Abstract
It is unclear whether the levels of Staphylococcus aureus colonization of hospital personnel with patient exposure are increased or whether personnel become colonized with more antibiotic-resistant strains. Differences in nasal and hand carriage of S. aureus between medical and nonmedical hospital personnel were examined. No differences in nasal carriage between the two groups were found; however, there was a trend that suggested differences in the rates of hand carriage of S. aureus (18% of nonmedical personnel and 10% of medical personnel). Medical personnel were colonized with more antibiotic-resistant isolates than nonmedical personnel (mean, 2.8 versus 2.1 isolates [P < 0.03]), and the strain profiles indicated that they tended to be more clonal in origin, suggesting that exposure to hospital isolates alters the colonization profile.
Staphylococcus aureus remains among the most important nosocomial pathogens because of both the diversity and the severity of the infections caused by these organisms (3, 11). Several studies have documented that these infections are most commonly caused by the patient's own commensal flora (9, 18, 20). The original reservoir(s) from which patients acquire these isolates remains unclear. While some infected patients are colonized with S. aureus at the time of hospitalization, others likely become colonized, often with more highly antibiotic-resistant isolates, during their hospital stays (3, 5, 7). Hospital personnel are among those implicated as possible sources of these potentially more antibiotic-resistant pathogens. Transmission of these strains to patients is then likely to occur during routine patient care. Despite the possible importance of this sequence of events, the questions of whether health care workers are more likely to be colonized with S. aureus and if these strains reflect those found in the hospital setting have received limited attention.
In the present investigation, S. aureus colonization rates among hospital personnel were compared to address three questions. (i) Are medical personnel (MP) with regular exposure to patients more likely to be colonized than nonmedical personnel (NMP)? (ii) Are MP at greater risk than NMP of becoming colonized with more antibiotic-resistant isolates? (iii) Do the staphylococcal isolates collected from MP tend to be more clonal than those collected from NMP?
Two hundred eighty employees of Montefiore Medical Center were screened for hand and nasal colonization with S. aureus between July 1999 and July 2000. Specimens for culture were collected at two sites: general medicine wards and an administrative site separate from the hospital. Only employees with direct patient contact were included in the MP group. Employees also completed a questionnaire concerning occupation, medical history, hand-washing behavior, and demographic information. The study was reviewed and approved by the Montefiore Medical Center Institutional Review Board.
Specimens for culture were collected from the anterior nares with cotton swabs (Becton Dickinson Culturette Systems, Sparks, Md.). The nasal swabs were cultured on mannitol salt agar. After a 48-h incubation at 37°C, positive colonies from each sample were isolated on 5% sheep blood agar plates (Becton Dickinson) and one to three colonies were selected for further analysis. Specimens were collected from the hands by having the subjects touch 150-mm-diameter mannitol salt agar plates (Becton Dickinson) with the palm and fingertips of the dominant hand. S. aureus species identification was confirmed by a coagulase production test and with a protein A detection kit (StaphAurex, Danford, United Kingdom).
Testing for susceptibility to antibiotics (amikacin, ampicillin, cephalothin, clindamycin, erythromycin, gentamicin, levofloxacin, oxacillin, penicillin, rifampin, trimethoprim-sulfamethoxazole, and vancomycin) was performed by the Kirby-Bauer disk diffusion method (12).
All isolates were typed by pulsed-field gel electrophoresis (PFGE). An aliquot of an overnight culture (0.5 ml) was centrifuged, the supernatant was decanted, and the remaining pellet was washed and resuspended in 10 mM Tris-1.0 M NaCl to a concentration of 2.5 × 108 cells/ml and processed as described previously (17). DNA samples were digested with SmaI (500 U/ml; Invitrogen Life Technologies) for 3 h at 25°C. Samples were run on a CHEF-DR III system (Bio-Rad) by using 1.0% agarose gels in TBE (50 mM Tris, 50 mM boric acid, 1 mM EDTA). The settings for PFGE were as follows: initial switch time, 1.0 s; final switch time, 30.0 s; included angle, 120°; current, 6.0V; and run time, 23 h. The buffer temperature was maintained at 14°C.
