Abstract
We describe a simple multiplex allele-specific (MAS)-PCR assay to detect mutations in the second base of the katG gene codon 315, including AGC→ACC and ACA (Ser→Thr) substitutions that confer resistance to isoniazid (INH) in Mycobacterium tuberculosis clinical isolates. The 315 ACC allele is found in the majority of Inhr strains worldwide, especially in areas with a high incidence of tuberculosis. The 315 ACA allele is characteristic of the New York City multidrug-resistant (MDR) strain W and its progenies in the United States. The mutations in katG315 are revealed depending on the presence or absence of an indicative fragment amplified from the wild-type allele of this codon. Initially optimized on the purified DNA samples, the assay was then tested on crude cell lysates and auramine-stained sputum slide preparations with the same reproducibility and interpretability of profiles generated by agarose gel electrophoresis. The MAS-PCR assay can be used for the detection of resistance to INH in clinical laboratories in regions with a high prevalence of MDR M. tuberculosis strains.
Presently, tuberculosis (TB) reemergence and spread are of worldwide concern. The situation is aggravated by the increasing circulation of multidrug-resistant (MDR) Mycobacterium tuberculosis strains that are defined as resistant to at least rifampin (RIF) and isoniazid (INH), which comprise the backbone of antitubercular chemotherapy. While RIF resistance is essentially mediated by the rpoB hot spot region mutations (18), INH resistance is apparently controlled by a more complex genetic system that involves several genes, namely, katG, inhA, kasA, and ahpC (16, 17). Finally, extensive studies have demonstrated that INH resistance is most frequently associated with mutations in katG. The INH used for therapy is a prodrug and necessitates catalytic activation to be converted into an active form. Catalase-peroxidase, the product of the katG gene, was shown to accomplish this function, and the mutations in this gene mediate INH resistance (4, 23). Early studies showed that the complete deletion of the gene is rare, since peroxidase activity in KatG is necessary to detoxify host antibacterial radicals (4, 8, 19; T. C. Victor, G. S. Pretorius, J. V. Felix, A. M. Jordaan, P. D. van Helden, and K. D. Eisenach, Letter, Antimicrob. Agents Chemother. 40:1572, 1996). For this reason, the predominant way of acquiring resistance via KatG alterations is through selection of those particular mutations that decrease catalase activity but still maintain the peroxidase activity of the enzyme at a certain level in viable Inhr organisms. Such mutations were found in up to 90% of Inhr strains, and one particular substitution in the codon 315, AGC→ACC (Ser→Thr), was reported as the most frequent, apparently providing the optimal balance between the decreased catalase and sufficiently high peroxidase activities of KatG (16). This mutation was reported to be associated with intermediate or high levels of resistance to INH (1 to 10 μg/ml [11, 16, 23]), though it has recently been suggested that the katG315 alteration alone may not always be clinically significant (1). The prevalence of the katG315 (AGC→ACC) mutation coding for the Ser→Thr substitution among MDR M. tuberculosis strains in the world, though generally high, varies in regions with low and high TB incidences, ranging from rare occurrences in Scotland (7) and Finland (11) to 26 to 30% in Singapore (10) and Madrid (15) to 52 to 64% in South Africa (9; Victor et al., letter) to 79% in Peru (6). In a previous study (13), the katG S315T mutation in M. tuberculosis isolates recovered from epidemiologically unlinked patients in northwestern Russia from 1996 to 2001 was screened by a PCR-restriction fragment length polymorphism (RFLP) assay. This mutation was found in 93.6% of 204 Inhr strains.
Particular methods described so far for the detection of katG315 changes include rather elaborate DNA sequencing (6, 9, 11), cleavase fragment length polymorphism (3), single-strand conformation polymorphism (19), and molecular beacon (15) analyses where mutations in different katG codons were screened, along with PCR-RFLP (13; Victor et al., letter) and dot blot hybridization (21, 24) strategies based on PCR, followed by either restriction endonuclease analysis or DNA hybridization, respectively. The aim of the present study was to develop a simple and rapid assay for detecting INH resistance in M. tuberculosis clinical isolates based exclusively on katG PCR and agarose gel electrophoresis without any supplementary sequence analysis.
