Abstract
Woodchucks (Marmota monax) have a high incidence of hepatocellular carcinoma (HCC) associated with chronic infection with woodchuck hepatitis virus (WHV) and serve as a model of hepatitis B virus-associated HCC in humans. Helicobacter hepaticus, an enterohepatic helicobacter in mice, is known to cause hepatocellular adenomas and carcinomas in susceptible mouse strains. In long-term chemical bioassays conducted with B6C3F1 mice, H. hepaticus has been regarded as a confounding factor because of its tumor-promoting activity. In order to determine if woodchucks harbor a Helicobacter sp. that might play a role in potentiating hepatic inflammation or neoplasia, a study was undertaken to determine whether woodchucks' livers were infected with a Helicobacter sp. Frozen liver samples from 20 (17 WHV-infected and 3 noninfected) woodchucks, 10 with WHV-associated hepatic tumors and 10 without tumors, were cultured by microaerobic techniques and analyzed by using genus- and species-specific helicobacter PCR primers. A 1,200-bp Helicobacter sp.-specific sequence was amplified from 14 liver samples. Southern hybridization confirmed the specific identity of the PCR products. Nine of the 10 livers with tumors had positive Helicobacter sp. identified by PCR, whereas 5 of the 10 livers without tumors were positive. By use of 16S rRNA species-specific primers for H. marmotae, two additional liver samples from the nontumor group had positive PCR amplicons confirmed by Southern hybridization. A urease-, catalase-, and oxidase-positive bacterium was isolated from one liver sample from a liver tumor-positive woodchuck. By 16S rRNA analysis and biochemical and phenotypic characteristics, the organism was classified as a novel Helicobacter sp. Subsequently, four additional bacterial strains isolated from feces of cats and characterized by biochemical, phenotypic, and 16S rRNA analysis were determined to be identical to the woodchuck isolate. We propose the name Helicobacter marmotae sp. nov. for these organisms. Further studies are required to ascertain if this novel Helicobacter sp. plays a tumor promotion role in hepadnavirus-associated tumors in woodchucks or causes enterohepatic disease in cats.
The genus Helicobacter is a rapidly expanding group of gram-negative, microaerophilic, spiral organisms that persistently colonize a variety of mammalian hosts and in some cases are responsible for significant clinical diseases (6). The type species, H. pylori, causes a persistent gastritis in the human stomach and is directly linked to peptic ulcer disease, gastric adenocarcinoma, and mucosa-associated lymphoid tissue lymphoma (15). Although gastric helicobacters have been the focus of experimental studies because of the importance of H. pylori-associated gastric disease in humans, the discovery of enterohepatic helicobacters in animals and humans has sparked an interest in exploring the pathogenic potential of these organisms as well.
Several years ago, two novel helicobacters, H. hepaticus and H. bilis, were isolated from livers of mice with hepatitis (8, 17). H. hepaticus infection is also associated with hepatic neoplasms in A/JCr and B6C3F1 mice, and this bacterium causes hepatitis and typhlitis in A/JCr inbred mice and AXB recombinant inbred mice (22, 38). Indeed, H. hepaticus infections in B6C3F1 mice used for carcinogenesis bioassays are considered a confounding factor in mouse liver tumorigenesis studies because of the bacteria's tumor-promoting ability (9, 14, 21). Other Helicobacter species, originally called “Flexispira rappini,” have also been isolated from the inflamed livers of aborted sheep fetuses, as well as livers of guinea pigs experimentally infected with “F. rappini” (2, 24). Additionally, novel helicobacters have been associated with enterohepatic disease in domesticated species. H. pullorum has been cultured from diseased chicken livers and from the feces of diarrheic humans (33). H. canis has been isolated from feces of diarrheic dogs, cats, and humans and the liver of a puppy with hepatitis (5, 34). H. cinaedi isolated from bacteremic humans has recently been isolated from the liver of a rhesus monkey with hepatitis and idiopathic colitis (10, 36).
In 1978, woodchuck hepatitis virus (WHV) was discovered in woodchucks, a large rodent native to North America. Chronic infection with WHV causes hepatitis and is associated with hepatocellular carcinoma (HCC) (35). This virus has been found in wild woodchuck populations in several eastern states of the United States. More than 90% of both wild-caught WHV-infected and colony-born experimentally infected woodchucks die of HCC within 2 to 3 years. These animals have chronic hepatitis with significant components of inflammation and regeneration, but cirrhosis does not occur (26). HCC also developed in up to 20% of woodchucks that had recovered from WHV infections (surface-antigen-negative, anti-HBc/anti-HBs-positive animals) but did not develop in any uninfected animals. These data support the view that WHV infection is necessary for HCC but may not be sufficient by itself to induce tumors. The possibility of cofactors that could contribute to tumor formation has been examined. Dietary aflatoxin B1 exposure has a synergistic effect with chronic WHV infection on hepatic tumorigenesis (1, 20). The presence of hepatic inflammation and biliary proliferation (both lesions are also noted in H. hepaticus-infected mouse livers) has been previously reported for WHV-seronegative woodchucks and suggested to be a response to chronic infection (27, 28). Since Helicobacter species can establish chronic infection and serve as a cocarcinogen in other rodents, and woodchucks without WHV infection have a background incidence of hepatitis, it is conceivable that a helicobacter might contribute to woodchuck HCC. However, a bacterial infection has not been previously entertained as a possible cofactor (26).