PFGE images were captured, archived, and analyzed with Diversity Database (version 1.0) software coupled with the GelDoc 1000 system (Bio-Rad). A tolerance of 1.0% was used in the comparison of bands from different strains (13). Dendrograms were constructed by the Dice coefficient (DC) method for calculation of relatedness and the unweighted pair group method of arithmetic averages for positioning. Two strains for which the DC was ≥70 were considered related. Strains for which the DC was ≥80 were considered subtypes of the same clone (6, 8). Degrees of relatedness were confirmed by cross-referencing of samples in the similarity matrix generated by the same Diversity Database population search that generated the dendrogram. Strains BK640, BK641, BK643, BK648, and BK700 (kindly provided by B. Kreiswirth, Public Health Research Institute, New York, N.Y.) were compared with the strains collected in this study.
Univariate relationships comparing data from the questionnaire and biological data for MP and NMP were examined by the use of chi-square statistics for categorical dependent variables. t tests were used to examine differences between the means of continuous dependent variables. Although some demographic differences were identified between the MP and the NMP groups, no demographic variables were associated with S. aureus colonization. Therefore, only the results of univariate analyses are presented. Two-tailed P values <0.05 were considered significant. P values obtained by Fisher's exact test are reported when cells had expected counts less than five. SAS (version 8.0) was used for data management and statistical analyses.
Two hundred eighty hospital employees (193 MP and 87 NMP) were enrolled in the study. The demographic and medical profiles of the study participants are summarized in Table 1. The nasal colonization rates for MP and NMP were virtually identical. MP were somewhat less likely than NMP to have positive hand cultures (P = 0.08). This decreased rate of carriage appeared to be related to a decreased interval since the time of the last hand washing for MP compared with that for NMP (P = 0.07). Among MP, 25.0% (9 of 36) of nasal carriers also had positive hand cultures, whereas among NMP, 50.0% (8 of 16) of nasal carriers had positive hand cultures. Seven of the paired isolates from MP were the same, and one pair was closely related, as determined by PFGE. Six of the pairs of isolates concurrently recovered from NMP were the same, and one pair was closely related.
TABLE 1.
Demographic and medical profiles of the hospital personnel screened for colonization with S. aureusa
| Characteristic | NMP (n = 87) | MP (n = 193) | P values |
|---|---|---|---|
| Sex (no. [%] of subjects)b | |||
| Male | 60 (69.8) | 127 (66.5) | 0.59 |
| Female | 26 (30.2) | 64 (33.5) | |
| Age (yr [mean ± SD]) | 38.5 ± 12.9 | 34.4 ± 8.9 | 0.01 |
| Ethnicity (no. [%] of subjects)c | |||
| Black | 30 (38.0) | 59 (36.0) | <0.001 |
| White | 27 (34.2) | 56 (34.1) | |
| Latino | 18 (22.8) | 8 (4.9) | |
| Asian | 4 (5.0) | 41 (25.0) | |
| No. (%) of subjects with: | |||
| S. aureus-positive nasal swabd | 16 (19.3) | 36 (19.7) | 0.94 |
| S. aureus-positive hand culturee | 15 (18.1) | 18 (10.2) | 0.08 |
| Time (min) since subject last washed hands (mean ± SD) | 83.2 ± 88.1 | 59.5 ± 70.8 | 0.07 |
| No. (%) of subjects who: | |||
| Were hospitalized in past 6 mo | 3 (3.5) | 8 (4.0) | 1.00 |
| Had taken antibiotics in past month | 3 (3.5) | 14 (7.3) | 0.22 |
| Had taken antibiotics in past 2 days | 1 (1.2) | 3 (1.6) | 1.00 |
| Had diabetes (types 1 and 2) | 1 (1.2) | 6 (3.2) | 0.44 |
| Had a skin rash | 1 (1.2) | 2 (1.0) | 1.00 |
| Had ever had an S. aureus infection | 4 (4.6) | 11 (5.7) | 1.00 |
| Had ever been treated for an S. aureus infection | 2 (2.3) | 4 (2.0) | 1.00 |
A total of 280 subjects were screened.
Data for three subjects (one control and two exposed subjects) were missing.
Data for 37 subjects (8 controls and 29 exposed subjects) were missing.
Fourteen subjects (4 controls and 10 exposed subjects) refused to be tested.
Twenty-one subjects (4 controls and 17 exposed subjects) refused to be tested.
