Abstract
Nucleic acid amplification assays such as the ligase chain reaction and PCR have encountered reproducibility problems. The initial extract and a newly extracted aliquot of urine specimens (n = 120) which had signal-to-cutoff (S/CO) ratios above 0.80 by the LCx Chlamydia assay were retested. Nucleic acid was extracted from an additional urine sample for testing by the AMPLICOR PCR Chlamydia assay. Fifteen percent (18 of 120) of the urine specimens were negative by all repeat tests (initial mean S/CO ratio by the LCx Chlamydia assay, 0.93; S/CO ratio range, 0.80 to 3.30). Repeat testing of the 102 specimens with possible positive results by the LCx Chlamydia assay by use of the initially extracted aliquot confirmed the results for 95 (93.1%) of the specimens; repeat testing of a newly extracted aliquot confirmed the results for 87 (85.3%) of the specimens. Twenty specimens had discordant results by the two repeat LCx Chlamydia assays. A total of 78 of 102 (76.5%) of the specimens were positive by the AMPLICOR PCR, and the AMPLICOR PCR confirmed the results for 82.1% (78 of 95) and 89.6% (78 of 87) of the specimens positive by the two repeat LCx Chlamydia assays, respectively. Some of the discrepancies observed by multiple repeat tests may have been due to specimen mislabeling or contamination during performance of the procedure rather than to the LCx Chlamydia assay. Both assays suffered from a lack of reproducibility on repeat testing with a small proportion of specimens, probably due to the presence of low levels of DNA, the presence of variable amounts of amplification inhibitors, and the loss of DNA during extraction.
For the past 8 years, clinical laboratories have become accustomed to using nucleic acid amplification (NAA) tests for the detection of Chlamydia trachomatis on swabs and in urine specimens from men and women (1-3, 5, 8, 10). These assays allow the effective management and treatment of C. trachomatis infections. The two NAA assays that have been in routine use the longest, the AMPLICOR PCR Chlamydia assay (Roche Diagnostics Systems, Branchburg, N.J.) and the LCx Chlamydia assay (Abbott Laboratories), have been reported to have reproducibility problems (4, 7).
By February 2001, the Abbott Laboratories Diagnostics Division had received customer complaints concerning high rates of positivity for negative controls, resulting in invalid assay runs of the LCx Chlamydia assay, and positive patient specimens which did not test positive upon retesting. Abbott issued a Device Correction letter which stated the following: the specificity of the assay for some on-market lots of the test kit had dropped as low as 92%, but the test sensitivity remained in the normal range. The letter instructed LCx Chlamydia assay users to take the following actions: (i) interpret the results for samples with signal-to-cutoff (S/CO) ratios less than 0.80 as negative and report that C. trachomatis plasmid DNA was not detected and that the sample could be presumed to be negative for C. trachomatis by ligase chain reaction (LCR) amplification and detection by microparticle enzyme immunoassay (MEIA), and (ii) retest all patient samples for which S/CO ratios are greater than or equal to 0.80. If the S/CO ratio by the repeat test was greater than or equal to 1.00, the sample should be considered LCx Chlamydia assay positive (C. trachomatis plasmid DNA was detected and the sample was reported to be positive for C. trachomatis by LCR amplification and detection by MEIA). If the S/CO ratio by the repeat test was less than 1.00, the sample should be considered LCx Chlamydia assay negative (plasmid DNA was not detected and the sample was presumed to be negative for C. trachomatis by LCR amplification and detection by MEIA). This repeat testing algorithm was developed to ensure that package insert claims for specificity were met.
