Abstract
Cryptosporidiosis is a significant cause of food-borne and waterborne outbreaks of diarrheal diseases. To better understand the route of transmission of Cryptosporidium parasites, a number of genotyping techniques have been developed, based on PCR-restriction fragment length polymorphism or sequencing analysis of antigen, structural, and housekeeping genes. In this study, a real-time assay for the detection of Cryptosporidium oocysts is described. This technique had a detection limit of five oocysts. By melting curve analysis of PCR products with fluorescence-labeled hybridization probes, this technique was able to differentiate five common Cryptosporidium parasites that are pathogenic for humans in a single PCR. We evaluated and validated the test using samples from presently known Cryptosporidium parasites that are pathogenic for humans. This technique provides an alternative molecular tool in epidemiologic studies of human cryptosporidiosis.
Cryptosporidium organisms are protozoan parasites that infect various vertebrate hosts. At least 10 species are recognized: C. parvum, C. baileyi, C. serpentis, C. muris, C. andersoni, C. wrairi, C. meleagridis, C. felis, C. canis, and C. saurophilum. Within C. parvum there are also different genotypes, some of which may be independent species (5, 27). Each Cryptosporidium sp. or genotype has a different host specificity, but at least five types of Cryptosporidium have been found to infect humans: C. parvum human genotype, C. parvum bovine genotype, C. meleagridis, C. felis, and C. canis (in the order of prevalence) (26, 27). Thus, the identification of Cryptosporidium species and genotypes is important for the assessment of the public health importance of Cryptosporidium oocysts of animal or environmental origins and for the tracking of infection or contamination sources.
Presently, the identification of Cryptosporidium spp. and genotypes is made mostly by PCR-restriction fragment length polymorphism or sequencing analysis of antigen, structural, and housekeeping genes (1-4, 6-21, 23, 24, 29). These procedures are usually time-consuming. We describe here a LightCycler PCR for the real-time detection and species identification of Cryptosporidium parasites. The technique takes advantage of the well-characterized genetic polymorphism in the small-subunit (SSU) rRNA, utilizes fluorescence-labeled probes for real-time detection of Cryptosporidium, and incorporates a melting curve analysis of PCR products for the differentiation of Cryptosporidium spp. and genotypes that are pathogenic for humans.
MATERIALS AND METHODS
Parasite specimens.
The Cryptosporidium parasites used in this study included C. parvum human, bovine, mouse, ferret, and marsupial genotypes; C. wrairi; C. meleagridis; C. felis; C. canis; C. baileyi; C. andersoni; and C. serpentis. All of the isolates used in this study were from naturally infected animals or humans, except for C. meleagridis, which was isolated from a turkey and passed through 1- to 2-week-old turkey poults. Identification of Cryptosporidium parasites was based on oocyst morphology, the infected host, and other traditional classification guidelines. All of the Cryptosporidium isolates used in the study were characterized previously at multiple genetic loci to confirm species and genotypes (22, 29, 30). Eimeria tenella and Eimeria acervulina from chickens and an Isospora sp. from a dog were used as controls. All stool samples containing Cryptosporidium, Isospora, and Eimeria oocysts were stored at 4°C in 2.5% potassium dichromate for less than 12 months before used in DNA extraction.
Oocyst isolation and DNA extraction.
For most samples, stools containing oocysts were used in DNA extraction, with the exception of one C. parvum bovine genotype isolate (isolate 6), for which oocysts purified by sucrose-Percoll centrifugation were used in DNA extraction. DNA was extracted from oocysts or stool samples by alkaline digestion (100 μl of stool pellets in 33.3 μl of 1 M KOH and 9.3 μl of 1 M dithiothreitol at 65°C for 15 min, followed by neutralization with 4.3 μl of 25% HCl and 80 μl of 2 M Tris-HCl, pH 8.3), phenol-chloroform-isoamyl alcohol extraction (Invitrogen, Carlsbad, Calif.), and DNA purification using a QIAamp DNA Stool Mini Kit (Qiagen, Valencia, Calif.).
Real-time PCR.