Twenty percent (7 of 35) of nasal isolates and one hand isolate from MP were methicillin resistant (Table 2). Two of 17 (12.5%) nasal isolates from NMP were methicillin-resistant S. aureus (MRSA) (one NMP was colonized with two distinct isolates), but no isolates from the hands of NMP were MRSA. In addition, 14.3% (5 of 35) of the nasal isolates from MP were resistant to trimethoprim-sulfamethoxazole, while no isolates from NMP were resistant to trimethoprim-sulfamethoxazole. Trimethoprim-sulfamethoxazole resistance was not limited to the MRSA isolates. Isolates from MP were resistant to significantly more antibiotics than those from NMP (2.8 versus 2.1 isolates [P < 0.03]).
TABLE 2.
Antibiotic susceptibilities of nasal isolates obtained from NMP or MPa
| Antibiotic | NMP (n = 16) | MP (n = 35) | P values |
|---|---|---|---|
| No. (%) of isolates resistant | No. (%) of isolates resistant | ||
| Amikacin | 0 (0.0) | 0 (0.0) | |
| Ampicillin | 15 (93.8) | 32 (91.4) | 1.00 |
| Cephalothin | 0 (0.0) | 4 (11.4) | 0.30 |
| Clindamycin | 1 (6.3) | 5 (14.3) | 0.65 |
| Erythromycin | 4 (25.0) | 17 (48.6) | 0.11 |
| Gentamicin | 0 (0.0) | 4 (11.4) | 0.30 |
| Levofloxacin | 0 (0.0) | 9 (25.7) | 0.04 |
| Oxacillin | 2 (12.5) | 7 (20.0) | 0.70 |
| Penicillin | 15 (93.8) | 32 (91.4) | 1.00 |
| Rifampin | 0 (0.0) | 0 (0.0) | |
| Trimethoprim-sulfamethoxazole | 0 (0.0) | 5 (14.3) | 0.17 |
| Vancomycin | 0 (0.0) | 0 (0.0) |
A total of 51 isolates were tested. One isolate was unavailable for susceptibility testing.
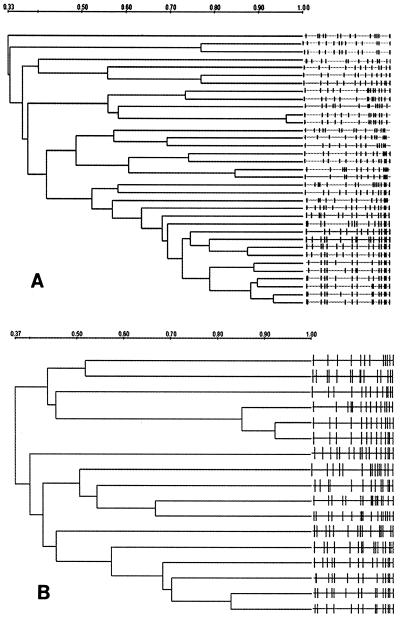
Dendrograms were generated from the PFGE profiles of the nasal isolates from MP and were compared with those generated from the PFGE profiles of the nasal isolates from NMP (Fig. 1). The isolates from MP and NMP did not segregate into separate populations (data not shown). Samples obtained from MP (22 of 35; 62.8%) had isolates that were more related (>70% similarity) than the isolates in samples obtained from NMP (5 of 17; 29.4%) (P < 0.05). This observation was true even with removal of the MRSA isolates from the analysis, although these clusters generally included isolates from no more than two subjects.
FIG. 1.
Dendrograms illustrating the relationship of S. aureus nasal isolates collected from MP (A) and NMP (B) and analyzed by PFGE. The right-hand sides of panels A and B illustrate the PFGE patterns for the individual isolates. The isolates were compared by use of the DC. Strains that were ≥70% similar were classified as related. One isolate from a member of the MP was unavailable for PFGE. Two isolates were from a single subject who was a member of the NMP. Note that the scales of the two panels differ.
Among the MRSA isolates detected, two groups of two isolates were greater than 80% related. These four MRSA isolates were also compared with the five MRSA clones (strains BK640, BK641, BK643, BK648, and BK700) that are prevalent in New York City. Strain BK640 was closely related to three of the study strains, and BK641 and BK700 were closely related to one isolate each (16). In this study, MP appeared to be at no greater risk of colonization with S. aureus than NMP. This finding is consistent with those from earlier studies that reported little difference between the colonization rates of hospitalized patients and the general population (9). Aries De Sousa et al. (2) found similar colonization rates among hospital personnel and patients in two Cape Verde hospitals, where infection control procedures are more limited than they are in the United States.
