Abstract
Medical imaging informatics must exceed the mere development of algorithms. The discipline is also responsible for the establishment of methods in clinical practice to assist physicians and improve health care. From our point of view, it is commonly accepted that model-based analysis of medical images is superior to other concepts, but only a few applications are found in daily clinical use. The gap between development of model-based image analysis and its routine application can be addressed by identifying four necessary transfer steps: formulation, parameterization, instantiation, and validation. Usually, computer scientists formulate the model and define its parameterization, i.e., configure a model to handle a selected subset of clinical data. During instantiation, the algorithm adapts the model to the actual data, which is validated by physicians. Since medical a priori knowledge and particular knowledge on technical details are required for parameterization and validation, these steps are considered to be bottlenecks. In this paper, we propose general schemes that allow an application- or image-specific parameterization to be performed by medical users. Combining noncontextual and contextual approaches, we also suggest a reliable scheme that allows application-specific validation, even if a gold standard is unavailable. To emphasize our point of view, we provide examples based on unsupervised segmentation in medical imagery, which is one of the most difficult tasks. Following the proposed schemes, an exact delineation of cells in micrographs is parameterized, validated, and successfully established in daily clinical use, while automatic determination of body regions in radiographs cannot be configured to support reliable and robust clinical use. The results stress that parameterization and validation must be based on clinical data that show all potential variations and artifact sources.
In the context of biomedical imaging and computer-assisted diagnosis, image analysis is an intense field of research and development. According to our experience, the most difficult part of medical image analysis is unsupervised segmentation, i.e., the automated localization and delineation of coherent structures of interest. Since relevant structures in biomedical images strongly differ in brightness, texture, and shape, sophisticated approaches include a priori knowledge. In order to incorporate this knowledge, model-based image analysis is frequently applied.
For segmentation, such a model provides the geometry for structures of interest and incorporates a priori knowledge on plausible geometry. The individual instance of the model best explaining the given image data is determined as a segmentation result. Here, plausibility can be determined, e.g., by laws of physics, or by probability density functions observed for large numbers of structures. The snake model, which was introduced in 1987 by Kass et al.,1 is one of the first models applied to segment medical images. In the succeeding 15 years, numerous models for image analysis have been developed and published. In our opinion, shape-based models2 and active contours3 are superior concepts for solving the localization and the delineation tasks in medical imaging, respectively.
Currently, a variety of research applications have been reported for the detection and measurement of tumors, vessels, and organs in a diversity of clinical indications such as diagnostics, treatment planning, and living organ donations. A few companies already provide some form of model-based image analysis (e.g., ImageChecker, R2 Technology, Sunnyvale, CA), while others integrate less sophisticated algorithms into their workstation software (e.g., syngo, Siemens AG Medical Solutions, Forchheim, Germany). Regardless of the complexity of the models used for image analysis, integration currently reaches only a semiautomatic level, where difficult and time-consuming interaction with physicians is still required.
For instance, the workflow for syngo Volume Evaluation, whereby the system “diagnoses” the volume of interest, is described in five steps in which the configuration (place seed points for a region-growing segmentation) of the segmentation algorithm as well as the validation and improvement of the result (mark and link unconnected regions of interest) is performed manually.4 As a result, the volume is quantified by the system. In computer-aided detection (note that computer-aided diagnosis and computer-aided detection are fundamentally different concepts providing the physician with qualitative assistance and quantitative measurements, respectively), the digital image is analyzed and annotated by the computer. For example, in the R2 software for mammography, most of the suspicious locations are marked in a so-called prompt image of lower resolution. After an initial (unprompted) search of the original mammogram, the reader consults this prompt image. Stepping through all the prompts, he or she checks the regions of the original mammogram, where mass and microcalcifications are delineated by the computer automatically, and modifies his or her judgment about the mammogram accordingly.5 This ensures that all significant regions of the mammogram are verified.