The most polymorphic region of the SSU rRNA gene (∼820 bp) was amplified from samples by PCR carried out in a LightCycler (Roche Molecular Biochemicals, Indianapolis, Ind.), using Cryptosporidium-specific forward primer 5′-GGAAGGGTTGTATTTATTAGATAAAG-3′ and reverse primer 5′-AAGGAGTAAGGAACAACCTCCA-3′ (29, 30). The PCR mixture contained 2 μl of Perkin-Elmer (Norwalk, Conn.) 10× buffer, 4 mM MgCl2, 100 μM (each) deoxynucleoside triphosphate, 400 nM forward and reverse primers, 200 nM (each) hybridization probes (probe 1, 5′-CCGTCTAAAGCTGATAGGTCAGAAACTTGAATG-flourescein-3′; probe 2, 5′-LCred705-GTCACATTAATTGTGATCCGTAAAG-3′), 0.5 μl of nonacetylated bovine serum albumin (10 mg/ml), 1 U of Taq polymerase, and 1 μl of DNA template in a total of 20 μl. Probe 1 was based on the sequence that is conserved among all Cryptosporidium parasites, whereas probe 2 was a sequence of the C. parvum human genotype and was designed to hybridize to all intestinal Cryptosporidium parasites with various extents of mismatches.
In initial assays, the manufacturer-suggested Master Hybridization Probe Kit (Roche) was also used. Each PCR mixture was then subjected to 55 cycles of denaturation at 94°C for 2 s, annealing at 50°C for 10 s, and extension at 72°C for 15 s, with an initial denaturation at 95°C for 3 min. Detection of the fluorescent signal was made after each cycle's annealing phase. For the determination of detection sensitivity, serial dilutions were made from the DNA extracted from a stock concentration of 10,000 C. parvum bovine oocysts/μl. Oocysts of the C. parvum bovine genotype were used in the sensitivity determination because of the easy acquisition of a large number of purified oocysts, even though one of the probes used in detection had two base mismatches to the C. parvum bovine genotype sequence. In some experiments, the PCR products were harvested from the LightCycler capillaries by centrifugation of the inverted capillaries after caps were removed. The PCR products were then visualized by staining of 1.2% agarose electrophoresis gels with ethidium bromide to determine whether the amplified product was the proper band size and to compare the sensitivities of gel electrophoresis and fluorescence-tagged detection.
Species differentiation and genotyping by melting curve analysis.
For identification of species and genotypes, known Cryptosporidium parasites as well as isolates of other apicomplexans were used in real-time PCR under the same conditions as described above. Species differentiation and genotyping were based on differences in melting temperatures of the PCR-probe complexes, which were determined by the extent of complementation of the probes to the target strand of PCR product. For the melting curve analysis, after the completion of the last PCR cycle, a quick denaturation was done at 95°C (0-s holding time), followed by a 30-s annealing step at 45°C with a slow ramp (0.1°C/s) up to 80°C with continuous detection throughout the ramp.
RESULTS
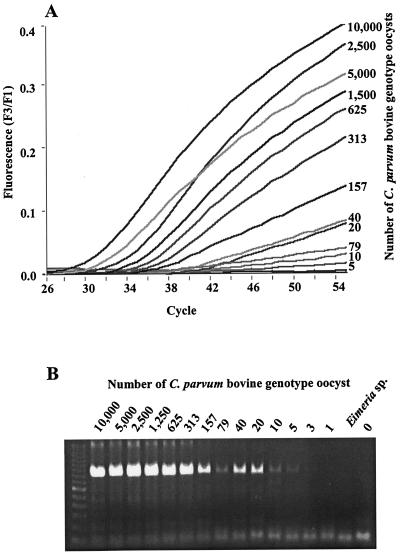
Most of the studies were done with the in-house master mixture described in Materials and Methods. With this master mixture, there was a direct relationship between the starting copies of the DNA template of the C. parvum bovine genotype and the cycle of PCR where detection of signal occurred (Fig. 1A). DNAs from the Eimeria spp. and the Isospora sp. never generated signals greater than background, demonstrating that the primers and probes are specific to Cryptosporidium spp. The electrophoresis gel for these samples (Fig. 1B) also showed no amplification for Eimeria, further supporting that the primers are Cryptosporidium specific. The detection limit for real-time PCR is about five oocysts of the C. parvum bovine genotype, which was about the same as that for gel electrophoresis.