This study describes the isolation and characterization by morphology, biochemistry, and 16S RNA analysis of a novel Helicobacter sp., H. marmotae, isolated from the liver of a woodchuck infected with WHV as well as the feces of commercially reared cats. This finding raises a testable hypothesis that novel Helicobacter spp. may be responsible for hepatitis in other hosts and that the presence of a persistent bacterial infection may be a cofactor in virally induced hepatic neoplasms.
MATERIALS AND METHODS
Animals.
Liver specimens from 20 wild-caught adult woodchucks maintained in captivity were evaluated. Their livers were collected aseptically at necropsy and stored at −70°C, until submitted for culture and PCR analysis.
Four cats (three received in one shipment in 1998 and one cat in a shipment received in 1999 from a specific-pathogen-free animal commercial source certified to have cats free of feline leukemia virus, feline immunodeficiency virus, and feline coronavirus) were part of a previous survey of enteric Helicobacter spp. and Campylobacter spp. (31). At the time of fecal culture, all cats were clinically healthy.
Slot blot analysis of serum WHV DNA.
For analysis of WHV DNA, whole blood was collected and stored at −70°C until analysis. WHV DNA was extracted from 200 μl of serum and eluted into 200 μl of Tris buffer (10 mM, pH 8.5) with a QIAMP blood kit (Qiagen, Valencia, Calif.). The probe was an EcoRI-digested WHV2 genome (3.32 kb) labeled with digoxigenin-II-dUTP (DIG High Prime DNA labeling detection starter kit II; Boehringer Mannheim, Indianapolis, Ind.). The WHV DNA standard used as a control was whole WHV2 genomic DNA linearized with EcoRI. Ten microliters of the extracted DNA solution was denatured with 30 μl of denaturing buffer at 65°C for 40 min and was loaded on each slot. Denatured DNA was fixed to a nylon filter (Nytran; Schleicher & Schuell, Keene, N.H.) by a UV cross-linking method. The filter and probe were incubated together, and after washing, the membranes were immediately scanned with the Lumi-Imager (Boehringer Mannheim) to generate a numerical value for the strength of the hybridization signal. Extracted negative and positive sera and several dilutions of standard DNA (0.01, 0.03, 0.06, and 0.1 ng of DNA) were included.
Bacterial isolation and biochemical characterization.
Samples from each liver were homogenized in 1.0 ml of phosphate-buffered saline. Each slurry was gently passed through a 0.45-μm-pore-size syringe filter, and the homogenate was streaked onto a Trypticase soy agar (with 5% sheep blood) plate (Remel Laboratories, Lenexa, Kans.) containing TVP (trimethoprim, vancomycin, and polymyxin) or CVA (cefoperzone, vancomycin, and amphotericin B) (Remel). In addition, a selective antibiotic medium was prepared as follows: blood agar base (Oxoid; Remel), 5% horse blood (Remel), 50 μg of amphotericin B/ml, 100 μg of vancomycin/ml, 3.3 μg of polymyxin B/ml, 200 μg of bacitracin/ml, and 10.7 μg of nalidixic acid/ml (all from Sigma Chemical Co., St. Louis, Mo.). Feces from the cats were directly streaked onto CVA medium. The cultures were then incubated at 37°C under microaerobic conditions in vented jars containing N2, H2, and CO2 (90:5:5). A detailed biochemical characterization analysis was performed on five isolates as previously described by Fox et al. (17).
Electron microscopy.
The isolate was examined by electron microscopy. Cells grown on blood agar for 48 h were gently suspended in 10 mM Tris-HCl buffer (pH 7.4) at a concentration of about 108 cells per ml. Samples were negatively stained with 1% (wt/vol) phosphotungstic acid (pH 6.5) for 20 to 30 s. Specimens were examined with a JEOL model JEM-1200EX transmission electron microscope operating at 100 kV.
Genomic DNA extraction for 16S rRNA sequencing.
Bacteria isolated from the liver of one woodchuck were cultured on blood agar plates, and cells were harvested and washed once with 1 ml of phosphate-buffered saline. The High Pure PCR template preparation kit was used for DNA extraction (Roche Molecular Biochemicals, Indianapolis, Ind.).
16S rRNA gene sequencing.
The sequences of the 16S rRNA gene of three isolates of H. marmotae MIT 98-6070, MIT 98-1705-1, and MIT 99-4759 were determined. For amplification of 16S rRNA cistrons, 16S rRNA gene sequencing, and 16S rRNA data analysis we used the methods described by Fox et al. (7). Briefly, primers C70 and B37 (7) were used to amplify the 16S rRNA genes. The amplicons were purified and directly sequenced by using a TAQuence cycle sequencing kit (U.S. Biochemicals, Cleveland, Ohio). The 16S rRNA gene sequences were entered into RNA, a program for analysis of 16S rRNA data in Microsoft Quickbasic for use with IBM PC-compatible computers, and were aligned as described previously (25). The database used contains approximately 100 Helicobacter, Wolinella, Arcobacter, and Campylobacter sequences and more than 900 sequences for other bacteria. Similarity matrices were constructed from the aligned sequences by using only those base positions for which 90% of the strains had data and were corrected for multiple base changes by the method of Jukes and Cantor (23). Phylogenetic trees were constructed by the neighbor-joining method (29).
