuberculosis complex isolates by gyrB DNA sequence polymorphism analysis. J. Clin. Microbiol. 38:3231-3234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pym, A. S., and R. Brosch. 2000. Tools for the population genomics of the tubercle bacilli. Genome Res. 10:1837-1839. [DOI] [PubMed] [Google Scholar]
- 27.Salfinger, M., L. B. Reller, B. Demchuk, and Z. T. Johnson. 1989. Rapid radiometric method for pyrazinamide susceptibility testing of Mycobacterium tuberculosis. Res. Microbiol. 140:301-309. [DOI] [PubMed] [Google Scholar]
- 28.Scorpio, A., and Y. Zhang. 1996. Mutations in pncA, a gene encoding pyrazinamidase/nicotinamidase, cause resistance to the antituberculous drug pyrazinamide in tubercle bacillus. Nat. Med. 2:662-667. [DOI] [PubMed] [Google Scholar]
- 29.Smid, I., M. Salfinger, and U. Vurma-Rapp. 1992. The use of radiometric tests in the speciation of mycobacteria—a review. Zentbl. Bakteriol. Parasitenkd. Infektkrankh. Hyg. Abt. 1 Orig. 276:493-501. [PubMed] [Google Scholar]
- 30.Sreevatsan, S., P. Escalante, X. Pan, D. A. Gillies II, S. Siddiqui, C. N. Khalaf, B. N. Kreiswirth, P. Bifani, L. G. Adams, T. Ficht, V. S. Perumaalla, M. D. Cave, J. D. van Embden, and J. M. Musser. 1996. Identification of a polymorphic nucleotide in oxyR specific for Mycobacterium bovis. J. Clin. Microbiol. 34:2007-2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sreevatsan, S., X. Pan, K. E. Stockbauer, N. D. Connell, B. N. Kreiswirth, T. S. Whittam, and J. M. Musser. 1997. Restricted structural gene polymorphism in the Mycobacterium tuberculosis complex indicates evolutionarily recent global dissemination. Proc. Natl. Acad. Sci. USA 94:9869-9874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Talbot, E. A., D. L. Williams, and R. Frothingham. 1997. PCR identification of Mycobacterium bovis BCG. J. Clin. Microbiol. 35:566-569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Taylor, G. M., M. Goyal, A. J. Legge, R. J. Shaw, and D. Young. 1999. Genotypic analysis of Mycobacterium tuberculosis from medieval human remains. Microbiology 145:899-904. [DOI] [PubMed] [Google Scholar]
- 34.Tsukamura, M., S. Mizuno, and H. Toyama. 1985. Taxonomic studies on the Mycobacterium tuberculosis series. Microbiol. Immunol. 29:285-299. [DOI] [PubMed] [Google Scholar]
- 35.van Soolingen, D., T. Hoogenboezem, P. E. de Haas, P. W. Hermans, M. A. Koedam, K. S. Teppema, P. J. Brennan, G. S. Besra, F. Portaels, J. Top, L. M. Schouls, and J. D. van Embden. 1997. A novel pathogenic taxon of the Mycobacterium tuberculosis complex, Canetti: characterization of an exceptional isolate from Africa. Int. J. Syst. Bacteriol. 47:1236-1245. [DOI] [PubMed] [Google Scholar]
- 36.van Soolingen, D., A. G. van der Zanden, P. E. de Haas, G. T. Noordhoek, A. Kiers, N. A. Foudraine, F. Portaels, A. H. Kolk, K. Kremer, and J. D. van Embden. 1998. Diagnosis of Mycobacterium microti infections among humans by using novel genetic markers. J. Clin. Microbiol. 36:1840-1845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Viana-Niero, C., C. Gutierrez, C. Sola, I. Filliol, F. Boulahbal, V. Vincent, and N. Rastogi. 2001. Genetic diversity of Mycobacterium africanum clinical isolates based on IS6110-restriction fragment length polymorphism analysis, spoligotyping, and variable number of tandem DNA repeats. J. Clin. Microbiol. 39:57-65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wayne, L. G., and G. P. Kubica. 1986. The mycobacteria, p. 1435-1457. In P. H. A. Sneath and J. G. Holt (ed.), Bergey's manual of systemic bacteriology, vol. 2. The Williams & Wilkins Co., Baltimore, Md.
- 39.Yates, M. D., C. H. Collins, and J. M. Grange. 1982. “Classical” and “Asian” variants of Mycobacterium tuberculosis isolated in South East England 1977-1980. Tubercle 63:55-61. [DOI] [PubMed] [Google Scholar]
- 40.Yates, M. D., J. M. Grange, and C. H. Collins. 1984. A study of the relationship between the resistance of Mycobacterium tuberculosis to isonicotinic acid hydrazide (isoniazid) and to thiophen-2-carboxylic acid hydrazide. Tubercle 65:295-299. [DOI] [PubMed] [Google Scholar]
- 41.Zumarraga, M. J., A. Bernardelli, R. Bastida, V. Quse, J. Loureiro, A. Cataldi, F. Bigi, A. Alito, M. Castro Ramos, S. Samper, I. Otal, C. Martin, and M. I. Romano. 1999. Molecular characterization of mycobacteria isolated from seals. Microbiology 145:2519-2526. [DOI] [PubMed] [Google Scholar]