Abstract
Objective: To help biomedical researchers recognize dynamically introduced abbreviations in biomedical literature, such as gene and protein names, we have constructed a support system called ALICE (Abbreviation LIfter using Corpus-based Extraction). ALICE aims to extract all types of abbreviations with their expansions from a target paper on the fly.
Methods: ALICE extracts an abbreviation and its expansion from the literature by using heuristic pattern-matching rules. This system consists of three phases and potentially identifies valid 320 abbreviation-expansion patterns as combinations of the rules.
Results: It achieved 95% recall and 97% precision on randomly selected titles and abstracts from the MEDLINE database.
Conclusion: ALICE extracted abbreviations and their expansions from the literature efficiently. The subtly compiled heuristics enabled it to extract abbreviations with high recall without significantly reducing precision. ALICE does not only facilitate recognition of an undefined abbreviation in a paper by constructing an abbreviation database or dictionary, but also makes biomedical literature retrieval more accurate. This system is freely available at http://uvdb3.hgc.jp/ALICE/ALICE_index.html.
It is essential for biomedical researchers to obtain knowledge from the MEDLINE database. However, numerous abbreviations such as gene and protein names, which are routinely used throughout the biomedical literature, hinder its efficient use. Abbreviations in the biomedical literature are highly ambiguous: one abbreviation may represent multiple expansions.1,2,3,4,5 For example, Liu et al.2 point out that 81.2% of abbreviations are ambiguous and had an average of 16.6 meanings. One typical example is the abbreviation PC. It may stand for personal computer, primary care, principal component, prostate cancer, etc. To make matters worse, the increasing number of biomedical papers in the MEDLINE database continues to incorporate new abbreviations into it.1,4 A support system is urgently needed to help researchers recognize the expansions of abbreviations.1,4,5,6,7,8
Here, we define abbreviations as “contractions of words or phrases that are used in place of their full versions” (we call these full versions expansions) and acronyms as “a type of abbreviations made up of the initial letters or syllables of other words.”9
Much effort has been expended to develop methods for extracting abbreviations and their expansions. For example, some algorithms use parentheses “()” to limit search criteria, while others use both parentheses and cue words such as “or” or “stands for.”6,10 Almost all algorithms use heuristic patterns to identify abbreviations and acronyms. To extract expansions, some algorithms use manually constructed heuristic pattern-matching rules, while others use automatically constructed statistical rules.1,3 Some heuristic algorithms use shallow parsing.7,11
Although some of these algorithms show good results, they have various limitations. For example, when identifying an abbreviation, some algorithms assume that an abbreviation consists only of one word or that it must be enclosed in parentheses. Supposing that the abbreviation is “AMI,” which stands for “acute myocardial infarction,” these algorithms can extract its expansion if the original expression is “acute myocardial infarction (AMI).” However, they cannot extract the expansion of the abbreviation if the original expression is “AMI (acute myocardial infarction)”. Moreover, when extracting an expansion, some algorithms assume that the initial letter of an expansion must be the same as that of its abbreviation. This means that these algorithms cannot extract the expansion of the abbreviation “AW,” for example, which stands for “water activity.”
Some researchers have noted that rarely occurring abbreviation types (or minor abbreviation types) such as those in the above examples can be safely ignored1,3,4 because minor abbreviation types have almost no impact on performance of an abbreviation extraction system, an abbreviation database, or an abbreviation dictionary. Furthermore, some abbreviations of minor types would possibly be extracted with major types. However, the existing algorithms are insufficient in meeting our goal to filter papers retrieved with a PubMed search.
We have been constructing a system to eliminate irrelevant papers for a query gene from PubMed search results. It is called PETER (PubMed Enhancer Toward Efficient Research). We found that when searching for biomedical literature in the MEDLINE database with a PubMed search, researchers are often bothered by the ambiguity of abbreviations, especially those of gene and protein names. To solve this problem, PETER needs an algorithm that can extract all types of abbreviations with their expansions from a target paper on the fly.
In this paper, we describe an algorithm called ALICE (Abbreviation LIfter using Corpus-based Extraction). It searches for parentheses and identifies and extracts pairs of abbreviations and their expansions by using heuristic pattern-matching rules. It uses the same strategy used by Yu et al.5 and Schwartz and Hearst.8 However, our algorithm uses additional manually expanded patterns, rules, and stop word lists, which are based on thorough investigation and heuristics. ALICE can potentially identify valid 320 abbreviation-expansion patterns as combinations of the rules. They include types that the previous algorithms do not cover; that is, our system overcame the above-mentioned limitations. As a result, ALICE achieved 95% recall and 97% precision on randomly selected titles and abstracts from the MEDLINE database. It indicates that it does not limit the scope of target literature to a specific biomedical research field for better performance. This system can help users construct not only a useful abbreviation database or dictionary, but also a system to retrieve papers from the MEDLINE database such as the PETER system. An abbreviation database or dictionary based on biomedical literature would help biomedical researchers recognize undefined abbreviations in a paper.