In summary, a variety of successful algorithms for computer-aided diagnosis by means of medical image analysis are presented in the literature, but robust use in clinical practice is still a major challenge for ongoing research in medical informatics.6 It is currently under discussion whether available models can solve clinical problems. However, this cannot be decided as long as various systems' models are not validated in clinical applications.7 In particular, unsupervised segmentation is not available for general use in the biomedical context,8 and fully automated image analysis is unlikely to provide a solution in the short term.5 Hence, medical image segmentation largely remains an unsolved research problem rather than a useful clinical tool. The discrepancy between intensity of research and lack of robust applications is striking. From our point of view, the transfer of models from algorithmic development into clinical application is the major bottleneck. We consider four steps to establish an application for medical image analysis:
Formulation means to define the data structures representing relevant biomedical objects and the algorithms determining a valid instance for given image values. Here, generality and robustness are the main criteria.9 Sufficient generality ensures that a model is applicable to different tasks and adaptable to various circumstances in routine use. Robustness is important because biomedical data are noisy, and artifacts are mostly inevitable.
Parameterization means to choose meaningful values for all technical parameters of the data structures and the algorithms and, therefore, to configure such a method to operate on specific clinical data. Hence, it is usually performed by the programmer. Specificity and target-group orientation are important. The model has to offer variables that can later be filled with application-specific information. However, this essential knowledge can be obtained only from biomedical experts, and most programmers are not experts.
Instantiation means to run the algorithm with the chosen parameters to find the individual model instance that best explains the input image.10 Here, the automated and reproducible detection of the solution instance is important. Model-based approaches that do not rely on continuous user interaction are preferred.
Validation has to be applied to ensure a trustworthy result from an unsupervised segmentation algorithm even if a gold standard is unavailable. This requires reliable computational measurements rather than a qualitative inspection by a human observer.
Configuration of model-based segmentation is crucial. For the family of active contours, which is evolved from the classic snake,1 relevant parameters include the weighting of image influences against smoothness constraints and an optional driving pressure, the number and distance of vertices, and all parameters of the various edge filters applied to compute image influences. For level sets, the model to determine the interface velocity requires configuration.11 Parameters need to be selected properly to determine the interface velocity relative to local shape properties, the location in the image domain, and the extension speed dependent on image values. Active shapes12 and all further development of such shape knowledge–based models require configuration regarding the number of landmarks in each shape, the number of considered modes of variation seen in training, the weighting of shape knowledge against image influences, and the determination of image influences for each landmark.
In all these examples, computer scientists are regularly involved in the development of the model and its parameterization, whereas the resulting instances are validated and used mostly by physicians. In contrast, a combination of technical and medical a priori knowledge is required for both steps. This transfer bottleneck is reflected also in the literature. In the majority of published papers, only the formulation and the instantiation of models are presented, but both parameterization and validation are not considered in serious detail. In this paper, we suggest schemes that are applicable in general to guide the transfer of model-based image analysis into particularly supporting biomedical applications. These schemes can serve as a road map for the difficult and complex tasks of parameterization and validation.
Classification of Existing Methods
In medicine, a novel diagnostic procedure is validated against the true diagnosis, which is termed the gold standard. Based on a definition from Wenzel and Hintze,13 a true gold standard in image processing must be reliable, i.e., its generation must follow an exactly determined and reproducible protocol; equivalent, i.e., it must equal real-life data with respect to structure, noise, or other relevant parameters; and independent, i.e., it must rely on a different procedure or another image than that to be evaluated.14
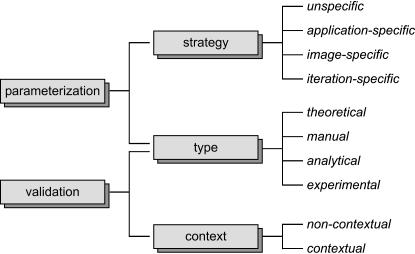
Such a gold standard is unavailable for many issues. Therefore, it is difficult to develop robust parameterization and appropriate validation schemes. These problems continuously plague medical image analysis research. ▶ gives an overview of the methods that have been suggested for parameterization and validation of model-based image analysis. The classification terms are adopted from Lehmann et al.9,15
Figure 1.
Categorization of methods for parameterization and validation applied to model-based image analysis.
Strategy of Parameterization
This class determines the point of time when the parameters for a model are set. Therefore, it addresses the flexibility of the model to operate on different image material.
Unspecific
Often the parameterization of a model is completed before it is transferred into a biomedical context. Such a strategic parameterization is characterized by a static choice of values for a parameter for all applications.
Application Specific
The parameterization is performed completely during the transfer of the model. It is the main step to adapt a model for the given task or to readjust it after the protocol of image acquisition was altered.
Image Specific
For several modalities, images can differ strongly in appearance and automated analysis requires an image-specific parameterization.16 Hence, at least some of the parameters are adapted individually to every image processed during application.