DNA extraction for PCR analysis.
DNA was extracted from the liver samples and the cultured organisms with the High Pure PCR template preparation kit (Roche Molecular Biochemicals) according to the manufacturer's directions. Briefly, the samples were lysed and incubated with 40 μl of proteinase K for 1 h at 55°C. Two hundred microliters of binding buffer was added to each sample and allowed to incubate for 10 min at 72°C before 100 μl of isopropanol was added. The samples were placed in a filter tube and centrifuged at 6,000 × g for 1 min. The flowthrough was discarded, and 500 μl of wash buffer was added to the samples and centrifuged as before. This washing step was repeated twice. Prewarmed (70°C) elution buffer (200 μl) was used to elute DNA from the filter.
PCR amplification of bacterial DNA.
One set of primer sequences chosen for PCR amplification recognize a region of the 16S rRNA gene specific for members of the Helicobacter genus. This set of primers produces an amplified product of 1.2 kb. PCR amplification was achieved by the method previously described (7). Briefly, 20 μl of the DNA preparation was added to 100 μl of a reaction mixture containing 1× Taq polymerase buffer (supplied by the manufacturer but supplemented with 1 M MgCl2 to a final concentration of 2.25 mM), 0.5 μM (each) primer, 200 μM (each) deoxynucleotide and 200 μg of bovine serum albumin per ml. Samples were heated at 94°C for 4 min, briefly centrifuged, and cooled to 58°C. At this time, 2.5 U of Taq polymerase (Roche Molecular Biochemicals) and 1.0 U of polymerase enhancer (Perfect Match; Stratagene, La Jolla, Calif.) were added, and then the samples were overlaid with 100 μl of mineral oil. The following conditions were used for amplification of the 462-bp fragment: 35 cycles of denaturation at 94°C for 1 min, annealing at 58°C for 2 min, and elongation at 72°C for 2 min, followed by an elongation step of 8 min at 72°C. For amplification of the 1.2-kb fragment the following conditions were used: 35 cycles of denaturation at 94°C for 1 min, annealing at 58°C for 2 min, and elongation at 72°C for 3 min, followed by an elongation step of 8 min at 72°C. Fifteen microliters of the sample was then electrophoresed through a 1% agarose gel followed by ethidium bromide staining and visualized by UV illumination.
Subsequent to obtaining the full sequences for one strain of the novel Helicobacter sp., PCR primers (forward primer G70, 5′-GCG GGT AAT TAA GTC AGA TG; reverse primer G69, 5′-TGT TTT CAA GCT CCC CAA AG) were designed for identification of H. marmotae in woodchuck liver tissues.
Southern blot analysis.
To confirm that the amplicons were of bacterial 16S rRNA rather than nonspecific mammalian genomic origin, Southern blot analysis was performed with a 16S ribosomal DNA helicobacter probe. Fifteen microliters of amplicons was electrophoresed through a 1% agarose gel and transferred onto a Hybond N nylon membrane as outlined by the manufacturer (Amersham, Arlington Heights, Ill.). DNA was then UV cross-linked. The fixed DNA was then hybridized with a helicobacter probe generated by PCR amplification of H. pylori with primers C97 and C05 or the PCR product amplified from the woodchuck isolate with H. marmotae-specific primers. The probe was labeled with horseradish peroxidase, hybridized overnight to the nylon membrane at 42°C, and exposed in the presence of Luminol to Hyperfilm-ECL as outlined by the manufacturer (Amersham).
Histopathology.
Ten formalin-fixed liver samples were processed by standard methods and embedded in paraffin. Six of the 10 samples were from woodchucks with liver tumors. Five-micrometer-thick sections were stained with either hematoxylin and eosin or Warthin-Starry silver stain. These sections were examined by light microscopy for evidence of inflammatory and degenerative lesions and for the presence of a bacterium with morphology consistent with members of the genus Helicobacter. Archival histologic sections of liver tissue from two WHV-negative woodchucks were also used for comparison. Frozen liver samples from these two archival woodchucks were not available for culture or PCR. Tissues were not available for the four cats.
Nucleotide sequence accession numbers.
The nucleotide sequence accession numbers for each strain studied are in Fig. 4, the phylogenetic tree. The accession no. for H. marmotae type strain MIT 98-6070 is AF333341.
FIG. 4.
Phylogenetic tree comparing H. marmotae with other closely related helicobacters.
RESULTS
WHV status.
Based on serum WHV DNA analysis, 17 of the woodchucks used in this study were naturally infected with WHV. At the time of necropsy, 10 of these woodchucks had WHV-associated hepatic tumors and 10 (3 of which were negative for WHV) had no grossly observable hepatic tumors.
Helicobacter sp. characterization.
Despite culturing all 20 livers, we were able to isolate a pure culture of the novel Helicobacter species from only one frozen liver from a tumor-positive woodchuck. In addition, we isolated pure cultures of the novel helicobacter from the feces of four cats. The bacteria were gram negative and had spiral morphology. The bacteria grew under microaerobic conditions as a spreading film on blood agar at 37 and 42°C but not at 25°C. The isolate was oxidase, catalase, urease, and alkaline phosphatase positive but did not hydrolyze indoxyl acetate and was γ-glutamyl transpeptidase negative. The bacterium did not reduce nitrate to nitrite; however, the bacterium grew in 1% glycine and was resistant to nalidixic acid and cephalothin (Table 1).