Background
Larkey et al.10 described an ad hoc algorithm called Acrophile to extract acronyms from Web pages. Their approach is based on the use of parentheses, cue words, and ad hoc rules. They tested four different extraction algorithms: Contextual, Canonical/Contextual, Canonical, and Simple Canonical. These algorithms differ from one another in terms of the types of acronyms, forms of expansions, and text patterns of acronym-expansion pairs they can identify. The four algorithms use different clues (e.g., parenthetical expressions, cue words such as “stands for” or “or”) to identify acronym-expansion pairs. Acrophile is one of a few systems that can identify acronyms introduced without parentheses. In addition, the Contextual algorithm pays special attention to digits. For example, if an acronym contains “3M” or “3D,” these are replaced with “MMM” or “three dimensional.” Because this system was constructed for Web pages, the performance of the system is not good for biomedical text, based on our preliminary experiment. It cannot extract pairs such as “14C-urea breath test (14C-UBT),” “granule membrane protein-140 (GMP-140),” “fibrinogen (Fg),” or “protein kinase C (PKC).”
Chang et al.1 used a supervised machine-learning algorithm to extract abbreviations and their expansions from MEDLINE abstracts. Their approach is based on the use of parentheses and the resemblance to a training set of human-annotated abbreviations. They assumed that an abbreviation was enclosed in parentheses. After scanning a text to find a candidate abbreviation inside parentheses, the system aligns the candidate with the words before the left parenthesis to match as many letters as possible in the two strings. Then, it converts the candidate abbreviation and its optimal alignments from the aligned words into a feature vector. Next, it applies a binary logistic regression classifier to generate a score from the feature vector. The algorithm had a maximum recall of 83% at 80% precision. The drawback is that an abbreviation must be defined within parentheses.
Wren and Garner4 developed a set of heuristics called Acronym Resolving General Heuristics (ARGH) to identify “acronym-definition pairs” in the MEDLINE database. To our knowledge, it is very similar to our approach; however, we could not fully compare their algorithm with ours because they evaluated ARGH with various rule sets (e.g., “term consists of one word only” and/or “require first letter match on abbreviation-type acronyms”), and none of the sets were the same as ours. They used systematic rates of precision and recall (refer to databases entries) and per-identification-event rates of precision and recall (refer to query texts). Although they mentioned that the systematic recall of the algorithm was around 93.0% and its systematic precision was around 96.5%, those are not per-identification-event rates that we used, and the heuristics for valid pairs are very limited as mentioned above.
Liu and Friedman3 proposed an algorithm based on the use of parentheses and statistical rules to extract a set of related terms from the biomedical literature. The system can extract not only abbreviations associated with their corresponding expansions, but also other semantically related terms such as synonyms, hyponyms, etc. This system is one of the systems that can identify synonymous terms besides abbreviations. First, it collects all parenthetical expressions from a large collection of texts. Next, it detects all outer-text strings that share the same inner-text. Then, it derives and assesses a set of pair-wise terms with frequency information. Finally, it separates these terms into a set of abbreviations and their expansions and a set of other related terms. The drawback is that it is not suitable for identifying expansions that occur only once in a text. The recall of the algorithm was around 88.5%, and its precision was 96.3%.
Schwartz and Hearst8 reported a simple algorithm based on the use of parentheses and ad hoc rules for identifying abbreviation definitions in biomedical texts. It extracts short-form, long-form pair candidates from a text and then it identifies the correct long-form among the candidates. Their system has more restrictions on the identifiable abbreviation types than ours. For example, correct short-forms must consist of at most two words and their length must be two to ten characters; correct long-forms must be adjacent to the short-form (i.e., they do not allow for an offset word6 between the short-form and long-forms) and include every letter of the short one, etc. They emphasized that their system was highly effective and less specific than other approaches that used carefully crafted rules for biomedical texts, and, above all, it was extremely simple. The algorithm had a recall of 82% and a precision of 96%. We consider their assumption is insufficient in covering those pairs appearing in the biomedical literature.