Iteration Specific
Even more crucial, parameters may be altered during the process of instantiation. For example, the optimal value of a certain parameter is made dependent on the number of iterations,17 properties of the model such as the size,18 or local image characteristics.19
Type of Parameterization and Validation
Beside the time of choice, we have observed and classified the sources from which the parameterization and validation originate (▶).
Table 1.
Nomenclature for the Type of Parameterization and the Type of Validation
| Parameterization/Validation Is Based on |
|||
|---|---|---|---|
| Type | Implementation | Quantitative Measure | Gold Standard |
| Theoretical | - | - | - |
| Manual | • | - | - |
| Analytical | • | • | - |
| Experimental | • | • | • |
Bullet or hyphen denotes used as input or not considered, respectively.
Theoretical
Especially in combination with unspecific parameterization, the theoretical approach is preferable. For this type, parameters are set only based on theoretical properties of the model. Regarding validation, neither experiments on synthetic or real data are performed nor is a gold standard applied. For instance, Haralick20 has proposed a theoretical scheme for the validation of image-processing algorithms. Note that in contrast to the terminology used in this paper, theoretical validation is also referred to as the analytical approach.21
Manual
In most applications, the parameters are adapted manually until model-based analysis for exemplary data conform with the subjective visual impression of an observer. Using this approach, even experienced users just guess the initial parameters and try to fight perceivable errors. Since reproducible rules for this type of parameterization do not exist, strong observer dependencies are obtained. Consequently, this method is called ad hoc22 or trial-and-error parameterization23 as well as empirical goodness.21 Although experiments are performed with an implemented model, neither a gold standard nor a quantitative measure is used for guidance of parameterization as well as validation.
Analytical
Similar to theoretical parameterization, the source of information is based on mathematical properties of an optimal parameterization but additionally includes exemplary observation data and computed measures. Analytical parameterization is based on the solution instance and additional properties of the input image. Since a gold standard is not used, it is also termed “parameter free.”24 Analytical validation is most often applied to pixel-based segmentation methods, but it has been also successfully used for the snake model.25,26 It can be improved if competing segmentation methods are available for comparison.27,28
Experimental
Experimental parameterization is based on the learning-from-examples paradigm.22 At least one gold standard image is needed. The paradigm states that if parameters are chosen such that the reference is reproduced in this example, the parameterization is also suitable to process similar images from the same series of acquisition. Experimental validation compares computational experiments to the gold standard. Note that in this taxonomy, manually created reference contours are not considered as a gold standard20 and consequently such strategies for validation are termed manual but not experimental.
Context of Validation
Regarding the published evaluation techniques, two different approaches have been proposed.15
Noncontextual
An algorithm is tested involving images with adjustable properties or systematic changes of parameters. Usually, they operate on synthetic images and quantify the quality of results. Noncontextual tests give reproducible and quantitative measures describing the behavior of a considered model under variable known image characteristics and changes in parameterization. Hence, this type of evaluation is directly related to the algorithm but not to its application in daily clinical use.
Contextual
The suitability of an algorithm is evaluated only for a certain application. Such validations rely on results of the target application. Usually, contextual tests directly apply a proposed model to an image analysis task from clinical research or routine. However, the lack of a gold standard often limits the experiments.
Proposition on Schemes for Parameterization
Although unspecific parameterization, i.e., one choice for all applications is desirable for model-based segmentation, it is unavailable for biomedical images because of the great variety in imaging modalities and object appearances due to inter- and/or intrasubject variabilities. Iteration-specific parameterization results in exhaustive computation and dramatically increases the complexity of automatic image analysis without the guarantee that it will improve a result. Therefore, we believe that application- or image-specific parameterizations are preferable strategies in biomedical imaging.
Furthermore, our viewpoint is that theoretical or analytical parameterization is not suitable for biomedical imaging informatics because the nature of biomedical data inhibits mathematical approaches in general. Manual parameterization is insufficiently reproducible. Technical experts are often unavailable when needed to adjust a model for a new set of images. Consequently, the necessity to combine technical knowledge in the model as well as medical knowledge in image content requires experimental schemes for parameterization.