TABLE 1.
Characteristics which differentiate H. marmotae from other nongastric Helicobacter speciesa
| Taxon | Catalase production | Nitrate reduction | Alkaline phosphatase hydrolysis | Urease | Indoxyl acetate hydrolysis | γ-Glutamyl transpeptidase | Growth
|
Susceptibility to:
|
Periplasmic fibers | No. of flagella | Distribution of flagella | G+C content (mol%) | ||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| At 42°C | With 1% glycine | Nalidixic acid (30-μg disk) | Cephalothin (30-μg disk) | |||||||||||
| H. marmotae | + (5/5) | − (5/5) | + (5/5) | + (5/5) | − (5/5) | − (5/5) | − (5/5) | + (5/5) | R (5/5) | R (5/5) | − | 2 | Bipolar | ND |
| H. canadensis | + | + | − | − | + | − | + | + | R | R | − | 2 | Bipolar | ND |
| H. rodentium | + | + | − | − | − | − | + | + | R | R | − | 2 | Bipolar | ND |
| H. pullorum | + | + | − | − | − | ND | + | − | R | S | − | 1 | Monopolar | 34-35 |
| H. fennelliae | + | − | + | − | + | − | − | + | S | S | − | 2 | Bipolar | 35 |
| H. trogontum | + | + | − | + | − | + | + | ND | R | R | + | 5-7 | Bipolar | ND |
| H. muridarum | + | − | + | + | + | + | − | − | R | R | + | 10-14 | Bipolar | 34 |
| H. hepaticus | + | + | − | + | + | − | − | + | R | R | − | 2 | Bipolar | ND |
| H. canis | − | − | + | − | + | + | + | − | S | I | − | 2 | Bipolar | 48 |
| H. bilis | + | + | − | + | − | + | + | + | R | R | + | 3-4 | Bipolar | ND |
| “H. rappini” | + | − | − | + | ND | + | + | − | R | R | + | 10-20 | Bipolar | 34 |
| H. cinaedi | + | + | − | − | − | − | − | + | S | I | − | 1-2 | Bipolar | 37-38 |
| H. pametensis | + | + | + | − | − | − | + | + | S | S | − | 2 | Bipolar | 38 |
| H. winghamensis | − | − | − | − | + | ND | − | ND | S | S | − | 1-2 | Bipolar | ND |
| H. mesocricatorum | + | + | + | − | ND | − | ND | − | S | R | − | 2 | Bipolar | ND |
| H. aurati | + | − | − | + | + | + | + | − | S | R | + | 7-10 | Bipolar | ND |
| H. typhlonius | + | +/− | − | − | − | − | + | + | S | R | − | 1 | Bipolar | ND |
| H. cholecystus | + | + | − | − | − | − | + | + | I | R | − | 2 | Bipolar | ND |
Ultrastructure.
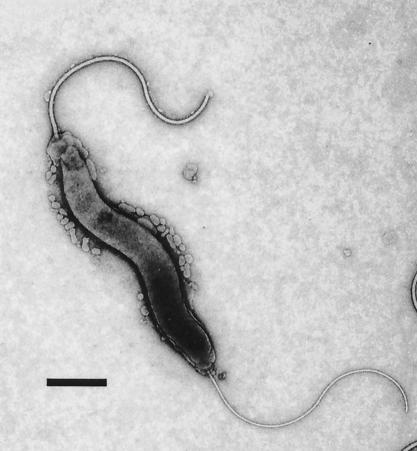
Bacterial cells had a spiral appearance and measured 0.2 by 1.5 μm. The cells had bipolar sheathed flagella typical of most enterohepatic helicobacters (Fig. 1).
FIG. 1.
Novel Helicobacter sp. has a curved to spiral appearance. The bacterium measures 0.2 by 1.5 to 5.0 μm and has bipolar sheathed flagella. Bar = 0.3 μm.
PCR identification in liver tissue.
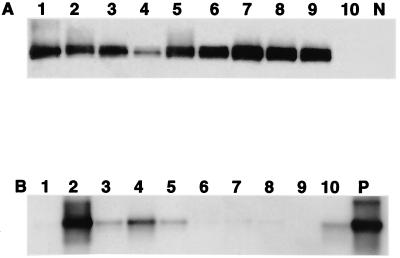
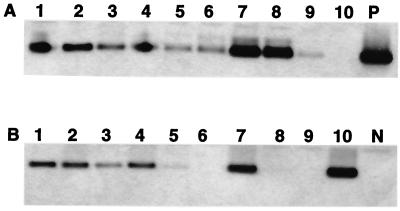
Nine of the 10 livers from tumor-positive woodchucks had positive Helicobacter spp. identified by PCR (with Helicobacter genus-specific primers) and confirmed by Southern blot hybridization (Fig. 2A), whereas 5 of 10 livers from tumor-negative woodchucks (3 of which were WHV negative) were Helicobacter spp. positive with the genus-specific PCR primers (Fig. 2B). By use of 16S rRNA species-specific primers for H. marmotae, two additional liver samples from the nontumor group had positive PCR amplicons confirmed by Southern hybridization (Fig. 3).
FIG. 2.