Methods
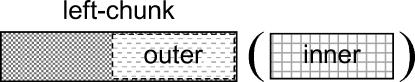
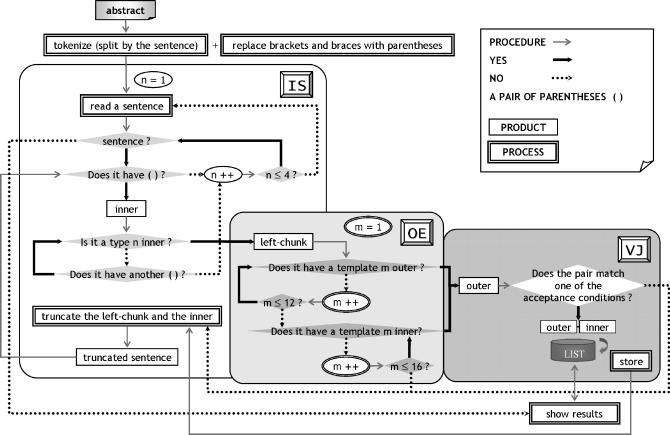
ALICE consists of three phases: Inner Search, Outer Extraction, and Validity Judgment. In this paper, we define a string inside a pair of parentheses as an inner, a string before the left parenthesis as a left-chunk, and an extracted string from a left-chunk as an outer. An inner is not necessarily the whole string inside the pair of parentheses (see the Inner Search phase, type 4 inner), and an outer may be identical to the left-chunk (see ▶). If an inner is an abbreviation, the outer is its expansion. Inversely, if an inner is an expansion, the outer is its abbreviation. For example, in the sentence “We used activating transcription factor 3 (ATF3) expression as a neuronal injury marker,” the inner is “ATF3,” the left-chunk is “We used activating transcription factor 3,” and the outer is “activating transcription factor 3.”
Figure 1.
Definitions of special expressions used in this paper. An inner is a string inside a pair of parentheses, a left-chunk is a string before the left parenthesis, and an outer is a string extracted from the left-chunk and is the correspondent of the inner as a pair of an abbreviation and its expansion.
In the Inner Search phase, ALICE searches for a pair of parentheses and identifies an inner. Once the inner is identified, the left-chunk is also determined. Then, its outer is extracted from the left-chunk in the Outer Extraction phase. Finally, in the Validity Judgment phase, the validity of the set of the inner and the outer as an abbreviation-expansion pair is judged. An overview of ALICE is shown in ▶, and this procedure along with examples is shown in detail in Appendixes 1 through 6.
Figure 2.
ALICE overview. In the Inner Search (IS) phase, ALICE searches for a pair of parentheses and identifies an inner. Once the inner is identified, its left-chunk is also determined. Then, its outer is extracted from the left-chunk in the Outer Extraction (OE) phase. Finally, in the Validity Judgment (VJ) phase, the validity of the set of the inner and its outer as an abbreviation-expansion pair is judged. If an inner is an abbreviation, the outer is its expansion. Inversely, if an inner is an expansion, the outer is its abbreviation.
Stop Word Lists
To adapt ALICE to the biomedical literature effectively, we manually crafted five stop word lists: (1) a list of inners, (2) a list of inner front words, (3) a list of inner first words, (4) a list of outers, and (5) a list of outer first words. Each location where those lists are applied is shown in ▶. The former three lists are used in the Inner Search phase, and the latter two are used in the Outer Extraction phase. These lists were results of our careful observation of false-positive errors during the construction. Some examples of the stop word lists are as follows: (1) “OH,” p+digits* (e.g., “p27”), CD+digits (e.g., “CD8”), etc. (stop words for inners); (2) “poly,” “oligo,” hyphen, etc. (inner front words); (3) preposition, wh adverb, etc. (inner first words); (4) “protein,” “cell,” etc. (outers); and (5) preposition, wh adverb, etc. (outer first words).
Figure 3.
Definitions of special expressions used for stop word lists. An inner front word is the word preceding a left parenthesis, an inner first word is the first inner word, and an outer first word is the first outer word. This figure shows an example that an outer and an inner consist of three and two words, respectively.
Safe Term List
In contrast to the stop word lists, we prepared a safe term list to cope with special abbreviations in the biomedical domain such as “in vitro,” “in vivo,” etc. Expansions beginning with a preposition can be extracted only if the first prepositional phrase is in the safe term list (see the Outer Extraction phase).
Preprocess
Before ALICE begins extraction, the title and the abstract of a paper are tokenized and split into sentences by using our tool called JASMINE (Just A Sentence-splitter Maximizing Intelligence of kNowledge Extraction) (JASMINE is freely available from http://uvdb3.hgc.jp/ALICE/program_download.html). It is based on the assumption that an abbreviation is defined within the same sentence. Then ALICE receives a sentence as an input and replaces all brackets “[]” and braces “{ }” with parentheses “().”