MATERIALS AND METHODS
Bacterial strains and susceptibility testing.
The 80 M. tuberculosis isolates were recovered from 80 unrelated adult (15 to 63 years) patients with newly diagnosed pulmonary TB. They originated from St. Petersburg and three neighboring provinces in northwestern Russia (Leningrad, Novgorod, and Pskov) and were admitted to the hospitals of the St. Petersburg Institute of Phthisiopulmonology and City Anti-Tuberculosis Dispensary of St. Petersburg between 1999 and 2001.
Species identification of the isolates was based on standard microbiological tests: colony morphology, acid-fast staining, and biochemical tests (27, 28). Löwenstein-Jensen medium was used for the cultivation of isolates, and susceptibility testing was performed by the absolute concentration method as recommended by the Russian Ministry of Health (order no. 558 of 28 June 1978) and has been described previously (25). A microbial suspension containing 5 × 108 organisms/ml was prepared according to the McFarland turbidity standards and was diluted 1:10; then, 0.2 ml of the dilution was added to Löwenstein-Jensen medium with or without a drug. The culture tubes were incubated at 37°C, and growth was monitored after 3 weeks of incubation and assessed as previously described (28). An isolate was considered resistant if bacterial growth occurred at a concentration of 1 μg of INH per ml, 20 μg of RIF per ml, 5 μg of streptomycin (STR) per ml, 2 μg of ethambutol (EMB) per ml, and 100 μg of pyrazinamide (PZA [pH 5.6]) per ml. The method of absolute concentration was previously shown in our laboratory to give results concordant with those generated by the proportion method in a comparative study with the National Mycobacterial Reference Laboratory in Turku, Finland (25).
DNA preparation.
DNA used for the PCR analysis was either extracted from cultured cells and purified as described by van Embden et al. (20) or obtained by resuspending a mycobacterial colony in 200 μl of 1× TE buffer (10 mM Tris-HCl and 1 mM EDTA [pH 7.4], with or without Chelex 100 [5%, wt/vol; Sigma]), followed by boiling for 20 min. After centrifugation, the DNA-containing supernatant (cell lysate) was harvested and stored at 4°C until used. The DNA of the strain M22 (W) with katG315 ACA sequence was kindly provided by Peter Small and Alexander Pym (Stanford University Medical Center).
The DNA samples from auramine-stained sputum smears (cell counts graded as 1+, 2+, or 3+ [27]) were obtained by scraping the material from the glass slide to a sterile Eppendorf tube and then resuspending it in a total volume of 75 μl of extraction buffer (15% Chelex 100 [Sigma], 0.5% Nonidet P-40, [Sigma], 0.5% Tween 20), followed by incubation for 20 min in a boiling water bath. After centrifugation, the removed supernatant was used for PCR.
Control for contamination during microbiological and genetic experiments was performed as recommended in references 28 and 5, respectively. In particular, a negative control sputum slide was processed along with the test slides. A control of possible contamination with previously amplified amplicons was performed by including a negative control sample (distilled water) in each PCR run; no contamination was detected.
PCR-RFLP analysis.