Southern blot hybridization of the 1,200-bp PCR products with genus-specific Helicobacter spp. primers. (A) Tumor-positive group. (B) Tumor-negative group. Lanes 1 to 10, liver tissue samples. P, Helicobacter DNA as positive control; N, reagent control.
FIG. 3.
Southern blot hybridization of PCR products with Helicobacter species-specific primers for the novel Helicobacter sp. (A) Tumor-positive group. (B) Tumor-negative group. Lanes 1 to 10, Liver tissue samples. P, Helicobacter DNA as positive control; N, reagent control.
Based on the cumulative PCR data with both sets of primers, 9 of 10 livers from tumor-positive woodchucks and 7 of 10 livers from tumor-negative woodchucks were helicobacter positive, for an overall prevalence of 80%. Because of the high prevalence of both helicobacter infection and tumors in the study group, no statistical association between the helicobacter PCR status and the presence of tumors could be determined.
Phylogenetic analysis.
Full 16S rRNA sequences (approximately 1,500 bases) were determined for three isolates (MIT 98-6070, MIT 98-1705-1, and MIT 99-4759). Comparison of these sequences with over 100 Helicobacter sequences in our database indicated that the isolates represent a new Helicobacter species, for which we propose the name H. marmotae. The sequence for H. marmotae is most closely related to sequences from helicobacters isolated from rhesus monkeys (AF333339 and AF333340 [11]) and cotton-top tamarins (AF107494 [30]) and H. fennelliae. The H. marmotae sequence differs from these sequences by 4 to 5% (Fig. 4).
Histopathology.
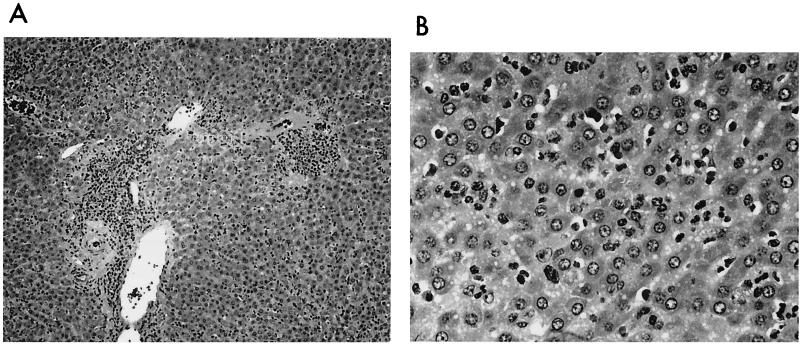
Liver samples from woodchucks with HCC and positive for Helicobacter spp. by PCR were characterized by portal tracts with moderate to marked degrees of oval cell or bile duct hyperplasia with a portal bridging pattern and periportal inflammation comprised primarily of a lymphoplasmacytic infiltrate. The parenchyma was characterized by multifocal necrosis of hepatocytes accompanied by phagocytic infiltration and lymphoplasmacytic inflammation. The liver samples from WHV-positive woodchucks without tumors, including the liver from which H. marmotae was isolated, had mild to moderate, chronic hepatitis centered on the portal areas. The hepatic parenchyma often contained small foci of necrotic hepatocytes accompanied by mild phagocytic and lymphoplasmacytic infiltrates. Mild peribiliary fibrosis and bile duct or oval cell proliferation were present in some specimens. Of interest, comparable histopathologic changes were observed in the liver samples from the three woodchucks that were negative for WHV but positive for Helicobacter spp. by PCR, consisting of chronic multifocal periportal hepatitis and biliary proliferation. Inflammation in the portal areas and parenchyma was comprised of lymphoplasmacytic infiltrates predominantly, with lesser numbers of macrophages and granulocytes (Fig. 5). Bacteria were not identified in Warthin-Starry-stained liver tissue.
FIG. 5.
(A) The portal areas are moderately infiltrated by a mixed inflammatory cell population, comprised chiefly of lymphocytes, plasma cells, and some macrophages. Inflammation abuts all portal structures, but a tendency to peribiliary condensation is present. Bile duct proliferation is present and extends into the adjacent parchyma. Hematoxylin and eosin stain. (B) An unstructured, mixed inflammatory infiltrate is present in the hepatic parenchyma, comprised chiefly of plasma cells, macrophages, and lymphocytes. Hematoxylin and eosin stain.
DISCUSSION
The identification of a novel Helicobacter sp., H. marmotae, from woodchuck liver by culture and H. marmotae-specific PCR and the feces of cats raises the number of enterohepatic Helicobacter spp. isolated from livers to eight. The others include H. bilis and H. hepaticus from mice, H. cholecystus in hamsters, H. pullorum from chickens, H. canis from dogs, H. cinaedi from monkeys (11), and Helicobacter sp. Flexispira taxa 2 and 5 from sheep (6). DNA of H. bilis and H. pullorum and Helicobacter sp. Flexispira taxon 8 has also been identified in Chilean patients with chronic cholecystitis (7).