The Inner Search Phase
A pair of parentheses is a trigger for ALICE to go into the Inner Search phase. After it locates parentheses, ALICE checks whether the inner is a candidate abbreviation or expansion. The rules for the check consist of nine conditions for discard (discard conditions) and four conditions for acceptance (acceptance conditions). The discard conditions were constructed for the following two purposes: (1) to see whether the inner, the inner front word, and the first inner word are included in their corresponding stop word lists, respectively, and (2) to check word categories (e.g., Is the inner composed only of digits? or Does the inner include a be-verb?) and the length of the words or the string in the inner (e.g., Is the inner composed only of one character? or Is the inner composed of more than five words?). The acceptance conditions were constructed based on the presence of a space before the left parenthesis and the characters of the inner words, as shown in ▶.
Table 1.
Acceptance Conditions for an Inner
| Type/Example | Target Appearance Pattern | Inner | The Initial Character of the First Inner Word | The Last Character of the Last Inner Word |
|---|---|---|---|---|
| 1 neutrophil peptide-1 (NP-1) | Outer(inner) | Alphanumeric characters, hyphens, spaces, under-bars, periods, primes, or commas | An alphanumeric | An alphanumeric or a period |
| 2 polyvinyl pyrrolidone (PVP) | Outer_(inner) | As above | As above | As above |
| 3 Secure Multipurpose Internet Mail Extensions (S/MIME) | Outer(inner) | Uppercase letters, digits, or slashes | An uppercase letter or a digit | An uppercase letter or a digit |
| Outer_(inner) | ||||
| 4 Liebowitz Social Anxiety Scale (LSAS; Liebowitz, 1987)‡ | Outer(inner; **) | A colon or a semicolon,† AND uppercase letters, digits, or hyphens | As above | As above |
| Outer(inner: **) | ||||
| Outer_(inner; **) | ||||
| Outer_(inner: **) |
‘_’: a space, ‘**’: any characters.
The inner is defined as a string before the colon or the semicolon.
In this case, only LSAS is the inner.
If the inner matches none of the discard and one of the acceptance conditions, the left-chunk is evaluated in the Outer Extraction phase. Otherwise, the left-chunk and the inner including the parentheses are truncated from the target sentence, and ALICE searches for another pair of parentheses in it. The accepted inner is called a type n inner, where n denotes the acceptance condition number (1 ≤ n ≤ 4). ALICE searches for inners of each type in a target sentence separately, and this means that each sentence is scanned four times.
The Outer Extraction Phase
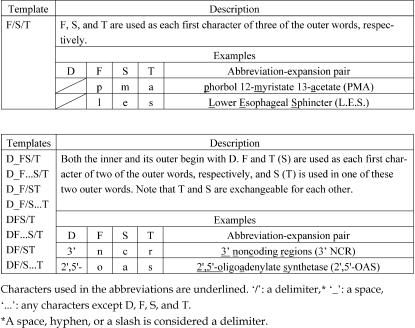
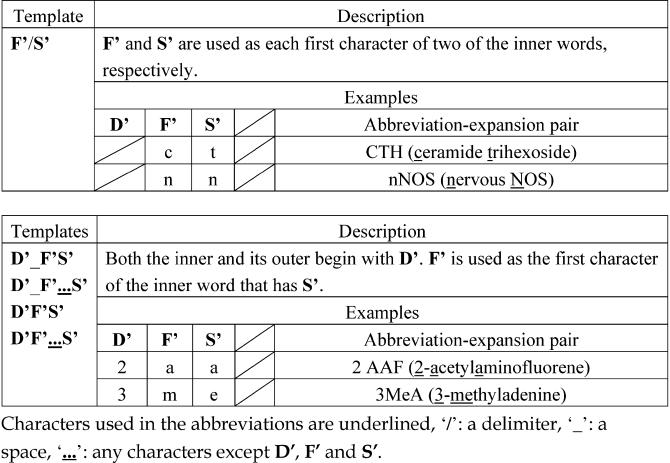
In the Outer Extraction phase, five discard conditions and 16 templates are used to extract an outer from its left-chunk. The discard conditions were constructed for the following two purposes: (1) to see whether the outer and the first outer word are included in their corresponding stop word lists, respectively, and (2) to check the length of the string in the outer (e.g., Is the outer composed of more than ten words?). Even if the first outer word is in its stop word list, it can be accepted if the first prepositional phrase is in the safe term list. The templates were constructed based on how abbreviations are formed from their expansions. Twelve of the templates (templates 1 through 12) are for an outer and to be used for extraction of those abbreviation-expansion pairs in which the parenthesized abbreviation follows its expansion (▶). On the other hand, the remaining templates (templates 13 through 16) are for an inner and to be used for extraction of those pairs in which the parenthesized expansion follows its abbreviation (▶).