Hospital personnel had a higher incidence of colonization with strains of MRSA, and in general, their isolates were more resistant to antibiotics than those collected from NMP. Isolates from MP also demonstrated greater clonality than those from NMP. The latter observation may reflect the limited diversity of MRSA (10). These findings show that while the colonization rates did not differ between the two groups, there were differences in the profiles of the strains from the two groups. This may reflect the different reservoirs of strains to which the two groups are exposed. Opal et al. (14) found that health care workers exposed to an environment with a high rate of endemic MRSA infection had a high incidence of either hand or nasal colonization with MRSA. The combination of an unchanged colonization rate coupled with higher levels of antibiotic resistance among the isolates from MP suggests that those subjects predisposed to nasal colonization, perhaps resulting from a genetic predisposition, are at greater risk of becoming colonized with hospital isolates (4, 15).
The trend toward the reduced levels of colonization of the hands with S. aureus among MP compared with that among the NMP correlated with how recently the hands had been washed. In addition to the demonstrated efficacy of hand washing, these data once again demonstrate that transient hand carriage of S. aureus occurs and may potentially serve as a means of S. aureus transmission to hospitalized patients. Although a recent study by von Eiff et al. (18) demonstrated that patients become infected with endogenous strains, the original source of these strains remained unclear (19, 20). In a different setting, Ahmed et al. (1) found that nasal carriage of S. aureus was not a risk factor for surgical wound infections in a Sudanese hospital. The ultimate reservoir for S. aureus isolates in the hospital setting remains an issue of considerable importance if more effective infection control interventions are to be developed.
When the PFGE profiles of the isolates were compared, the majority of isolates from subjects positive for both hand and nasal isolates had identical profiles, with few persons harboring more than one strain. Of interest, there was a greater likelihood for strains from MP to cluster than for strains from NMP to cluster, and this pattern persisted even when the MRSA isolates were removed from the analysis.
This study therefore suggests that the S. aureus colonization rates of MP are unaffected by patient exposure since MP are no more likely to be colonized than NMP. However, an important finding is that the strain profiles, most notably, the antibiotic susceptibility profiles, between strains from MP and NMP are different. If MP are identified as vectors of transmission, their isolates are likely to reflect the antibiotic susceptibility profile prevalent in the hospital setting. An interesting speculation is whether genetic or environmental factors are responsible for this observation.
Acknowledgments
We gratefully acknowledge the cooperation of the Montefiore Medical Center health care workers.
F. D. Lowy is supported by grants (grants DA09656 and DA11868) from the National Institute on Drug Abuse.
REFERENCES
- 1.Ahmed, A. O., A. van Belkum, A. H. Fahal, A. E. Elnor, E. S. Abougroun, M. F. VandenBergh, E. E. Zijlstra, and H. A. Verbrugh. 1998. Nasal carriage of Staphylococcus aureus and epidemiology of surgical-site infections in a Sudanese university hospital. J. Clin. Microbiol. 36:3614-3618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aires De Sousa, M., I. Santos Sanches, M. L. Ferro, and H. De Lencastre. 2000. Epidemiological study of staphylococcal colonization and cross-infection in two West African hospitals. Microb. Drug Resist. 6:133-141. [DOI] [PubMed] [Google Scholar]
- 3.Archibald, L., L. Phillips, D. Monnet, J. E. McGowan, F. Tenover, and R. Gaynes. 1997. Antimicrobial resistance in isolates from inpatients and outpatients in the United States: increasing importance of the intensive care unit. Clin. Infect. Dis. 24:211-215. [DOI] [PubMed] [Google Scholar]