Whether the H. marmotae isolated from a woodchuck is responsible for the hepatitis noted in these animals and other woodchucks not infected with the woodchuck hepadnavirus is at present unknown and will require further studies. The isolation of H. marmotae from only one liver despite the presence of helicobacters in DNA from a large percentage of the woodchuck livers can be partially explained by the fact that Helicobacter spp. often do not survive freezing unless samples are stored in glycerol broth medium (37). Though similar in morphology to H. canis, which is also isolated from cats, H. marmotae can be differentiated from H. canis biochemically by being urease positive and indoxyl acetate hydrolysis and γ-glutamyl transpeptidase negative (5, 31). We have previously analyzed the 1.2-kb products from Helicobacter genus-specific PCR analysis of 19 cat helicobacter isolates by AluI enzyme digestion (31). Five patterns were observed. Three patterns grouped taxonomically with H. bilis or Helicobacter (“Flexispira”) taxon 8, one was similar to H. canis, and the remaining pattern matched that of the novel H. marmotae isolated from woodchucks (31). Thus, restriction fragment length polymorphism analysis clearly supports our data that H. marmotae can be taxonomically distinguished from other feline enteric helicobacters.
Interestingly, H. marmotae experimentally inoculated into A/JCr mice produces chronic hepatitis and typhlocolitis (J. G. Fox, unpublished observations). Given that cats are often diagnosed with chronic cholangiohepatitis of unknown etiology, it will be interesting to ascertain whether this novel helicobacter causes liver disease in cats as well as other mammals (19).
A body of evidence is accumulating which supports the conjecture that enterohepatic helicobacters may act as cofactors in carcinogenesis. We have previously shown that H. mustelae, a natural gastric pathogen in ferrets, strongly promotes gastric adenocarcinoma initiated by the gastric carcinogen N-methyl-N′-nitrso-N-nitrosoguanidine (16). Experimentally, nitrosamine-induced gastric adenocarcinoma is promoted by H. pylori infection in gerbils (15). Others have demonstrated that bile duct cancer is strongly promoted by liver fluke infection in hamsters (4). It is interesting to speculate as to whether these hamsters were also infected with H. cholecystus, a novel helicobacter known to colonize the bile of hamsters (18). Also, several recent studies of A/J and B6C3F1 mice infected with H. hepaticus suggest that the bacterium is a tumor promoter in the mouse liver (3, 13, 14, 21, 32, 38). Additional studies are required to ascertain whether H. marmotae acts as a tumor promoter in WHV-infected woodchucks and whether H. marmotae colonizes the lower bowel of woodchucks, the site of colonization of other enterohepatic helicobacters.
In summary, the finding of a novel Helicobacter sp., H. marmotae, in an animal model used to study virus-induced liver cancer raises intriguing questions as to whether similar Helicobacter spp. may be present in humans infected with hepatitis virus as well as cats with chronic cholangiohepatitis (19). If present, it will be important to determine whether these persistent helicobacters play a role in the development of hepatocellular cancers.
Description of H. marmotae sp. nov.
(mar′mo.tae. F marmotte, fusion of OF marmotte, a monkey, and Romansch murmont, a marmot [L mus, muris, a mouse, plus montis, a mountain], relating to a short-tailed burrowing rodent species found in North America.) Cells are slender, curved to spiral rods (0.2 by 1.5 to 5 μm), which have one to three spirals. The bacterium is gram negative and nonsporulating; it is motile by means of sheathed, single unipolar or bipolar flagella. Cultures grown on solid agar media appear as spreading layers. Cells exhibit microaerobic but not aerobic or anaerobic growth. Growth occurs at 37 and 42°C. The bacteria are oxidase, catalase, urease, and alkaline phosphatase positive but γ-glutamyl transpeptidase negative. The organism does not hydrolyze indoxyl acetate nor reduce nitrate to nitrite. Cells are resistant to nalidixic acid and cephalothin. Bacteria have been isolated from the liver of woodchucks and feces of clinically normal cats. The type strain is MIT 98-6070 (ATCC BAA-546). The 16S rRNA sequence accession no. for the type strain is AF333341.
Acknowledgments
We thank Hans G. Trüper for his advice in naming the novel helicobacter, H. marmotae.
This work was supported by NIH grants R01-CA67529 (J.G.F.), P01 CA26723 (J.G.F.), R01A150952 (J.G.F.), RR07036 (J.G.F.), and R01 DE10374 (F.E.D. and B.J.P.).