Table 2.
Representative Templates for Outers and Examples of Abbreviation-Expansion Pairs
Table 3.
Representative Templates for Inners and Examples of Abbreviation-Expansion Pairs
All the templates are represented as sequence patterns of symbols that denote characters of abbreviations or delimiters of words that compose expansions. As for the templates 1 thorough 12, D matches the initial characters of an inner where the first one is a digit and the following are any characters except alphabetic characters; D is null if the initial character of an inner is not a digit. It can be expressed as the following regular expression: |^\d[^a-zA-Z]*^. F, S, and T match the first, the second, and the third characters of an inner. F must be alphabetical, and S and T must be alphanumeric. Characters after the third are ignored.
As for the other templates, D' matches the initial characters of an outer in the same manner as those of an inner (D). F' and S' match the first and the second characters of an outer. It should be noted that ALICE identifies only one word just before the left parenthesis as an outer in the templates 13 through 16. Although it is not always true that an abbreviation consists of only one word, we observed that almost all abbreviations preceding their expansions did. Some representative templates are shown in ▶▶.
If the outer matches none of the discard conditions and one of the templates, the inner-outer pair is evaluated in the Validity Judgment phase. Otherwise, the left-chunk and the inner including the parentheses are truncated from the target sentence, and ALICE returns to the Inner Search phase to search for another pair of parentheses in the sentence. The accepted outer is called a template m outer, where m denotes the matched template number (1 ≤ m ≤ 16).
The Validity Judgment Phase
In the Validity Judgment phase, 14 discard and five acceptance conditions are used to judge the validity of a set of an inner and its outer through 11 steps. Each step consists of one or multiple discard or acceptance conditions. If a pair meets the step k conditions, it is judged by the conditions; otherwise, it is to be judged at the step k + 1 (1 ≤ k ≤ 11). When the pair matches none of the discard and one of the acceptance conditions, the set is judged to be valid and stored in a valid pair list. Regardless of the pair's validity, the left-chunk and the inner including the parentheses are truncated from the target sentence, and ALICE returns to the Inner Search phase to search for another pair of parentheses in the sentence.
The ALICE System
As shown in ▶, ALICE searches for a type n inner (1 ≤ n ≤ 4) in a target sentence in the Inner Search phase. The initial n is 1 (n = 1). If the system finds a type 1 inner, it checks if the left-chunk includes a template m outer (1 ≤ m ≤ 16) in the Outer Extraction phase. The system applies the templates in the order from 1 to 16 until finding a template m outer. It then judges the validity of the extracted set of the inner and the outer in the Validity Judgment phase with the five acceptance conditions. Accordingly, ALICE has 320 abbreviation-expansion pattern combinations for the validity judgement (4 Inner Search × 16 Outer Extraction × 5 Validity Judgment patterns). If the pair is judged to be valid, it is stored for output. After finishing the evaluation of the inner, the system truncates the left-chunk and the inner including the parentheses from the target sentence and searches for another type 1 inner in it. After completing the search for all type 1 inners in the sentence, the system searches for all type 2 inners in it. Type 3 and type 4 inners are searched for in the same manner. If there is no sentence to be processed, ALICE outputs the stored results.
Figure 4.
ALICE flowchart. ALICE scans each sentence four times in the Inner Search (IS) phase (1 ≤ n ≤ 4), and it checks each string before the left parenthesis up to 16 times in the Outer Extraction (OE) phase (1 ≤ m ≤ 16). The extracted inner-outer pair is evaluated with the five acceptance conditions in the Validity Judgment (VJ) phase.
Results
ALICE was tested on a corpus of 1,000 abstracts with titles that were randomly selected from the MEDLINE database (PMID: 12500000–12599999). We call it ALICE Corpus. ALICE Corpus was manually tagged with pairs of abbreviations and their expansions by three biologists. The first author is one of them and organized the corpus construction. The others are independent of the ALICE system and helped the construction. There were 1,095 tagged abbreviation-expansion pairs, and ALICE identified 1,070 pairs. Among them, 1,039 pairs were correct. Thus, the recall was 95% and the precision was 97% (▶). We define recall and precision as follows:
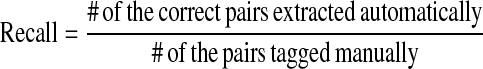 |
 |
Table 4.