FIG. 1.
(A) Real-time detection of fluorescent hybridization probes to PCR product formation of a 1:2 serial dilution of a C. parvum bovine standard from 10,000 oocysts per reaction down to a single oocyst. The detection limit is five oocysts. (B) Agarose electrophoresis gel stained with ethidium bromide. PCR products of serial dilutions of the C. parvum bovine genotype were visualized under UV light after ethidium bromide staining.
Melting curve analysis showed that all PCR products of the C. parvum bovine genotype had the same melting curve profile irrespective of the amount of template used. The calculated melting temperatures for PCR products from the DNA of 3 to 10,000 oocysts were all around 59.3°C. Melting curve analysis further confirmed that the PCR product from five oocysts was Cryptosporidium specific, because it had the same melting temperature as other PCR products of the C. parvum bovine genotype. Melting temperatures calculated from different PCR runs were stable, with interassay variations generally smaller than 0.25°C (data not shown).
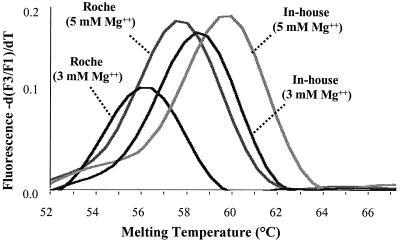
The magnesium chloride concentration, however, was found to directly affect the melting temperatures of the probes. As shown in Fig. 2, when the magnesium chloride concentration in the reaction tube was increased from 3 to 5 mM, a difference of greater than 1°C in melting temperature was observed with DNA from the C. parvum bovine genotype, no matter whether the commercial (in the Roche Master Hybridization Probe Kit) or the in-house PCR buffers were used. In subsequent studies, we used 4 mM magnesium chloride, because it gave the most consistent PCR amplification for all Cryptosporidium parasites (data not shown).
FIG. 2.
Effect of magnesium chloride on melting temperature in two PCR buffer systems. Roche, amplification of the C. parvum bovine genotype using Roche's Master Hybridization Kit, with 3 or 5 mM MgCl2. In-house, amplification of the C. parvum bovine genotype with the PCR master mix described in Materials and Methods, with 3 or 5 mM MgCl2.
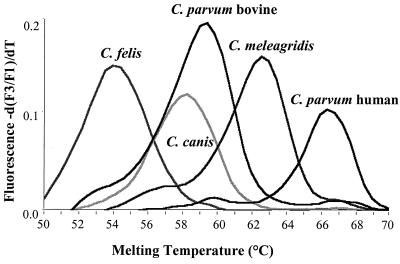
The real-time PCR did not detect the Isospora sp., the Eimeria spp., C. andersoni, or C. serpentis. However, DNA samples of all of the intestinal Cryptosporidium parasites tested resulted in signals in real-time PCR (Table 1). Agarose gel electrophoresis of the PCR products revealed that DNA samples of C. andersoni and C. serpentis generated PCR products of the expected size, even though no signals were seen in real-time PCR (Table 1). In contrast, DNA samples of the Eimeria spp. and the Isospora sp. were not amplified by the PCR. There was a wide range of melting temperatures (54.23 to 66.50°C) for the probe set for different Cryptosporidium parasites. Different Cryptosporidium spp. and genotypes generally had different melting curves and melting temperatures (Fig. 3 and Table 1). The following exceptions were noticed: (i) C. wrairi and the C. parvum ferret genotype had similar melting temperatures, (ii) the C. parvum bovine and C. parvum mouse genotypes had similar melting temperatures, and (iii) the Cryptosporidium dog genotype and C. baileyi had a melting temperature difference of less than 0.5°C (Table 1).
TABLE 1.