REFERENCES
- 1.Bannasch, P., N. I. Khoshkhou, H. J. Hacker, S. Radaeva, M. Mrozek, U. Zillmann, A. Kopp-Schneider, U. Haberkorn, M. Elgas, T. Tolle, et al. 1995. Synergistic hepatocarcinogenic effect of hepadnaviral infection and dietary aflatoxin B1 in woodchucks. Cancer Res. 55:3318-3330. [PubMed] [Google Scholar]
- 2.Bryner, J. H., A. E. Ritchie, L. Pollet, C. A. Kirkbride, and J. E. Collins. 1987. Experimental infection and abortion of pregnant guinea pigs with a unique spirillum-like bacterium isolated from aborted ovine fetuses. Am. J. Vet. Res. 48:91-97. [PubMed] [Google Scholar]
- 3.Diwan, B. A., J. M. Ward, D. Ramljak, and L. M. Anderson. 1997. Promotion by Helicobacter hepaticus-induced hepatitis of hepatic tumors initiated by N-nitrosodimethylamine in male A/JCr mice. Toxicol. Pathol. 25:597-605. [DOI] [PubMed] [Google Scholar]
- 4.Flavell, D. J., and S. B. Lucas. 1983. Promotion of N-nitrosodimethylamine-initiated bile duct carcinogenesis in the hamster by the human liver fluke, Opisthorchis viverrini. Carcinogenesis 7:927-930. [DOI] [PubMed] [Google Scholar]
- 5.Foley, J. E., S. Marks, L. Munson, A. Melli, D. E. Dewhirst, S. Yu, Z. Shen, and J. G. Fox. 1999. Isolation of Helicobacter canis from a colony of Bengal cats with endemic diarrhea. J. Clin. Microbiol. 37:3271-3275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fox, J. G. 1997. The expanding genus of Helicobacter: pathogenic and zoonotic potential. Semin. Gastrointestinal Dis. 8:124-141. [PubMed] [Google Scholar]
- 7.Fox, J. G., F. E. Dewhirst, Z. Shen, Y. Fen, N. S. Taylor, B. Paster, R. L. Erickson, C. N. Lau, P. Correa, J. C. Araya, and I. Roa. 1998. Hepatic Helicobacter species identified in bile and gallbladder tissue from Chileans with chronic cholecystitis. Gastroenterology 114:755-763. [DOI] [PubMed] [Google Scholar]
- 8.Fox, J. G., F. E. Dewhirst, J. G. Tully, B. J. Paster, L. Yan, N. S. Taylor, M. J. Collins, Jr., P. L. Gorelick, and J. M. Ward. 1994. Helicobacter hepaticus sp. nov., a microaerophilic bacterium isolated from livers and intestinal mucosal scrapings from mice. J. Clin. Microbiol. 32:1238-1245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fox, J. G., R. Drolet, R. Higgins, S. Messier, L. Yan, B. E. Coleman, B. J. Paster, and F. E. Dewhirst. 1996. Helicobacter canis isolated from a dog liver with multifocal necrotizing hepatitis. J. Clin. Microbiol. 34:2479-2482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fox, J. G., L. Handt, B. J. Sheppard, S. Xu, F. E. Dewhirst, S. Motzel, and H. Klein. 2001. Isolation of Helicobacter cinaedi from the colon, liver and mesenteric lymph node of a rhesus monkey with chronic colitis and hepatitis. J. Clin. Microbiol. 39:1580-1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fox, J. G., L. Handt, S. Xu, Z. Shen, F. E. Dewhirst, C. Dangler, K. Lodge, S. Motzel, and H. Klein. 2001. Novel Helicobacter spp isolated from colonic tissue of rhesus monkeys with chronic idiopathic colitis. J. Med. Microbiol. 50:421-429. [DOI] [PubMed] [Google Scholar]
- 12.Fox, J. G., and A. Lee. 1997. The role of Helicobacter species in newly recognized gastrointestinal tract diseases of animals. Lab. Anim. Sci. 47:222-255. [PubMed] [Google Scholar]
- 13.Fox, J. G., X. Li, L. Yan, R. J. Cahill, R. Hurley, R. Lewis, and J. C. Murphy. 1996. Chronic proliferative hepatitis in A/JCr mice associated with persistent Helicobacter hepaticus infection: a model of Helicobacter-induced carcinogenesis. Infect. Immun. 64:1548-1558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fox, J. G., J. MacGregor, Z. Shen, X. Li, R. Lewis, and C. A. Dangler. 1998. Comparison of methods to identify Helicobacter hepaticus in B6C3F1 mice used in a carcinogenesis bioassay. J. Clin. Microbiol. 36:1382-1387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fox, J. G., and T. C. Wang. 2000. Overview of Helicobacter pylori, p. 371-388. In J. J. Goedert (ed.), Infectious causes of cancer: targets for invention. Humana Press, Totowa, N.J.
- 16.Fox, J. G., J. S. Wishnok, J. C. Murphy, S. R. Tannenbaum, and P. Correa. 1993. MNNG-induced gastric carcinoma in ferrets infected with Helicobacter mustelae. Carcinogenesis 14:1957-1961. [DOI] [PubMed] [Google Scholar]
- 17.Fox, J. G., L. L. Yan, F. E. Dewhirst, B. J. Paster, B. Shames, J. C. Murphy, A. Hayward, J. C. Belcher, and E. N. Mendes. 1995. Helicobacter bilis sp. nov., a novel helicobacter isolated from bile, livers, and intestines of aged, inbred mice. J. Clin. Microbiol. 33:445-454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Franklin, C. L., C. S. Beckwith, R. S. Livingston, L. K. Riley, S. V. Gibson, C. L. Besch-Williford, and R. R. Hook, Jr. 1996. Isolation of a novel Helicobacter species, Helicobacter cholecystus sp. nov., from the gallbladders of Syrian hamsters with cholangiofibrosis and centrilobular pancreatitis. J. Clin. Microbiol. 34:2952-2958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gagne, J. M., D. J. Weiss, and P. J. Armstrong. 1996. Histopathologic evaluation of feline inflammatory liver disease. Vet. Pathol. 33:521-526. [DOI] [PubMed] [Google Scholar]
- 20.Gemechu-Hatewu, M., K. L. Platt, F. Oesch, H. J. Hacker, P. Bannasch, and P. Steinberg. 1997. Metabolic activation of aflatoxin B1 to aflatoxin B1-8,9-epoxide in woodchucks. Int. J. Cancer 73:587-591. [DOI] [PubMed] [Google Scholar]
- 21.Hailey, J. R., J. K. Haseman, J. R. Bucher, A. E. Radovsky, D. E. Malarkey, R. T. Miller, A. Nyska, and R. P. Maronpot. 1998. Impact of Helicobacter hepaticus infection in B6C3F1 mice from twelve National Toxicology Program two-year carcinogenesis studies. Toxicol. Pathol. 26:602-611. [DOI] [PubMed] [Google Scholar]
- 22.Ihrig, M., M. Schrenzel, and J. G. Fox. 1999. Differential susceptibility to hepatic inflammation and proliferation in AXB recombinant inbred mice chronically infected with Helicobacter hepaticus. Am. J. Pathol. 155:571-582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jukes, T. H., and C. R. Cantor. 1969. Evolution of protein molecules, p. 21-132. In H. N. Munro (ed.), Mammalian protein metabolism. Academic Press, New York, N.Y.