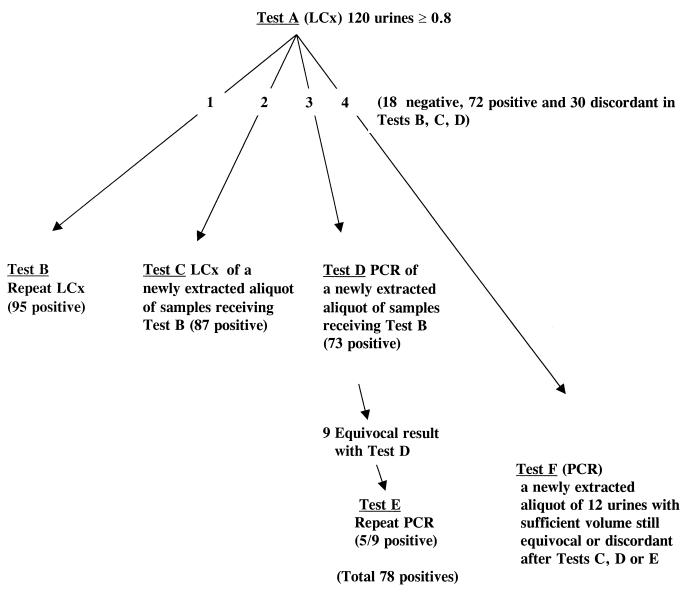
We initiated a study of urine samples (the algorithm used is illustrated in Fig. 1) to record and analyze the specificity of the LCx Chlamydia assay for positive samples, as outlined by the directive, with the following objectives: (i) to determine whether the results of testing of a sample newly extracted from the original urine specimen conducted on the next day (test C) were similar to those obtained with the original extract (test A) and to those obtained by repeat testing of the initially processed urine specimen (test B) and (ii) to test on the second day an additional aliquot extracted from the original urine specimen by the AMPLICOR PCR (test D). All tests were performed by experienced technologists according to the instructions for the testing of urine provided in the package inserts of each of the commercial tests. Samples were tested by the AMPLICOR PCR without knowledge of the repeat testing results obtained by the LCx Chlamydia assay. When an equivocal result was achieved by the AMPLICOR PCR, the test was repeated in duplicate (test E), as outlined in the package insert. Discordant results were further investigated by testing a new aliquot neat and/or at a dilution of 1:4 by the AMPLICOR PCR (test F) (Fig. 1). Five different lots of the LCx Chlamydia assay (lots 74588 M300, 77305 M300, 76962 M300, 75310 M400, and 75314 M400) were used in this study.
FIG. 1.
Algorithm for testing of urine specimens for C. trachomatis by the LCx Chlamydia assay and the AMPLICOR PCR.
Of 1,040 urine specimens tested from 4 April to 7 August 2001, the results for a total of 120 (from 59 men and 61 women) fulfilled the criteria for retesting described in the Device Correction letter. These were processed by use of the testing algorithm (Fig. 1), and 18 (15%) were negative by all repeat tests (tests B, C, and D), leaving 102 samples with possible true-positive results. For these 102 specimens, repeat testing of the initially extracted aliquot by the LCx Chlamydia assay (test B) confirmed the positive results for 95 (93.1%) of the specimens, and repeat testing of a newly extracted sample by the LCx Chlamydia assay on the next day (test C) confirmed the positive results for 87 (85.3%) of the specimens. Testing by the AMPLICOR PCR confirmed the positive results for 78 of 95 (82.1%) of the specimens positive by test B and 78 of 87 (89.6%) specimens positive by test C.
Of the 102 specimens possibly positive, the results for 72 were concordant by all repeat tests, leaving 30 specimens with discordant results among tests A, B, C, and D. Twenty samples had discordant results between the two repeat LCx Chlamydia assays. Three of the six samples negative by test B and positive by test C were positive by the AMPLICOR PCR. Analysis by the AMPLICOR PCR of the 14 samples which were positive by test B and negative by test C showed that 12 were negative and 2 were positive. Some of these samples with discordant results may have been contaminated in the initial round of testing (test A) and therefore would have remained positive by test B because the aliquot already extracted would have been stabilized. Alternatively, the second and third aliquots, because of the presence of low numbers of DNA copies, had no amplifiable nucleic acids after extraction.