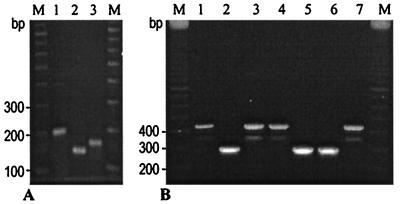
A primer pair, katg1F (5′-AGCTCGTATGGCACCGGAAC) and katg4R (5′-AACGGGTCCGGGATGGTG) (Fig. 1), was used to amplify a katG315 fragment (katG positions 904 to 1103 in strain H37Rv [GenBank accession number X68081, positions 2882 to 3081]) as described previously (13). The amplified fragment was digested with HapII (Amersham Pharmacia Biotech); the obtained restriction fragments were electrophoresed in 1.5% standard agarose gels (Quantum Bioprobe) and visualized under UV light. The katG codon 315 missense mutation AGC-Ser→ACC-Thr created an additional HapII site (C↑CGG) and could be detected by cleavage with this restriction endonuclease (or its isoschizomer, like MspI). As a result, the size of the largest digest was 132 bp for the mutated (315 AGC→ACC) allele, the INH-resistant strain, and 153 bp for the katG315 wild-type or differently mutated alleles (Fig. 2A, lanes 2 and 3, respectively). The shorter 10- to 21-bp digests ran outside of the gel and were not considered.
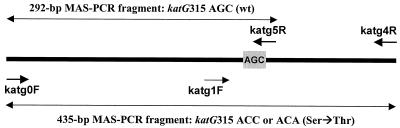
FIG. 1.
Schematic view of the katG gene fragment targeted by MAS-PCR assay using three primers (katg0F, katg5R, and katg4R [boldface arrows]). Short arrows indicate primers; long double-headed arrows depict the indicated PCR fragments. The katG codon 315 AGC is in the shaded box. The katg1F and katg4R primers are used in the PCR-RFLP assay.
FIG. 2.
katG 315 PCR-RFLP (A) and MAS-PCR (B) profiles of the M. tuberculosis strains. (A) Lanes: 1, 200-bp PCR fragment used for HapII-RFLP analysis; 2, strain with katG 315 AGC→ACC mutation; 3, strain with katG315 wild-type allele. (B) Lanes: 1, 3, and 4, strains with the katG315 AGC→ACC mutation; 2, 5, and 6, strains with the katG315 wild-type allele; 7, strain with the AGC→ACA mutation (strain W); M, 100-bp DNA ladder (Amersham Pharmacia Biotech).
MAS-PCR assay.
The multiplex allele-specific (MAS)-PCR assay was previously developed for embB306 mutational analysis (12). In the present study, we applied this strategy to distinguish between the wild-type and mutated alleles of katG315 (Fig. 1). The assay used three primers. The inner reverse primer katg5R was positioned so that its 3′ end paired with the second base (G) of the codon 315 wild-type allele (AGC) (Fig. 1). Consequently, in the absence of mutation in this position in katG315, a 292-bp fragment was amplified by the outer forward primer katg0F and the inner reverse primer katg5R (Fig. 2B, lanes 2, 5, and 6). If a mutation (e.g., AGC→ACC) occurred, this resulted in a mismatch at the 3′ end of this wild-type inner primer and, under the appropriate stringent PCR conditions, in the absence of the 292-bp PCR product. Two outer primers, katg0F and katg4R, flanked the entire katG315 region under investigation (Fig. 1). In the case of the katG315 wild-type allele, amplification of this 435-bp fragment was prevented under selected reaction conditions due to the concurrent action of the katg5R inner primer. The 435-bp fragment was amplified in only the strains with katG315 mutations (Fig. 2B, lanes 1, 3, 4, and 7) and was therefore indicative of the INH resistance phenotype.
The following primers were used for a single tube PCR targeting a portion of the katG gene (katG positions 669 to 1103 in strain H37Rv [accession number X68081, positions 2647 to 3081]): two outer primers, forward primer katg0F (5′-GCAGATGGGGCTGATCTACG) and reverse primer katg4R, and one inner reverse primer, katg5R (5′- ATACGACCTCGATGCCGC). A purified DNA sample (0.1 μl) or 10 μl of lysate or sputum slide preparation was added to a PCR mixture (final volume of 30 μl) that contained 30 pmol each of katg0F and katg5R, 40 pmol of katg4R, 1.5 mM MgCl2, 1 U of recombinant Taq DNA polymerase (MBI Fermentas), and 200 μM concentrations of each deoxynucleoside triphosphate. The reaction was performed in a PTC-100 thermal controller (MJ Research, Inc.) under the following conditions: initial denaturation at 96°C for 3 min; 5 cycles at 95°C for 1 min, 64°C for 1 min, and 72°C for 30 s; 5 cycles at 95°C for 1 min, 62°C for 40 s, and 72°C for 30 s; 20 cycles (25 cycles for sputum slide or lysate probes) at 94°C for 1 min, 60°C for 40 s, and 72°C for 30 s; and final elongation at 72°C for 3 min. The amplified fragments were electrophoresed in 1.5% agarose gels and visualized under UV light.