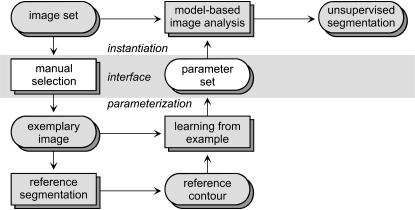
Application-specific Parameterization
▶ shows the proposed scheme for application-specific parameterization. Since the instantiation of the model cannot take place until technical parameters are set appropriately, the task of instantiation is connected to the parameterization by an interface (▶, shaded in gray). The input side of this interface is the choice of one image from the image set while the output side contains the required set of parameters. During parameterization, a manual segmentation is created by the physician. Since a manual reference comprises high inter- and intraobserver dependence, it is used only indirectly by the learning-from-examples paradigm.22 The resulting parameters are passed back to the instantiation via the interface and then allow unsupervised segmentation of all images in a homogeneous set. We refer the reader interested in more technical details to an exemplary implementation for a general-purpose active contour model.29
Figure 2.
Proposed scheme for automated application-specific parameterization using the learning-from-examples paradigm.
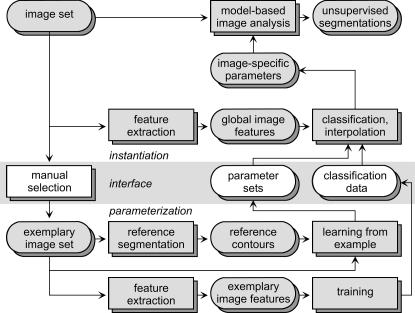
Image-specific Parameterization
Image-specific parameterization changes the parameters of the model according to the appearance of each actual image. Again, a set of images and a model for segmentation are given and model-based image analysis is applied to yield unsupervised segmentations (▶, upper row). For the interface, a set of exemplary images is chosen by the physicians incorporating task-specific knowledge. These images must represent the variety of the object's appearances. For each exemplary image, a reference segmentation is required, and a parameterization is created using the learning-from-examples paradigm. User interaction for reference segmentation is reduced substantially using computer-assisted methods such as live wires.30 Global image features are extracted automatically from the references (▶, bottom row). These features describe the overall appearance composed from all structures in the image. For our purposes, texture features that allow computation of a quantitative measure of similarity between images are used.31 During application, unsupervised segmentation is enabled by first extracting the global image features for every image. Then, the similarities of appearance between the current image and all exemplary prototypes are computed. The image-specific parameterization results from interpolation between the parameter sets of the prototypes16 or from just adopting the parameters of the nearest neighbor reference. An implementation of this scheme using a robust co-occurrence–based global texture classification with a respective interpolation of prototype parameter sets is given in Lehmann et al.16
Figure 3.
Proposed scheme for image-specific parameterization that is based on a priori extracted global image features.
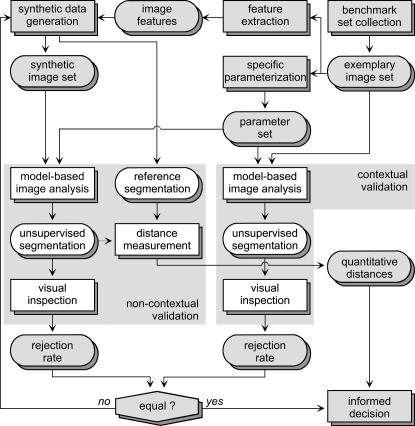
Proposition on Schemes for Validation
▶ shows the proposed scheme for validation. The basic idea is to support an informed decision of the physician about the reliability and quality of image processing applied to routine data. Therefore, noncontextual experiments are related to contextual validation schemes performed on data without a ground truth or gold standard. Based on the selected benchmark collection of real data without known references, features are extracted and used to generate a synthetic data set with a priori known reference. This set is designed to be reliable and independent. Model-based image analysis is applied to both data compilations and inspected visually, whereas the manual decision is limited to acceptance or rejection. The synthetic imagery is modified iteratively (e.g., by adding noise) until the number of rejected samples is equal in both data sets. This indicates reliably that the synthetic data are equivalent to the real-life data. In other words, the synthetic data now have similar properties as a gold standard. Note that applying the scheme of ▶, interaction is reduced to decide failure, but manual delineation is avoided. For the required creation of realistic synthetic data, we refer to a method based on Fourier decomposition and synthesis of textures for contours extracted from clinical data.32
Figure 4.
Proposed scheme for experimental validation of application-specific segmentation.
Demonstration Example
To exemplify the process of scheme-guided parameterization and validation, we use a balloon-based deformable model as “contestant.” We use a simplified planar version with fixed topology of a general discrete contour model for topology-adaptive multichannel segmentation in two, three, and four dimensions.33 This contestant is transferred to different applications.