- 24.Kirkbride, C. A., C. E. Gates, and J. E. Collins. 1986. Abortion in sheep caused by a non-classified anaerobic, flagellated bacterium. Am. J. Vet. Res. 47:259-262. [PubMed] [Google Scholar]
- 25.Paster, B. J., and F. E. Dewhirst. 1988. Phylogeny of Campylobacter, wolinellas, Bacteroides gracilis, and Bacteroides ureolyticus by 16S ribosomal ribonucleic acid sequencing. Int. J. Syst. Bacteriol. 38:56-62. [Google Scholar]
- 26.Popper, H., L. Roth, R. H. Purcell, B. C. Tennant, and J. L. Gerin. 1987. Hepatocarcinogenicity of the woodchuck hepatitis virus. Proc. Natl. Acad. Sci. USA 84:866-870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Roth, L., J. M. King, W. E. Hornbuckle, H. J. Harvey, and B. C. Tennant. 1985. Chronic hepatitis and hepatocellular carcinoma associated with persistent woodchuck hepatitis virus infection. Vet. Pathol. 22:338-343. [DOI] [PubMed] [Google Scholar]
- 28.Roth, L., J. M. King, and B. C. Tennant. 1991. Hepatic lesions in woodchucks (Marmota monax) seronegative for woodchuck hepatitis virus. J. Wildl. Dis. 27:281-287. [DOI] [PubMed] [Google Scholar]
- 29.Saitou, N., and M. Nei. 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4:406-425. [DOI] [PubMed] [Google Scholar]
- 30.Saunders, K. E., Z. Shen, F. Dewhirst, B. Paster, C. A. Dangler, and J. Fox. 1999. Novel intestinal Helicobacter species isolated from cotton-top tamarins (Saguinus oedipus) with chronic colitis. J. Clin. Microbiol. 37:146-151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shen, Z., Y. Feng, F. E. Dewhirst, and J. G. Fox. 2001. Coinfection with enteric Helicobacter spp. and Campylobacter spp. in cats. J. Clin. Microbiol. 39:2166-2172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sipowicz, M. A., C. M. Weghorst, Y. H. Shiao, G. S. Buzard, R. J. Calvert, M. R. Anver, L. M. Anderson, and J. M. Rice. 1997. Lack of p53 and ras mutations in Helicobacter hepaticus-induced liver tumors in A/JCr mice. Carcinogenesis 18:233-236. [DOI] [PubMed] [Google Scholar]
- 33.Stanley, J., D. Linton, A. P. Burens, F. E. Dewhirst, S. L. W. On, A. Porter, R. J. Owen, and M. Costas. 1994. Helicobacter pullorum sp. nov.—genotype and phenotype of a new species isolated from poultry and from human patients with gastroenteritis. Microbiology 140:3441-3449. [DOI] [PubMed] [Google Scholar]
- 34.Stanley, J., D. Linton, A. P. Burens, F. E. Dewhirst, R. J. Owen, A. Porter, S. L. W. On, and M. Costas. 1993. Helicobacter canis sp. nov., a new species from dogs: an integrated study of phenotype and genotype. J. Gen. Microbiol. 139:2495-2504. [DOI] [PubMed] [Google Scholar]
- 35.Summers, J., J. M. Smolec, and R. Snyder. 1978. A virus similar to human hepatitis B virus associated with hepatitis and hepatoma in woodchucks. Proc. Natl. Acad. Sci. USA 75:4533-4537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Vandamme, P., E. Falsen, B. Pot, K. Kersters, and J. D. Ley. 1990. Identification of Campylobacter cinaedi isolated from blood and feces of children and adult females. J. Clin. Microbiol. 28:1016-1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Versalovic, J., and J. G. Fox. 2001. Taxonomy and phylogeny of Helicobacter, p. 15-28. In M. Achtman and S. Suerbaum (ed.), Helicobacter pylori: molecular and cellular biology. Horizon Scientific Press, Wymondham, United Kingdom.
- 38.Ward, J. M., J. G. Fox, M. R. Anver, D. C. Haines, C. V. George, M. J. Collins, P. L. Gorelick, K. Nagashima, M. A. Gonda, R. V. Gilden, J. G. Tully, R. J. Russell, R. E. Benveniste, B. J. Paster, F. E. Dewhirst, J. C. Donovan, L. M. Anderson, and J. M. Rice. 1994. Chronic active hepatitis and associated liver tumors in mice caused by a persistent bacterial infection with a novel Helicobacter species. J. Natl. Cancer Inst. 86:1222-1227. [DOI] [PubMed] [Google Scholar]