Sequencing of the katG fragment.
The 500-bp katG fragment was amplified with the KG1 (5′-AATCGATGGGCTTCAAGACG) and KG2 (5′-CTCGTAGCCGTACAGGATCTCG) primers. The PCR (35 cycles) was performed as follows: 94°C for 1 min, 56°C for 1 min, and 72°C for 1 min. PCR products were analyzed in 2% agarose gels, purified with a Qiagen purification kit (Qiagen), and sequenced with the [32P]-end-labeled KG2 primer as described in reference 26.
RESULTS AND DISCUSSION
The M. tuberculosis clinical strains studied included 15 Inhs and 65 Inhr isolates from new TB cases in the northwestern region of the Russian Federation from 1999 to 2001 (13). The Inhs strains were pan-susceptible. The Inhr strains were also resistant to RIF and STR (n = 57); to RIF, STR, and EMB (n = 2); to RIF, STR, and PZA (n = 4); and to RIF, STR, EMB, and PZA (n = 2).
First, the sample under investigation was assessed by the katG315 PCR-RFLP assay (13). None of the 15 Inhs strains had the katG315 AGC→ACC mutation. Since other, rarely described mutations in codon 315 confer INH resistance (11), we assumed that these Inhs strains had a wild-type allele of codon 315 (AGC). The katG315 AGC→ACC alteration was observed in all 65 Inhr strains. Sequence analysis of six randomly selected strains confirmed the PCR-RFLP assay results: the katG315 mutation (AGC→ACC) was found in three Inhr strains, and the katG315 wild-type allele (AGC) was identified in three Inhs strains.
The MAS-PCR assay was evaluated to detect katG codon 315 alteration in the strains studied. All 15 Inhs strains produced a single MAS-PCR 292-bp band (Fig. 2B, lanes 2, 5, and 6), as expected, implying no mutation in the katG315 second base. A single MAS-PCR 435-bp fragment was amplified in all the 65 Inhr strains harboring the AGC→ACC mutation (Fig. 2B, lanes 1, 3, and 4). The generation of an either 292- or 435-bp fragment provided a PCR quality confirmation so as to rule out eventual false-negative results due to lack of amplification. In general, the MAS-PCR results on the katG315 variation were concordant with those generated by PCR-RFLP analysis for all the strains tested. A mutation in the katG315 second base, one other than AGC→ACC, should have also resulted in the amplification of a single 435-bp fragment, though such mutations are rare (8, 16) and were not found in our isolates.
The high prevalence of the katG315 ACC mutant allele among Inhr M. tuberculosis clinical isolates in northwestern Russia (93.6% for all isolates), especially among the strains of the Beijing family (100% for the Beijing isolates from new TB cases from 1996 to 2001), was previously shown (13). The strains of this family are known to be highly prevalent in East Asia and in the countries of the former Soviet Union, and they have also been reported in other geographical regions, including North America (2, 14, 22). The notorious New York City strain W, which caused several MDR-TB outbreaks in the United States in the nineties, also belongs to the Beijing family (2). However, unlike other members of this family, one of the specific features of the W (New York City) strain and its progenies is that their INH resistance is associated with a rare mutation in the katG codon 315, AGC→ACA (Ser→Thr) (16). This double mutation does not create a HapII site (CCGG), and PCR-RFLP analysis cannot distinguish it from the wild-type allele. In contrast, the MAS-PCR assay that we used to detect the AGC→ACC mutation allowed us to also identify the AGC→ACA mutation. In such a case, a mismatch at the 3′ end of the inner wild-type primer katg5R (Fig. 1) is even more dramatic, as it involves two bases of codon 315. MAS-PCR was performed on one available W strain that harbored the katG315 ACA allele. This analysis unambiguously identified the presence of this mutation and hence the INH resistance phenotype of the W strain (Fig. 2B, lane 7).