Delineation of Cells in Micrographs
Suppose a system designed for the automated quantification of micrographs showing immunohistochemically stained motor neurons from the spinal cord of adult rats.34 In this system, a segmentation of the cell soma is needed to quantify structures located next to the cell membrane. In routine application, a series consists of several hundreds of similar micrographs, each showing one cell for quantification. This task is characterized by a basically reproducible imaging system and is used to demonstrate application-specific parameter sets. To check the scheme proposed in ▶, the following steps were performed:
To test the reproducibility, 12 observers created a manual segmentation for one example cell. Comparing all combinations resulted in an mean overlap of 96.6 ± 1.1% pixels and the cumulative overlap given by the quotient of the intersection of all 12 segmentations divided by their union was 90.1%. In other words, the observers agreed in the overall location of relevant structures in the image but showed disagreement in the precise delineation.
Then, parameter sets were computed automatically for each manual segmentation. Segmenting the input as well as three other cells from this series with the resulting different parameter sets, the automated segmentations showed a mean overlap of 97.3 ± 1.7%, 96.1 ± 1.9%, 97.6 ± 0.9%, and 98.3 ± 0.6% (overall mean 97.3%). Therefore, we conclude that an automated parameterization using the learning-from-examples paradigm results in reproducible segmentations and is even able to slightly decrease interobserver variabilities.
Following the validation scheme (▶), 200 synthetic images were created to obtain quantitative quality measures. These images are composed of an object with varying contours that copy real cell shapes, noisy image values with textures similar to real micrographs, and blurred object borders to ensure failures in the segmentation (▶).
The subjective visual validation identified three failures (98.5% accepted). For the accepted segmentations, a mean Cartesian distance from automated segmentation to the known true contours of dC = 1.67 ± 0.77 pixels was measured.
In the respective benchmark set of 681 cells, 27 segmentations were rejected in the subjective visual validation (96% accepted).
Figure 5.
To validate the model-based segmentation of immunohistochemically stained motor neurons (a), synthetic images that mimic the appearance of these micrographs have been created (b–e).
The acceptance rates are close enough so that secured quantitative information results from this validation: Applied to micrographs of motor neurons, the deformable balloon model in combination with application-specific parameterization gives accepted fully automated segmentations in 96% of all images processed with this stain. The average distance between cell border and segmentation result is then estimated to 1.67 pixels. The users participated in the acceptance and rejection of the benchmark set segmentations and were additionally provided with quantitative information regarding the system's quality and possible failures. In compliance with ▶, this enabled an “informed decision” to use the system in a study.34
Determination of Body Regions in Radiographs
Content-based image retrieval in medical applications (IRMA) is a challenging task of medical image analysis.35 Within the IRMA project (http://irma-project.org), automatic categorization of images with respect to the imaged body region is one of the first processing steps. Model-based segmentation is employed to delineate relevant structures in radiographs, and the outlines are used for classification. A total of 1,616 plain radiographs were arbitrarily selected from clinical records, differing in imaged body part and imaging direction. The imaging properties varied strongly. Therefore, this retrospective analysis is an example of image-specific parameter sets. To assess the impact of image-specific analysis and to check the scheme proposed in ▶, the following steps were performed:
Synthetic phantoms were generated from sinusoid curves. In addition to noise and unsharp object borders, the image intensity and contrast was varied.16 One, four, and eight different examples were used to set respective parameters for the model.
The results of unsupervised segmentation were compared considering the mean overlap and the number of accepted segmentations with O>85% (▶). Using eight prototypes, the mean overlap as well as the number of accepted images is about 90%. In comparison, only 33% of the segmentations are accepted if fixed parameterization is used.
In the benchmark set of radiographs, application- and image-specific parameterizations were used competitively. The segmentations were inspected visually by a trained radiologist. Applying the application-specific parameterization, only 496 (31%) results were accepted. With an image-specific parameterization based on 19 prototypes, the acceptance rate was increased to 71% (1,145).
Table 2.