Melting temperatures of the fluorescent hybridization probes for different isolates of Cryptosporidium parasites
| Species/genotype | Sample | Origin | Result of PCR and gel electrophoresis | Melting temp (°C) |
|---|---|---|---|---|
| C. parvum/human | 120 | Human | + | 66.50 |
| C. parvum/human | 503 | Human | + | 66.50 |
| C. parvum/bovine | 6 | Bovine | + | 59.30 |
| C. parvum/bovine | 11 | Bovine | + | 59.30 |
| C. parvum/mouse | 350 | Mouse | + | 59.20 |
| C. parvum/marsupial | 428 | Kangaroo | + | 56.60 |
| C. parvum/ferret | 351 | Ferret | + | 55.61 |
| C. wrairi | 517 | Guinea pig | + | 55.55 |
| C. canis | 244 | Dog | + | 58.04 |
| C. canis | 715 | Dog | + | 58.04 |
| C. meleagridis | 295 | Turkey | + | 62.40 |
| C. felis | 288 | Cat | + | 54.23 |
| C. baileyi | 764 | Chicken | + | 58.38 |
| C. andersoni | 20 | Bovine | + | No detection |
| C. serpentis | 63 | Lizard | + | No detection |
| E. tenella | 353 | Chicken | − | No detection |
| E. acervulina | 354 | Chicken | − | No detection |
| Isospora sp. | 865 | Dog | − | No detection |
FIG. 3.
Melting curve profiles of the fluorescent probes for the five Cryptosporidium parasites that are pathogenic for humans (C. parvum human and bovine genotypes, C. meleagridis, C. felis, and C. canis). The differences in the melting temperatures are due to DNA sequence polymorphism in the probing region of the SSU rRNA gene.
DISCUSSION
The results of this study show the potential of real-time PCR in rapid diagnosis of human cryptosporidiosis. In preliminary evaluations, it detected and differentiated all common human Cryptosporidium parasites in a single PCR. It had sensitivity compatible to that of traditional PCR but had the advantage of real-time detection of PCR products and did not need restriction digestion or sequence analysis for species differentiation or genotyping. With the use of different fluorescent dyes, multiplex PCR potentially can be developed to allow simultaneous detection of multiple pathogens (Cryptosporidium and other enteropathogens). Thus, in clinical laboratories with real-time PCR machines, real-time PCR can be an ideal screening tool for the detection and genotyping of Cryptosporidium parasites in stool samples. The only requirement is the use of stools that have not been preserved in formalin, which is one of the commonly used stool preservatives but is problematic for all PCR-based diagnostic tools.
The use of a biprobe format for real-time detection of PCR products in this assay increases the specificity of the detection. The specificity of the detection came from the use of specific primers in the PCR and the use of specific probes in the real-time detection of PCR products. The specificity of primers used in this assay has been evaluated extensively, and they can amplify all Cryptosporidium parasites (25, 26, 28-30). The two probes used for the real-time detection added another layer of specificity, because they were chosen to be Cryptosporidium specific based on multiple alignment of SSU rRNA sequences of various apicomplexan parasites. The latter was confirmed by the failure to detect PCR products of C. andersoni and C. serpentis during real-time detection, even though the primers amplified DNAs of C. andersoni and C. serpentis. This is because one of the two probes, GTCACATTAATTGTGATCCGTAAAG, was designed to hybridize only with PCR products of intestinal Cryptosporidium parasites (C. parvum, C. wrairi, C. meleagridis, C. saurophilum, C. felis, C. canis, and C. baileyi, etc.). The use of melting curve analysis offers another layer of specificity of detection to the assay, because nonspecific PCR products are likely to have wrong melting temperatures even if they hybridize to the probes.