The initial S/CO ratios for the 18 urine specimens with negative results ranged from 0.80 to 3.30 (mean S/CO ratio, 0.93), and 61% (11 of 18) had S/CO ratios less than 1. The initial S/CO ratios for the 30 specimens with discordant results ranged from 0.88 to 4.14 (mean S/CO ratio, 1.95). The initial S/CO ratios for the samples which were positive by at least one of the repeat tests ranged from 1.01 to 6.79 (mean S/CO ratio, 3.62). None of the 72 samples positive by tests B, C, and D had an initial S/CO ratio by the LCx Chlamydia assay that was less than 1. Thus, except for a few outliers, samples with confirmed positive results had higher S/CO ratios by the initial LCx Chlamydia test.
The study shows that repeat testing of the initially extracted samples by the LCx Chlamydia assay (test B) would have enabled reporting of positive results for 95 specimens, of which some may have been false-positive results. The false-positive results were probably attributed to laboratory procedures, not the assay. Testing of a newly extracted aliquot by the LCx Chlamydia assay would have led to the reporting of positive results for 87 specimens, 3 of which needed further repeat testing with another aliquot of urine to be found positive by the AMPLICOR PCR (Table 1, specimens 80, 86, and 103).
TABLE 1.
Variability of results of LCx Chlamydia assay and AMPLICOR PCR for 12 urine specimens on repeat testing and testing of a newly extracted aliquot
| Specimen no. | S/CO ratio (result) by LCx Chlamydia assaya
|
AMPLICOR PCR ODb
|
||||
|---|---|---|---|---|---|---|
| Initial extraction
|
New extraction (test C) | Initial extraction
|
New extraction (test F) | |||
| First test (test A) | Repeat test (test B) | First test (test D) | Repeat test (test E) | |||
| 51 | 1.27 (+) | 0.03 (−) | 0.03 (−) | 2.546 (+)c | NDd | 0.016 (−) |
| 65 | 1.72 (+) | 2.60 (+) | 0.77 (−) | 0.226 (e)e | 0.014 (−), 0.015 (−) | 0.144 (−) |
| 70 | 1.45 (+) | 2.89 (+) | 3.73 (+) | 0.344 (e) | 0.625 (+), 0.012 (−) | 1.237 (+) |
| 78 | 2.85 (+) | 3.57 (+) | 3.34 (+) | 0.088 (−) | ND | 0.492 (e) |
| 80 | 3.39 (+) | 3.35 (+) | 3.39 (+) | 0.128 (−) | ND | 3.188 (+) |
| 86 | 3.70 (+) | 3.54 (+) | 3.58 (+) | 0.031 (−) | ND | 0.975 (+) |
| 87 | 1.50 (+) | 1.11 (+) | 0.01 (−) | 0.017 (−) | ND | 0.209 (e) |
| 103 | 2.47 (+) | 0.08 (−) | 2.17 (+) | 0.787 (e) | 0.012 (−), 0.006 (−) | 1.868 (+) |
| 64 | 0.88 (e) | 0.83 (−) | 1.40 (+) | 0.009 (−) | ND | 0.005 (−) |
| 81 | 3.08 (+) | 3.19 (+) | 2.77 (+) | 0.071 (−) | ND | 0.012 (−) |
| 90 | 3.54 (+) | 3.71 (+) | 3.81 (+) | 0.019 (−) | ND | 0.002 (−) |
| 91 | 3.43 (+) | 3.34 (+) | 3.67 (+) | 0.403 (e) | 0.027 (−), 0.011 (−) | 0.551 (e) |
By the LCx Chlamydia assay, an S/CO ratio of ≥1.0 is considered a positive result.
By the AMPLICOR PCR an optical density (OD) of <0.2 is a negative result for C. trachomatis after duplicate tests.
An optical density ≥0.8 is a positive result.
ND, not done.
An optical density ≥0.2 but <0.8 is equivocal (e) and the sample should be retested in duplicate. The final result was interpreted by using three values with a new cutoff of >0.2.