Noncontextual Validation of Image-specific Parameterization
| Method | No. of Prototypes | Mean Pixel Overlap | No. of Acceptance (O>85%) |
|---|---|---|---|
| Application specific | 1 | 45.1 ± 38.5 | 27 (33.3%) |
| Image specific | 4 | 83.7 ± 22.7 | 64 (79.0%) |
| Image specific | 8 | 90.9 ± 9.3 | 72 (88.9%) |
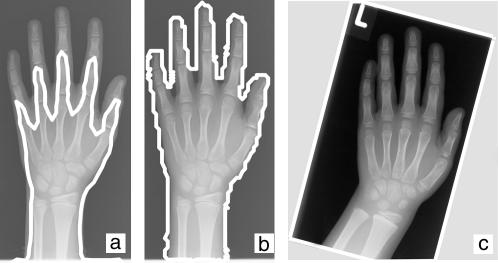
These figures correspond to the tests on synthetic images (▶). The potential of image-specific parameterization was reproduced for clinical data (▶a,b). However, even the acceptance rate for only four prototypes using synthetic data (79%) is still too high in comparison to the 71% that are obtained for the benchmark set of clinical images. The reasons for the high rejection rate in the benchmark set were identified. As a major problem, the segmentation is disturbed by collimator fields in the radiographs that were not manually cut after digitalization (▶c). These artifacts were not contained in the synthetic images. In consequence, further research and development activities were initiated to automatically detect collimators in digitized film radiographs.36 Based on the difference in rejection rates, the informed decision was made to delay the application and wait for the availability of the collimator detection.
Figure 6.
The image-adaptive configuration of previously unsuccessful segmentation (a) gives a robust contour in most radiographs of the test set (b). However, the system cannot cope with shutters in the radiographs (c), which were not considered in the configuration and transfer of this application.
As an important result for configuration strategies, this finding stresses the imperative that all variations and potential artifact sources that can appear in clinical data need to be considered during parameterization and validation of image analysis algorithms.
Discussion
Since a priori knowledge on image characteristics and object appearances is incorporated into each individual task, we argue that model-based image analysis is the preferred concept in biomedical image analysis. In contrast to the considerable improvements in the research field of image analysis,2,3,37 we do not observe a similar number of robust applications in the clinics. The stepwise transfer of the models from development to application has been identified as the main bottleneck. While generic schemes for the formulation of models already exist,9 parameterization and validation are still crucial. For instance, Ji and Yan38 have claimed that there are no straightforward rules for parameter settings of model-based segmentation and that most settings are obtained by trial and error, which makes a model vulnerable to inconsistencies assigned by users. Nonetheless, manual parameterization is principally proposed in the literature. As a result, model-based image analysis is restricted to images that are available for parameterization and validation during development and often fails if slightly altered images are acquired in clinical routine. In agreement, Chen and Sun39 have emphasized the impact of automated parameterization of routine applicability of model-based segmentation methods. This bottleneck is widened by the proposed schemes for application- and image-specific setting of model parameters, which are designed for heterogeneous image sets and applicable to different models for image analysis.
However, finding the optimal set of model parameters is not the only problem of biomedical imaging informatics. Similar to the information about the impacts and risks of a certain procedure, which the patient is given by the physician before he or she consents to treatment, the physicians routinely using model-based image analysis must be informed about the quality of automatic image processing. Hence, engineers who want to transfer an image analysis system into daily clinical use should act according to the suggestion of Feynman (Hutchings et al.40) of how to communicate scientific discovery: “The idea is to try to give all the information to help others to judge the value of your contribution; not just the information that leads to judgment in one particular direction or another.” In other words, careful evaluation of the parameterized model is required and must be communicated in detail to the physician.
Furthermore, the validation must address the combination of the model and its parameterization. To fulfill these constraints, a gold standard is required.15,20,32 However, in medical image processing, a gold standard frequently is unavailable8 as it either requires invasive surgery to implant secured markers or the extraction of the tissue shortly after imaging. Applying the hybrid scheme proposed in this paper, noncontextual tests allow the extraction of reproducible quantitative measures for the quality of model-based analysis methods, and these measures are reliably propagated to contextual tests, even without a gold standard. In agreement with Brown,41 our scheme avoids theoretical validation methods, and we agree with Haralick20 that manual validation is also inappropriate.
In biomedical image analysis, the users of model-based approaches are physicians rather than engineers, software developers, and image analysis scientists. Consequently, they do not have sufficient technical knowledge to adapt parameters. Using our schemes, user interaction is minimized. Even for image-specific parameterization, the remaining interaction is tolerable if a method for unsupervised segmentation does not otherwise exist but a large set of images must be analyzed. As a result, clinical workflow integration of image analysis is massively supported. Once the parameterization and evaluation is performed according to the schemes suggested, a site-specific adaptation of the image-processing algorithm is obtained, taking implicitly into account the site-specific rules for image acquisition. Specialized to the site, image analysis then operates automatically.