The MAS-PCR assay was initially performed on the purified DNA preparations with which the PCR conditions and concentrations were optimized. The assay was then tested on a selection of 22 DNA samples from crude cell lysates and sputum slide preparations, all from the same patients (seven strains, Inhs, katG315 AGC; 15 strains, Inhr, katG315 ACC). The MAS-PCR profiles obtained from smear preparations and purified DNA from cultured cells were equally unambiguous in interpretation and concordant for each of the 22 patients examined (data not shown).
A limited number of gene codons has recently been suggested to reliably predict resistance to the major drugs in the majority of M. tuberculosis strains in areas with high TB incidence (21). However, for RIF, a minimum of three codons of rpoB should be analyzed and neither wild-type nor mutant alleles can be detected by any restriction endonuclease. On the other hand, the so noticeably prevalent katG315 AGC→ACC or ACA mutations (6, 9, 13, 15; Victor et al., letter) present markers of choice for INH resistance detection. The katG315 MAS-PCR assay can therefore be suggested as a simple and rapid tool for detecting with high probability the INH resistance in M. tuberculosis clinical strains. It is also applicable for direct detection in stained sputum smear preparations, which should facilitate the adequate and timely choice of antituberculosis therapy. The assay is easy to perform and to interpret; it is based on a single tube PCR and minigel electrophoresis without any further extension and can be implemented into the routine practices of clinical laboratories in areas with a high prevalence of MDR M. tuberculosis strains. However, preliminary evaluation of the katG315 mutations' distribution is a prerequisite for each particular setting prior to large-scale implementation.
Acknowledgments
We are thankful to Peter Small and Alexander Pym for kindly providing us with DNA of the W strain. We are grateful to Alessandra Riva for critical reading of the manuscript and English language corrections.
This study was partly supported by the Délégation Générale au Réseau International des Instituts Pasteur et Instituts Associés, Institut Pasteur, Paris, France.
REFERENCES
- 1.Abate, G., S. E. Hoffner, V. O. Thomsen, and H. Miörner. 2001. Characterization of isoniazid-resistant strains of Mycobacterium tuberculosis on the basis of phenotypic properties and mutations in katG. Eur. J. Clin. Microbiol. Infect. Dis. 20:329-333. [DOI] [PubMed] [Google Scholar]
- 2.Bifani, P. J., B. Mathema, N. E. Kurepina, and B. N. Kreiswirth. 2002. Global dissemination of the Mycobacterium tuberculosis W-Beijing family strains. Trends Microbiol. 10:45-52. [DOI] [PubMed] [Google Scholar]
- 3.Brow, M. A., M. C. Oldenburg, V. Lyamichev, L. M. Heisler, N. Lyamicheva, J. G. Hall, N. J. Eagan, D. M. Olive, L. M. Smith, L. Fors, and J. E. Dahlberg. 1996. Differentiation of bacterial 16S rRNA genes and intergenic regions and Mycobacterium tuberculosis katG genes by structure-specific endonuclease cleavage. J. Clin. Microbiol. 34:3129-3137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dobner, P., S. Rusch-Gerdes, G. Bretzel, K. Feldmann, M. Rifai, T. Loscher, and H. Rinder. 1997. Usefulness of Mycobacterium tuberculosis genomic mutations in the genes katG and inhA for the prediction of isoniazid resistance. Int. J. Tuberc. Lung Dis. 1:365-369. [PubMed] [Google Scholar]
- 5.Dragon, E. A., J. P. Spadoro, and R. Madej. 1993. Quality control of polymerase chain reaction, p. 160-168. In D. H. Persing, T. F. Smith, F. C. Tenover, and T. J. White (ed.), Diagnostic molecular microbiology. American Society for Microbiology, Washington, D.C.