Results of the ALICE Evaluation on ALICE Corpus
It should be noted that not all parenthetical structures are involved in abbreviations. There were 4,573 parenthetical expressions in ALICE Corpus, and 1,070 pairs were finally extracted from it (▶). This means that ALICE extracted approximately 23% (1,070/4,573) of all the parenthetical expressions in the corpus, i.e., if a system identifies all the expressions as abbreviations, more than 75% of them would be invalid. In the Inner Search phase, approximately 53% (2,433/4,573) of all the parenthetical expressions in the corpus were extracted as inners (262 type 1, 2,117 type 2, 36 type 3, and 18 type 4). In the Outer Extraction phase, 1,257 inners or 52% (1,257/2,433) of all the inners extracted in the Inner Search phase were paired up with their outers. Then, 1,070 sets of the inner-outer or approximately 85% (1,070/1,257) of all the pairs extracted in the Outer Extraction phase were identified as valid abbreviation-expansion pairs in the Validity Judgment phase. About 15% of all the sets of the inner-outer identified in the Outer Extraction phase were discarded in the Validity Judgment phase.
Table 5.
The Number of the Extracted Candidates in Each Phase
Next, we compared ALICE with three downloadable algorithms or seven conditions: the Larkey et al.10 algorithms (obtained from http://ciir.cs.umass.edu/irdemo/acronym/getacros.html) of (i) Canonical, (ii) Canonical/Contextual, and (iii) Simple Canonical; the Chang et al.1 algorithm (obtained from http://bionlp.stanford.edu/webservices.html) at three score cutoffs (the three score cutoffs are labeled in ▶ of their paper) of (iv) 0.88, (v) 0.14, and (vi) 0.03; and the Schwartz and Hearst8 algorithm (obtained from http://biotext.berkeley.edu/software.html). When we tested the seven conditions using ALICE Corpus, the recalls and the precisions were as follows: (i) 13% and 70%, (ii) 13% and 68%, (iii) 34% and 67%, (iv) 45% and 96%, (v) 88% and 91%, (vi) 90% and 86%, and (vii) 89% and 93%, respectively (▶). To investigate the accuracy of ALICE using existing gold standard, we then used the original DEVELOPMENT (▶) and EVALUATION (▶) corpora of Medstract Gold Standards (obtained from http://scylla.cs.brandeis.edu/gold-standards.html). Because of the errors in the original ones, the previously reported results were not comparable.1,7,8 Accordingly, we prepared modified standards (▶▶). The modifications we did were (1) elimination of all the synonyms (e.g., “alpha-Tocopherol (vitamin E)” and “estradiol-17 beta (E2)”), (2) revision of several abbreviation-expansion pairs (e.g., “cAMP-dependent protein kinase A (PKA)” was replaced with “protein kinase A (PKA)”), and (3) addition of some abbreviations that should have been included in the original version (e.g., “primary ethylene response element (PERE)”). The corpora used are available from http://uvdb3.hgc.jp/ALICE/corpus_download.html.
Table 6.
Results Obtained Using ALICE Corpus
| Algorithm | Recall (%) | Precision (%) | F-measure |
|---|---|---|---|
| Larkey et al. (Canonical) | 13 | 70 | 22 |
| Larkey et al. (Canonical/Contextual) | 13 | 68 | 22 |
| Larkey et al. (Simple Canonical) | 34 | 67 | 45 |
| Chang et al. (score = 0.88) | 45 | 96 | 61 |
| Chang et al. (score = 0.14) | 88 | 91 | 89 |
| Chang et al. (score = 0.03) | 90 | 86 | 88 |
| Schwartz and Hearst | 89 | 93 | 91 |
| ALICE | 95 | 97 | 96 |
Table 7.
Results Obtained Using the Original DEVELOPMENT Corpus
| Algorithm | Recall (%) | Precision (%) | F-measure |
|---|---|---|---|
| Larkey et al. (Canonical) | 3 | 5 | 4 |
| Larkey et al. (Canonical/Contextual) | 3 | 5 | 4 |
| Larkey et al. (Simple Canonical) | 3 | 6 | 4 |
| Chang et al. (score = 0.88) | 38 | 62 | 47 |
| Chang et al. (score = 0.14) | 57 | 55 | 56 |
| Chang et al. (score = 0.03) | 60 | 54 | 57 |
| Schwartz and Hearst | 61 | 58 | 59 |
| ALICE | 63 | 62 | 62 |
Table 8.