Although segmentation was selected as an example of a task of biomedical image analysis, the proposed schemes are more generally applicable. By the nature of applied sciences such as medical informatics, general strategies are obtained inductively but cannot be derived deductively. Consequently, the usefulness of the schemes proposed in this viewpoint paper is not “proven” by the two examples given. Nonetheless, they demonstrate that the concepts for automatic parameterization and validation are tenable. Also, they emphasize how to adopt these schemes to establish other tasks of model-based image analysis in a routine setting. Therefore, they may contribute to establish biomedical imaging informatics in health care.
References
- 1.Kass M, Witkin A, Terzopoulos D. Snakes: active contour models. Int J Comput Vision. 1987;1:321–31. [Google Scholar]
- 2.Jain AK, Zhong Y, Dubuisson-Jolly MP. Deformable template models: a review. Signal Process. 1998;71:109–29. [Google Scholar]
- 3.McInerney T, Terzopoulos D. Deformable models in medical image analysis: a survey. Med Image Anal. 1996;1:91–108. [DOI] [PubMed] [Google Scholar]
- 4.Sennst DA, Kachelriess M, Leidecker C, Schmidt B, Watzke O, Kalender WA. An extensible software-based platform for reconstruction and evaluation of CT images. Radiographics. 2004;24:601–13. [DOI] [PubMed] [Google Scholar]
- 5.Astley SM, Gilbert FJ. Review: computer-aided detection in mammography. Clin Radiol. 2004;59:390–9. [DOI] [PubMed] [Google Scholar]
- 6.Duncan JS, Ayache N. Medical image analysis: progress over two decades and the challenges ahead. IEEE Trans Pattern Anal Machine Intell. 2000;22:85–106. [Google Scholar]
- 7.Lehmann TM, Meinzer HP, Tolxdorff T. Advances in biomedical image analysis: past, present and future challenges. Methods Inf Med. 2004;43:308–14. [PubMed] [Google Scholar]
- 8.Haynor DR. Performance evaluation of image processing algorithms in medicine: a clinical perspective. Procs SPIE. 2000;3979:18. [Google Scholar]
- 9.Lehmann TM, Bredno J, Spitzer K. On the design of active contours for medical image processing: a scheme for classification and construction. Methods Inf Med. 2003;43:89–98. [PubMed] [Google Scholar]
- 10.Undrill PE, Delibasis K, Cameron GG. An application of genetic algorithms to geometric model-guided interpretation of brain anatomy. Pattern Recogn. 1997;30:217–27. [Google Scholar]
- 11.Sethian JA. Level set methods and fast marching methods: evolving interfaces in computational geometry, fluid mechanics, computer vision, and materials science. Cambridge monographs on applied and computational mathematics. Cambridge, UK: Cambridge University Press, 1999.
- 12.Cootes TF, Taylor CJ, Cooper DH, Graham J. Active shape models: their training and application. Image Vision Comput. 1995;61:38–59. [Google Scholar]
- 13.Wenzel A, Hintze H. The choice of gold standard for evaluation tests for caries diagnosis. Dentomaxillofac Radio. 1999;28:132–6. [DOI] [PubMed] [Google Scholar]
- 14.Lehmann TM. From plastic to gold: a unified classification scheme for reference standards in medical image processing. Procs SPIE. 2002;4684:1819–27. [Google Scholar]
- 15.Nguyen TB, Ziou D. Contextual and non-contextual performance evaluation of edge detectors. Pattern Recogn Lett. 2000;21:805–16. [Google Scholar]
- 16.Lehmann TM, Bredno J, Spitzer K. Texture-adaptive active contour models. Lect Notes Comput Sci. 2001;2013:387–96. [Google Scholar]
- 17.Wang J, Li X. A system for segmenting ultrasound images. Proc ICAPR. 1998:456–61.