The use of a second C. trachomatis NAA assay such as the AMPLICOR PCR allowed us to assess its performance during repeat testing. Aliquots newly extracted from 12 of the samples were retested by two LCx Chlamydia assays (tests A and C) and two AMPLICOR PCR assays (tests D and F) (Table 1). For eight samples the result was different from the original result (positive, negative, or equivocal) when they were tested by the AMPLICOR PCR assay, and for four samples the results between tests A and C, for which the LCx Chlamydia assay was used, were different. The data illustrate the variability in the results that can be seen for clinical samples which may contain low levels of nucleic acid (3) and also illustrate how sampling may influence whether a selected aliquot of a specimen is positive (9). Superimposed on this is the realization that some nucleic acids may become lost during the extraction process. This loss of nucleic acids for amplification and the variable levels of inhibitors present in a sample (6) may be capable of dropping the result for a sample from the positive to the equivocal zone or from the borderline positive to the negative zone. We were unable to perform spiking experiments to detect inhibitors because of the limited volumes of urine specimens available. As the newer amplification assays become used and repeat testing is performed, we may learn that more of them are prone to variable reproducibilities. Proficiency testing programs are an important, but limited, way of monitoring performance. A system of routine recording of results and the use of controls, calibrators, and specimens known to be positive but with variable strengths should enable clinical laboratories to identify problems and act quickly to rectify them.
REFERENCES
- 1.Bauwens, J. E., A. M. Clark, M. J. Loeffelholz, S. A. Herman, and W. E. Stamm. 1993. Diagnosis of Chlamydia trachomatis urethritis in men by polymerase chain reaction assay of first-catch urine. J. Clin. Microbiol. 31:3013-3016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chernesky, M. A., D. Jang, H. Lee, J. D. Burczak, H. Hu, J. Sellors, S. J. Tomazic-Allen, and J. B. Mahony. 1994. Diagnosis of Chlamydia trachomatis infections in men and women by testing first-void urine by ligase chain reaction. J. Clin. Microbiol. 32:2682-2685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Coutlee, F., M. de Ladurantaye, C. Tremblay, J. Vincelette, L. Labrecque, and M. Roger. 2000. An important proportion of genital samples submitted for Chlamydia trachomatis detection by PCR contain small amounts of cellular DNA as measured by β-globin gene amplification. J. Clin. Microbiol. 38:2512-2515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gronowski, A. M., S. Copper, D. Baorto, and P. R. Murray. 2000. Reproducibility problems with the Abbott Laboratories LCx assay for Chlamydia trachomatis and Neisseria gonorrhoeae. J. Clin. Microbiol. 38:2416-2418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jaschek, G., C. A. Gaydos, L. E. Welsh, and T. C. Quinn. 1993. Direct detection of Chlamydia trachomatis in urine specimens from symptomatic and asymptomatic men by using a rapid polymerase chain reaction assay. J. Clin. Microbiol. 31:1209-1212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mahony, J., S. Chong, D. Jang, K. Luinstra, M. Faught, D. Dalby, J. Sellors, and M. Chernesky. 1998. Urine specimens from pregnant and nonpregnant women inhibitory to amplification of Chlamydia trachomatis nucleic acid by PCR, ligase chain reaction, and transcription-mediated amplification: identification of urinary substances associated with inhibition and removal of inhibitory activity. J. Clin. Microbiol. 36:3122-3126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Peterson, E. M., V. Darrow, J. Blanding, S. Aarnaes, and L. M. de la Maza. 1997. Reproducibility problems with the AMPLICOR PCR Chlamydia trachomatis test. J. Clin. Microbiol. 35:957-959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schachter, J., W. E. Stamm, T. C. Quinn, W. W. Andrews, J. D. Burczak, and H. H. Lee. 1994. Ligase chain reaction to detect Chlamydia trachomatis infection of the cervix. J. Clin. Microbiol. 32:2540-2543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Smieja, M., J. B. Mahony, C. H. Goldsmith, S. Chong, A. Petrich, and M. Chernesky. 2001. Replicate PCR testing and probit analysis for detection and quantitation of Chlamydia pneumoniae in clinical specimens. J. Clin. Microbiol. 39:1796-1801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Stary, A., E. Schuh, M. Kerschbaumer, B. Götz, and H. Lee. 1998. Performance of transcription-mediated amplification and ligase chain reaction assays for detection of chlamydial infection in urogenital samples obtained by invasive and noninvasive methods. J. Clin. Microbiol. 36:2666-2670. [DOI] [PMC free article] [PubMed] [Google Scholar]