Results Obtained Using the Original EVALUATION Corpus
| Algorithm | Recall (%) | Precision (%) | F-measure |
|---|---|---|---|
| Larkey et al. (Canonical) | 1 | 2 | 1 |
| Larkey et al. (Canonical/Contextual) | 1 | 2 | 1 |
| Larkey et al. (Simple Canonical) | 1 | 2 | 1 |
| Chang et al. (score = 0.88) | 21 | 88 | 34 |
| Chang et al. (score = 0.14) | 60 | 73 | 66 |
| Chang et al. (score = 0.03) | 65 | 71 | 68 |
| Schwartz and Hearst | 64 | 76 | 69 |
| ALICE | 63 | 75 | 68 |
Table 9.
Results Obtained Using the Modified DEVELOPMENT Corpus
| Algorithm | Recall (%) | Precision (%) | F-measure |
|---|---|---|---|
| Larkey et al. (Canonical) | 4 | 6 | 5 |
| Larkey et al. (Canonical/Contextual) | 4 | 6 | 5 |
| Larkey et al. (Simple Canonical) | 4 | 7 | 5 |
| Chang et al. (score = 0.88) | 54 | 92 | 68 |
| Chang et al. (score = 0.14) | 77 | 78 | 77 |
| Chang et al. (score = 0.03) | 80 | 76 | 78 |
| Schwartz and Hearst | 82 | 81 | 81 |
| ALICE | 85 | 87 | 86 |
Table 10.
Results Obtained Using the Modified EVALUATION Corpus
| Algorithm | Recall (%) | Precision (%) | F-measure |
|---|---|---|---|
| Larkey et al. (Canonical) | 1 | 2 | 1 |
| Larkey et al. (Canonical/Contextual) | 1 | 2 | 1 |
| Larkey et al. (Simple Canonical) | 1 | 2 | 1 |
| Chang et al. (score = 0.88) | 25 | 93 | 39 |
| Chang et al. (score = 0.14) | 75 | 80 | 77 |
| Chang et al. (score = 0.03) | 80 | 78 | 79 |
| Schwartz and Hearst | 80 | 83 | 81 |
| ALICE | 82 | 86 | 84 |
Discussion
ALICE has been developed to extract pairs of abbreviations and their expansions from MEDLINE titles and abstracts. Our system overcame several limitations that existing systems have; that is, (1) an abbreviation must consist of only one word, (2) it must include at least one uppercase letter, (3) it must be inside parentheses, (4) its first character must be the same as that of the expansion, (5) every character in it must be used within the expansion in the same order, (6) an expansion must not contain special characters such as (), [], etc., and (7) there must be a space just before a left parenthesis. Consequently, our algorithm can extract such pairs as “oestrogen receptor (ER),” “rt-PA-APSAC patency study (TAPS),” “brain Po2 (Pbro2),” etc. Furthermore, separate searches for type 1 and type 2 inners enable it to extract expansions containing parentheses (e.g., “(S)-2-amino-3-(3-hydroxy-5-methyl-4-isoxazole)propionic acid (AMPA)”).
The system developed by Schwartz and Hearst8 shows the same performance as ALICE concerning Medstract Gold Standards (▶, ▶, ▶, and ▶); however, the difference between theirs and ours becomes clear when the systems are run on a large corpus (▶). Recall and precision obtained using small data sets do not always reflect those using large data sets.4 Our heuristic algorithm based on the large amount of literature in MEDLINE (approximately 1,500 papers apart from ALICE Corpus) can extract abbreviations with high recall without significantly reducing precision for any size of data sets. We believe that in order to develop a high-performance system, we need specially crafted rules for the target domain such as biomedical papers or newspaper articles. ALICE achieved further accuracy by using the five stop word lists and the safe term list.
One of the limitations of ALICE is that it cannot make a clear distinction between synonyms (e.g., “authentic PPAR/RXR binding element (Aco-PPRE)” and “3-nitrotyrosine (3-NO2-Tyr)”) and expansions (e.g., “N2 partial pressure (PN2)” and “skin temperature (Tsk)”). Some long forms are quite difficult even for experts to judge whether they are synonyms or expansions. Accordingly, we regarded a long form as an expansion if all the characters in its short form were included in the long form. However, there are some exceptions in ALICE Corpus (e.g., “percutaneous septal ablation (PTSMA)” and “congenital stationary night blindness (CSNBX)”) and ALICE cannot extract those examples. Another limitation is that it is impossible to retrieve expansions divided by enumeration (e.g., in the string “topoisomerase I (topo I) or II (topo II),” only “topoisomerase I (topo I)” can be identified). In addition, when a sentence has words between an abbreviation and its expansion (offset words6), ALICE cannot remove them (e.g., in “Orthopedic Advisory Committee of the World Federation of Hemophilia (OAC),” the words “of the World Federation of Hemophilia” are offset words). These problems must be solved.