- 6.Escalante, P., S. Ramaswamy, H. Sanabria, H. Soini, X. Pan, O. Valiente-Castillo, and J. M. Musser. 1998. Genotypic characterization of drug-resistant Mycobacterium tuberculosis isolates from Peru. Tuber. Lung Dis. 79:111-118. [DOI] [PubMed] [Google Scholar]
- 7.Fang, Z., C. Doig, A. Rayner, D. T. Kenna, B. Watt, and K. J. Forbes. 1999. Molecular evidence for heterogeneity of the multiple-drug-resistant Mycobacterium tuberculosis population in Scotland (1990 to 1997). J. Clin. Microbiol. 37:998-1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Haas, W. H., K. Schilke, J. Brand, B. Amthor, K. Weyer, R. B. Fourie, G. Bretzel, V. Sticht-Groh, and H. J. Bremer. 1997. Molecular analysis of katG gene mutations in strains of Mycobacterium tuberculosis complex from Africa. Antimicrob. Agents Chemother. 41:1601-1603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kiepiela, P., K. S. Bishop, A. N. Smith, L. Roux, and D. F. York. 2000. Genomic mutations in the katG, inhA, and ahpC genes are useful for the prediction of isoniazid resistance in Mycobacterium tuberculosis isolates from Kwazulu Natal, South Africa. Tuber. Lung Dis. 80:47-56. [DOI] [PubMed] [Google Scholar]
- 10.Lee, A. S. G., I. H. K. Lim, L. L. H. Tang, A. Telenti, and S. Y. Wong. 1999. Contribution of kasA analysis to detection of isoniazid-resistant Mycobacterium tuberculosis in Singapore. Antimicrob. Agents Chemother. 43:2087-2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Marttila, H. J., H. Soini, P. Huovinen, and M. K. Viljanen. 1996. katG mutations in isoniazid-resistant Mycobacterium tuberculosis isolates recovered from Finnish patients. Antimicrob. Agents Chemother. 40:2187-2189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mokrousov, I., O. Narvskaya, E. Limeschenko, T. Otten, and B. Vyshnevskiy. 2002. Detection of ethambutol-resistant Mycobacterium tuberculosis strains by multiplex allele-specific PCR assay targeting embB306 mutations. J. Clin. Microbiol. 40:1617-1620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mokrousov, I., O. Narvskaya, T. Otten, E. Limeschenko, L. Steklova, and B. Vyshnevskiy. 2002. High prevalence of KatG Ser315Thr substitution among isoniazid-resistant Mycobacterium tuberculosis clinical isolates from northwestern Russia, 1996 to 2001. Antimicrob. Agents Chemother. 46:1417-1424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Narvskaya, O., I. Mokrousov, T. F. Otten, and B. I. Vyshnevskiy. 1999. Genetic marking of polyresistant Mycobacterium tuberculosis strains isolated in the north-west of Russia. Probl. Tuberk. 1999(3):39-41. [PubMed]
- 15.Piatek, A. S., A. Telenti, M. R. Murray, H. El-Hajj, W. R. Jacobs, Jr., F. R. Kramer, and D. Alland. 2000. Genotypic analysis of Mycobacterium tuberculosis in two distinct populations using molecular beacons: implication for rapid susceptibility testing. Antimicrob. Agents Chemother. 44:103-110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ramaswami, S., and J. M. Musser. 1998. Molecular genetic basis of antimicrobial agent resistance in Mycobacterium tuberculosis: 1998 update. Tuber. Lung Dis. 79:3-29. [DOI] [PubMed] [Google Scholar]
- 17.Slayden, R. A., and C. E. Barry III. 2000. The genetics and biochemistry of isoniazid resistance in Mycobacterium tuberculosis. Microbes Infect. 2:659-669. [DOI] [PubMed] [Google Scholar]