As shown in this study, most Cryptosporidium species and C. parvum genotypes could be differentiated from each other by melting curve analysis, judged by the melting temperatures calculated. The C. parvum human genotype had a melting temperature different from that of the C. parvum bovine genotype. Likewise, C. meleagridis and C. felis also had melting temperatures different from those of each other and other Cryptosporidium parasites. Thus, all five Cryptosporidium parasites that are pathogenic for humans (the C. parvum human and bovine genotypes, C. meleagridis, C. felis, and C. canis) could be differentiated from each other by melting curve analysis. A few other Cryptosporidium parasites had melting temperatures similar to those of the Cryptosporidium parasites that are pathogenic for humans. For example the C. parvum mouse genotype had a melting temperature similar to that of the C. parvum bovine genotype, and C. baileyi had melting temperature close to that of C. canis. These Cryptosporidium parasites have not been detected in humans, and thus this should not present a problem in real-time PCR analysis of clinical samples.
The melting curves of the PCR products from various Cryptosporidium parasites are directly related to the sequence diversity in the region of the SSU rRNA gene covered by the LCred probe. Because the probe sequence was based on the C. parvum human genotype, isolates of the C. parvum human genotype had the highest melting temperature (66.5°C) during melting curve analysis. C. meleagridis had one base mismatch and thus had a somewhat lower melting temperature (62.4°C). The bovine and mouse genotypes of C. parvum had two mismatches and thus had even lower melting temperatures (59.2 to 59.3°C). C. felis had the most sequence divergence and thus had the lowest melting temperature (54.2°C). The position of mismatches also affected the melting temperature; C. canis had a melting temperature lower than those of the C. parvum bovine and mouse genotypes (58.0 versus 59.2 to 59.3°C), even though they all had two base mismatches with the probe sequence.
In summary, the real-time PCR developed in this study can detect and differentiate all five common Cryptosporidium parasites that are pathogenic for humans. The sensitivity was similar to that of conventional PCR, but the specificity was increased because of the use of two hybridization probes during the detection phase and melting curve analysis during the differentiation phase. It is possible to increase the sensitivity of detection by further standardization and reducing the size of amplicon. Real-time PCR has a more rapid turnaround time for reporting results and with further development can be quantitative, both of which will be important in investigations of waterborne outbreaks of cryptosporidiosis.
REFERENCES
- 1.Awad-el-Kariem, F. M., D. C. Warhurst, and V. McDonald. 1994. Detection and species identification of Cryptosporidium oocysts using a system based on PCR and endonuclease restriction. Parasitology 109:19-22. [DOI] [PubMed] [Google Scholar]
- 2.Bonnin, A., M. N. Fourmaux, J. F. Dubremetz, R. G. Nelson, P. Gobet, G. Harly, M. Buisson, D. Puygauthier-Toubas, G. Gabriel-Pospisil, M. Naciri, and P. Camerlynck. 1996. Genotyping human and bovine isolates of Cryptosporidium parvum by polymerase chain reaction-restriction fragment length polymorphism analysis of a repetitive DNA sequence. FEMS Microbiol. Lett. 137:207-211. [DOI] [PubMed] [Google Scholar]