- 4.Cole, A. M., P. Dewan, and T. Ganz. 1999. Innate antimicrobial activity of nasal secretions. Infect. Immun. 67:3267-3275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Diekema, D. J., M. A. Pfaller, F. J. Schmitz, J. Smayevsky, J. Bell, R. N. Jones, and M. Beach. 2001. Survey of infections due to Staphylococcus species: frequency of occurrence and antimicrobial susceptibility of isolates collected in the United States, Canada, Latin America, Europe, and the Western Pacific region for the SENTRY Antimicrobial Surveillance Program, 1997-1999. Clin. Infect. Dis. 32(Suppl. 2):S114-S132. [DOI] [PubMed] [Google Scholar]
- 6.Dominguez, M. A., H. de Lencastre, J. Linares, and A. Tomasz. 1994. Spread and maintenance of a dominant methicillin-resistant Staphylococcus aureus (MRSA) clone during an outbreak of MRSA disease in a Spanish hospital. J. Clin. Microbiol. 32:2081-2087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fluit, A. C., C. L. Wielders, J. Verhoef, and F. J. Schmitz. 2001. Epidemiology and susceptibility of 3,051 Staphylococcus aureus isolates from 25 university hospitals participating in the European SENTRY study. J. Clin. Microbiol. 39:3727-3732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gerner-Smidt, P., L. M. Graves, S. Hunter, and B. Swaminathan. 1998. Computerized analysis of restriction fragment length polymorphism patterns: comparative evaluation of two commercial software packages. J. Clin. Microbiol. 36:1318-1323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kluytmans, J., A. van Belkum, and H. Verbrugh. 1997. Nasal carriage of Staphylococcus aureus: epidemiology, underlying mechanisms, and associated risks. Clin. Microbiol. Rev. 10:505-520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kreiswirth, B., J. Kornblum, R. D. Arbeit, W. Eisner, J. N. Maslow, A. McGeer, D. E. Low, and R. P. Novick. 1993. Evidence for a clonal origin of methicillin resistance in Staphylococcus aureus. Science 259:227-230. [DOI] [PubMed] [Google Scholar]
- 11.Lowy, F. D. 1998. Staphylococcus aureus infections. N. Engl. J. Med. 339:520-532. [DOI] [PubMed] [Google Scholar]
- 12.National Committee for Clinical Laboratory Standards. 1997. Performance standards for antimicrobial disk susceptibility tests. NCCLS document M2-A6, 6th ed. National Committee for Clinical Laboratory Standards, Wayne, Pa.
- 13.Nimmo, G. R., J. Schooneveldt, G. O'Kane, B. McCall, and A. Vickery. 2000. Community acquisition of gentamicin-sensitive methicillin-resistant Staphylococcus aureus in southeast Queensland, Australia. J. Clin. Microbiol. 38:3926-3931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Opal, S. M., K. H. Mayer, M. J. Stenberg, J. E. Blazek, D. J. Mikolich, D. L. Dickensheets, L. W. Lyhte, R. R. Trudel, and J. M. Musser. 1990. Frequent acquisition of multiple strains of methicillin-resistant Staphylococcus aureus by healthcare workers in an endemic hospital environment. Infect. Control Hosp. Epidemiol. 11:479-485. [DOI] [PubMed] [Google Scholar]
- 15.Peacock, S. J., I. de Silva, and, F. D. Lowy. 2001. What determines nasal carriage of Staphylococcus aureus? Trends Microbiol. 9:605-610. [DOI] [PubMed] [Google Scholar]
- 16.Roberts, R. B., A. de Lencastre, W. Eisner, E. P. Severina, B. Shopsin, B. N. Kreiswirth, A. Tomasz, et al. 1998. Molecular epidemiology of methicillin-resistant Staphylococcus aureus in 12 New York hospitals. J. Infect. Dis. 178:164-171. [DOI] [PubMed] [Google Scholar]
- 17.van Belkum, A., W. van Leeuwen, M. E. Kaufmann, B. Cookson, F. Forey, J. Etienne, R. Goering, F. Tenover, C. Steward, F. O'Brien, W. Grubb, P. Tassios, N. Legakis, A. Morvan, N. El Solh, R. de Ryck, M. Struelens, S. Salmenlinna, J. Vuopio-Varkila, M. Kooistra, A. Talens, W. Witte, and H. Verbrugh. 1998. Assessment of resolution and intercenter reproducibility of results of genotyping Staphylococcus aureus by pulsed-field gel electrophoresis of SmaI macrorestriction fragments: a multicenter study. J. Clin. Microbiol. 36:1653-1659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.von Eiff, C., K. Becker, K. Machka, H. Stammer, G. Peters, et al. 2001. Nasal carriage as a source of Staphylococcus aureus bacteremia. N. Engl. J. Med. 344:11-16. [DOI] [PubMed] [Google Scholar]
- 19.Williams, R. E. O. 1963. Healthy carriage of Staphylococcus aureus: its prevalence and its importance. Bacteriol. Rev. 27:56-71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Williams, R. E. O., M. P. Jevons, R. A. Shooter, C. J. W. Hunter, J. A. Girling, J. D. Griffiths, and G. W. Taylor. 1959. Nasal staphylococci and sepsis in hospital patients. Br. Med. J. 2:658-662. [DOI] [PMC free article] [PubMed] [Google Scholar]