- 18.Garrido A, De La Blanca NP. Physically-based active shape models: initialization and optimization. Pattern Recogn. 1998;31:1003–17. [Google Scholar]
- 19.Ivins J, Porrill J. Constrained active region models for fast tracking in color image sequences. Comput Vision Image Understand. 1998;72:54–71. [Google Scholar]
- 20.Haralick RM. Validating image processing algorithms. Proc SPIE. 2000;3979:2–27. [Google Scholar]
- 21.Zhang YJ. A survey on evaluation methods for image segmentation. Pattern Recogn. 1996;29:1335–46. [Google Scholar]
- 22.Cagnoni S, Dobrzeniecki AB, Poli R, Yanch JC. Genetic algorithm-based interactive segmentation of 3D medical images. Image Vision Comput. 1999;17:881–95. [Google Scholar]
- 23.Gang X, Segawa E, Tsuji S. Robust active contours with insensitive parameters. Pattern Recogn. 1994;27:879–84. [Google Scholar]
- 24.Borsotti M, Campadelli P, Schettini R. Quantitative evaluation of color image segmentation results. Pattern Recogn Lett. 1998;19:741–7. [Google Scholar]
- 25.Gennert MA, Yuille AL. Determining the optimal weights in multiple objective function optimization. Proc 2nd Int Conf ComputVision. 1988:87–9.
- 26.Lai KF, Chin RT. Deformable contours: modeling and extraction. IEEE Trans Pattern Anal Machine Intell. 1995;17:1084–90. [Google Scholar]
- 27.Henkelman RM, Kay I, Bronskill MJ. Receiver operator characteristic (ROC) analysis without truth. Med Decis Making. 1990;10:24–9. [DOI] [PubMed] [Google Scholar]
- 28.Beiden SV, Champbell G, Meier KL, Wagner RF. On the problem of ROC analysis without truth: the EM algorithm and the information matrix. Proc SPIE. 2000;3981:126–34. [Google Scholar]
- 29.Bredno J, Lehmann TM, Spitzer K. Automatic parameter setting for balloon models. Proc SPIE. 2000;3979:1185–94. [Google Scholar]
- 30.Falcao AX, Udupa JK, Samarasekera S, Sharma S, Hirsch BE, De A, et al. User-steered image segmentation paradigms: live wire and live lane. Graphical Models Image Process. 1998;60:233–60. [Google Scholar]
- 31.Randen T, Husøy JH. Filtering for texture classification: a comparative study. IEEE Trans Pattern Anal Machine Intell. 1999;21:291–310. [Google Scholar]
- 32.Lehmann TM, Bredno J, Spitzer K. Silver standards obtained from Fourier-based texture synthesis to evaluate segmentation procedures. Proc SPIE. 2001;4322:214–25. [Google Scholar]
- 33.Bredno J, Lehmann TM, Spitzer K. A general discrete contour model in 2, 3, and 4 dimensions for topology-adaptive multi-channel segmentation. IEEE Trans Pattern Anal Machine Intell. 2003;25:550–63. [Google Scholar]
- 34.Lehmann TM, Bredno J, Metzler V, Brook G, Nacimiento W. Computer-assisted quantification of axo-somatic boutons at the cell membrane of motoneurons. IEEE Trans Biomed Eng. 2001;48:706–17. [DOI] [PubMed] [Google Scholar]
- 35.Lehmann TM, Güld MO, Thies C, Fischer B, Spitzer K, Keysers D, et al. Content-based image retrieval in medical applications. Methods Inf Med. 2004;43:354–61. [PubMed] [Google Scholar]
- 36.Lehmann TM, Goudarzi S, Linnenbrügger NI, Keysers D, Wein BB. Automatic localization and delineation of collimation fields in digital and film-based radiographs. Proc SPIE. 2002;4684:1215–23. [Google Scholar]
- 37.Xu C, Pham DL, Prince JL. Image segmentation using deformable models. In: Sonka M, Fitzpatrick JM, editors. Handbook of Medical Imaging, Vol. 2. Bellingham, WA: SPIE Press, 2000.
- 38.Ji L, Yan H. Robust topology-adaptive snakes for image segmentation. Image Vision Comput. 2002;20:147–64. [Google Scholar]
- 39.Chen DH, Sun YN. A self-learning contour finding algorithm for echocardiac analysis. Proc SPIE. 1998;3338:971–81. [Google Scholar]
- 40.Hutchings E, Leighton R, Feynman RP, Hibbs A, editors. Surely you're joking, Mr. Feynman!: adventures of a curious character. New York: WW Norton, 1997.
- 41.Brown CW. Building a medical image processing algorithm verification database. Proc SPIE. 2000;3979:772–80. [Google Scholar]