- 3.Carraway, M., S. Tzipori, and G. Widmer. 1997. A new restriction fragment length polymorphism from Cryptosporidium parvum identifies genetically heterogeneous parasite populations and genotypic changes following transmission from bovine to human hosts. Infect. Immun. 65:3958-3960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Deng, M. Q., and D. O. Cliver. 1998. Differentiation of Cryptosporidium parvum isolates by a simplified randomly amplified polymorphic DNA technique. Appl. Environ. Microbiol. 64:1954-1957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fayer, R., U. Morgan, and S. J. Upton. 2000. Epidemiology of Cryptosporidium: transmission, detection and identification. Int. J. Parasitol. 30:1305-1322. [DOI] [PubMed] [Google Scholar]
- 6.Gasser, R. B., X. Zhu, S. Caccio, R. Chalmers, G. Widmer, U. M. Morgan, R. C. Thompson, E. Pozio, and G. F. Browning. 2001. Genotyping Cryptosporidium parvum by single-strand conformation polymorphism analysis of ribosomal and heat shock gene regions. Electrophoresis 22:433-437. [DOI] [PubMed] [Google Scholar]
- 7.Gibbons, C. L., B. G. G. Gazzard, M. Ibrahim, S. Morris-Jones, C. S. L. Ong, and F. M. Awad-El-Kariem. 1998. Correlation between markers of strain variation in Cryptosporidium parvum: evidence of clonality. Parasitol. Int. 47:139-147. [Google Scholar]
- 8.Gobet, P., and S. Toze. 2001. Sensitive genotyping of Cryptosporidium parvum by PCR-RFLP analysis of the 70-kilodalton heat shock protein (HSP70) gene. FEMS Microbiol. Lett. 200:37-41. [DOI] [PubMed] [Google Scholar]
- 9.Kimbell, L. M., D. L. Miller, W. Chavez, and N. Altman. 1999. Molecular analysis of the 18S rRNA gene of Cryptosporidium serpentis in a wild-caught corn snake (Elaphe guttata guttata) and a five-species restriction fragment length polymorphism-based assay that can additionally discern C. parvum from C. wrairi. Appl. Environ. Microbiol. 65:5345-5349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Leng, X., D. A. Mosier, and R. D. Oberst. 1996. Differentiation of Cryptosporidium parvum, C. muris, and C. baileyi by PCR-RFLP analysis of the 18S rRNA gene. Vet. Parasitol. 62:1-7. [DOI] [PubMed] [Google Scholar]
- 11.Lowery, C. J., J. E. Moore, B. C. Millar, D. P. Burke, K. A. McCorry, E. Crothers, and J. S. Dooley. 2000. Detection and speciation of Cryptosporidium spp. in environmental water samples by immunomagnetic separation, PCR and endonuclease restriction. J. Med. Microbiol. 49:779-785. [DOI] [PubMed] [Google Scholar]
- 12.Morgan, U. M., C. C. Constantine, D. A. Forbes, and R. C. Thompson. 1997. Differentiation between human and animal isolates of Cryptosporidium parvum using rDNA sequencing and direct PCR analysis. J. Parasitol. 83:825-830. [PubMed] [Google Scholar]
- 13.Ong, C. S. L., D. L. Eisler, S. H. Goh, J. Tomblin, F. M. Awad-El-Kariem, C. B. Beard, L. H. Xiao, I. Sulaiman, A. Lal, M. Fyfe, A. King, W. R. Bowie, and J. L. Isaac-Renton. 1999. Molecular epidemiology of cryptosporidiosis outbreaks and transmission in British Columbia, Canada. Am. J. Trop. Med. Hyg. 61:63-69. [DOI] [PubMed] [Google Scholar]
- 14.Pedraza-Diaz, S., C. Amar, G. L. Nichols, and J. McLauchlin. 2001. Nested polymerase chain reaction for amplification of the Cryptosporidium oocyst wall protein gene. Emerg. Infect. Dis. 7:49-56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Peng, M. M., L. Xiao, A. R. Freeman, M. J. Arrowood, A. A. Escalante, A. C. Weltman, C. S. Ong, W. R. MacKenzie, A. A. Lal, and C. B. Beard. 1997. Genetic polymorphism among Cryptosporidium parvum isolates: evidence of two distinct human transmission cycles. Emerg. Infect. Dis. 3:567-573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rochelle, P. A., E. M. Jutras, E. R. Atwill, R. De Leon, and M. H. Stewart. 1999. Polymorphisms in the beta-tubulin gene of Cryptosporidium parvum differentiate between isolates based on animal host but not geographic origin. J. Parasitol. 85:986-989. [PubMed] [Google Scholar]
- 17.Spano, F., L. Putignani, S. Guida, and A. Crisanti. 1998. Cryptosporidium parvum: PCR-RFLP analysis of the TRAP-C1 (thrombospondin-related adhesive protein of Cryptosporidium-1) gene discriminates between two alleles differentially associated with parasite isolates of animal and human origin. Exp. Parasitol. 90:195-198. [DOI] [PubMed] [Google Scholar]