- 18.Telenti, A., P. Imboden, F. Marchesi, D. Lowrie, S. Cole, M. J. Colston, L. Matter, K. Schopfer, and T. Bodmer. 1993. Detection of rifampicin-resistance mutations in Mycobacterium tuberculosis. Lancet 341:647-650. [DOI] [PubMed] [Google Scholar]
- 19.Temesgen, Z., K. Satoh, J. R. Uhl, B. C. Kline, and F. R. Cockerill III. 1997. Use of polymerase chain reaction single-strand conformation polymorphism (PCR-SSCP) analysis to detect a point mutation in the catalase-peroxidase gene (katG) of Mycobacterium tuberculosis. Mol. Cell. Probes 11:59-63. [DOI] [PubMed] [Google Scholar]
- 20.van Embden, J. D. A., M. D. Cave, J. T. Crawford, J. W. Dale, K. D. Eisenach, B. Gicquel, P. Hermans, C. Martin, R. McAdam, T. M. Shinnik, and P. Small. 1993. Strain identification of Mycobacterium tuberculosis by DNA fingerprinting: recommendations for a standardized methodology. J. Clin. Microbiol. 31:406-409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.van Rie, A., R. Warren, I. Mshanga, A. M. Jordaan, G. D. van der Spuy, M. Richardson, J. Simpson, R. P. Gie, D. A. Enarson, N. Beyers, P. D. van Helden, and T. C. Victor. 2001. Analysis for a limited number of gene codons can predict drug resistance of Mycobacterium tuberculosis in a high-incidence community. J. Clin. Microbiol. 39:636-641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.van Soolingen, D., L. Qian, P. E. W. de Haas, J. T. Douglas, H. Traore, F. Portaels, Z. Quing, D. Enkhasaikan, P. Nymadawa, and J. D. A. van Embden. 1995. Predominance of a single genotype of Mycobacterium tuberculosis in countries of East Asia. J. Clin. Microbiol. 33:3234-3238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.van Soolingen, D., P. E. W. de Haas, H. R. van Doorn, E. Kuijper, H. Rinder, and M. W. Borgdorff. 2000. Mutations in amino acid position 315 of the katG gene are associated with high-level resistance to isoniazid, other drug resistance, and successful transmission of Mycobacterium tuberculosis in The Netherlands. J. Infect. Dis. 182:1788-1790. [DOI] [PubMed] [Google Scholar]
- 24.Victor, T. C., A. M. Jordaan, A. van Rie, G. D. van der Spuy, M. Richardson, P. D. van Helden, and R. Warren. 1999. Detection of mutations in drug resistance genes of Mycobacterium tuberculosis by a dot-blot hybridization strategy. Tuber. Lung Dis. 79:343-348. [DOI] [PubMed] [Google Scholar]
- 25.Viljanen, M. K., B. I. Vyshnevskiy, T. F. Otten, E. Vyshnevskaya, M. Marijamaki, H. Soini, P. J. Laippala, and A. V. Vasilyef. 1998. Survey of drug-resistant tuberculosis in northwestern Russia from 1984 through 1994. Eur. J. Clin. Microbiol. Infect. Dis. 17:177-183. [DOI] [PubMed] [Google Scholar]
- 26.Wang, B., Q. Fang, W. B. Williams, and D. B. Weiner. 1992. Double-stranded DNA sequencing by linear amplification with Taq DNA polymerase. BioTechniques 13:172-176. [PubMed] [Google Scholar]
- 27.World Health Organization. 1998. Part II. Microscopy, p. 43. Laboratory services in tuberculosis control. World Health Organization, Geneva, Switzerland.
- 28.World Health Organization. 1998. Part III. Culture, p. 77. Laboratory services in tuberculosis control. World Health Organization, Geneva, Switzerland.