- 18.Spano, F., L. Putignani, J. McLauchlin, D. P. Casemore, and A. Crisanti. 1997. PCR-RFLP analysis of the Cryptosporidium oocyst wall protein (COWP) gene discriminates between C. wrairi and C. parvum, and between C. parvum isolates of human and animal origin. FEMS Microbiol. Lett. 150:209-217. [DOI] [PubMed] [Google Scholar]
- 19.Sturbaum, G. D., C. Reed, P. J. Hoover, B. H. Jost, M. M. Marshall, and C. R. Sterling. 2001. Species-specific, nested PCR-restriction fragment length polymorphism detection of single Cryptosporidium parvum oocysts. Appl. Environ. Microbiol. 67:2665-2668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sulaiman, I. M., L. Xiao, C. Yang, L. Escalante, A. Moore, C. B. Beard, M. J. Arrowood, and A. A. Lal. 1998. Differentiating human from animal isolates of Cryptosporidium parvum. Emerg. Infect. Dis. 4:681-685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sulaiman, I. M., A. A. Lal, M. J. Arrowood, and L. H. Xiao. 1999. Biallelic polymorphism in the intron region of beta-tubulin gene of Cryptosporidium parasites. J. Parasitol. 85:154-157. [PubMed] [Google Scholar]
- 22.Sulaiman, I. M., U. M. Morgan, R. C. Thompson, A. A. Lal, and L. Xiao. 2000. Phylogenetic relationships of Cryptosporidium parasites based on the 70-kilodalton heat shock protein (HSP70) gene. Appl. Environ. Microbiol. 66:2385-2391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Vasquez, J. R., L. Gooze, K. Kim, J. Gut, C. Petersen, and R. G. Nelson. 1996. Potential antifolate resistance determinants and genotypic variation in the bifunctional dihydrofolate reductase-thymidylate synthase gene from human and bovine isolates of Cryptosporidium parvum. Mol. Biochem. Parasitol. 79:153-165. [DOI] [PubMed] [Google Scholar]
- 24.Widmer, G., L. Tchack, C. L. Chappell, and S. Tzipori. 1998. Sequence polymorphism in the beta-tubulin gene reveals heterogeneous and variable population structures in Cryptosporidium parvum. Appl. Environ. Microbiol. 64:4477-4481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Xiao, L., K. Alderisio, J. Limor, M. Royer, and A. A. Lal. 2000. Identification of species and sources of Cryptosporidium oocysts in storm waters with a small-subunit rRNA-based diagnostic and genotyping tool. Appl. Environ. Microbiol. 66:5492-5498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Xiao, L., C. Bern, J. Limor, I. Sulaiman, J. Roberts, W. Checkley, L. Cabrera, R. H. Gilman, and A. A. Lal. 2001. Identification of 5 types of Cryptosporidium parasites in children in Lima, Peru. J. Infect. Dis. 183:492-497. [DOI] [PubMed] [Google Scholar]
- 27.Xiao, L., U. M. Morgan, R. Fayer, R. C. Thompson, and A. A. Lal. 2000. Cryptosporidium systematics and implications for public health. Parasitol. Today 16:287-292. [DOI] [PubMed] [Google Scholar]
- 28.Xiao, L., A. Singh, J. Limor, T. K. Graczyk, S. Gradus, and A. Lal. 2001. Molecular characterization of Cryptosporidium oocysts in samples of raw surface water and wastewater. Appl. Environ. Microbiol. 67:1097-1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Xiao, L. H., L. Escalante, C. F. Yang, I. Sulaiman, A. A. Escalante, R. J. Montali, R. Fayer, and A. A. Lal. 1999. Phylogenetic analysis of Cryptosporidium parasites based on the small-subunit rRNA gene locus. Appl. Environ. Microbiol. 65:1578-1583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Xiao, L. H., U. M. Morgan, J. Limor, A. Escalante, M. Arrowood, W. Shulaw, R. C. A. Thompson, R. Fayer, and A. A. Lal. 1999. Genetic diversity within Cryptosporidium parvum and related Cryptosporidium species. Appl. Environ. Microbiol. 65:3386-3391. [DOI] [PMC free article] [PubMed] [Google Scholar]