Conclusion
We have developed an algorithm called ALICE that extracts abbreviations and their expansions from the biomedical literature by using heuristic pattern-matching rules. Our exhaustive study of abbreviations enables ALICE to address 320 pattern combinations for extracting valid abbreviation-expansion pairs. As a result, it can extract abbreviations with high recall without significantly reducing precision; it achieved 95% recall and 97% precision on randomly selected titles and abstracts from the MEDLINE database. ALICE helps construct not only a useful abbreviation database or dictionary, but also a system to retrieve papers from the MEDLINE database. We have been constructing a system called PETER to select relevant papers from PubMed search results containing a large number of irrelevant papers. ALICE is one of the components of PETER.
Appendix 1. Discard conditions for inners
(1) The word just before the left parenthesis is in the inner front word list (e.g., “poly,” “oligo,” hyphen).
(2) Both the initial character of the first inner word and the last character of the last inner word are digits.
(3) The first word of the inner is in the inner first word list (e.g., preposition, wh-adverb).
(4) The inner consists of only one word in the inner list (e.g., “OH,” p+digits (e.g., “p27”), CD+digits (e.g., “CD8”)).
(5) The inner includes a be-verb.
(6) The inner consists of one character.
(7) The inner consists of more than five words.
Appendix 2. Acceptance conditions for inners
- i) There is no space just before the left parenthesis; and,
- ii) a) The inner consists of alphanumeric characters, hyphens, spaces, under-bars, periods, primes, or commas;
- b) the initial character of the first inner word is an alphanumeric character; and,
- c) the last character of the last inner word is an alphanumeric character or a period.
- e.g., neutrophil peptide-1 (NP-1)
- i) There is a space just before the left parenthesis; and,
- ii) The components of the inner are the same as those of type 1.
- e.g., polyvinyl pyrrolidone (PVP)
- i) The inner consists of uppercase letters, digits, or slashes; and,
- ii) Both the initial character of the first inner word and the last character of the last inner word are uppercase letters or digits.
- e.g., Secure Multipurpose Internet Mail Extensions (S/MIME)
- i) One of the characters inside the parentheses is a colon or a semicolon, and the inner is defined as a string before it; and,
- ii) a) The inner consists of uppercase letters, digits, or hyphens; and,
- b) Both the initial character of the first inner word and the last character of the last inner word are uppercase letters or digits.
- e.g., Liebowitz Social Anxiety Scale (LSAS; Liebowitz, 1987).
Appendix 3. Discard conditions for outers
(a) The first word of the outer is in the outer first word list (e.g., preposition, wh-adverb).
(b) The outer consists of only one word in the outer list (e.g., “protein,” “cell”).
(c) The outer is more than ten words.
(d) The outer is more than five words when extracted with templates 3, 4, 5, 6, 8, 9, and 10, and the inner contains a space.
(e) The outer is more than five words when extracted with template 12.
Appendix 4. Templates for and examples of inner-outer pairs
- Legends
- /: a delimiter
- _: a space
- |: or
- pp: a preposition
- al: alphabetic characters
- …: any characters except D, F, S, and T
- …: any characters except D', F', and S'
- 1) D_F/S/T | DF/S/T‡
- Both the inner and its outer begin with D. F, S, and T are used as each first character of three of the outer words, respectively.
- e.g., 3′ long terminal repeat (3′ LTR)
- 2-h plasma glucose (2hPG)
- 2) F/S/T
- F, S, and T are used as each first character of three of the outer words, respectively.
- e.g., phorbol 12-myristate 13-acetate (PMA)
- Lower Esophageal Sphincter (L.E.S.)
- 3) D_FS/T | D_F…S/T | D_F/ST | D_F/S…T | DFS/T | DF…S/T | DF/ST | DF/S…T‡
- Both the inner and its outer begin with D. F and T (S) are used as each first character of two of the outer words, respectively, and S (T) is used in one of these two outer words. Note that T and S are exchangeable for each other.
- e.g., 3' noncoding regions (3′ NCR)
- 2',5′-oligoadenylate synthetase (2',5′-OAS)
- 4) FS/T | F…S/T | F/ST | F/S…T
- F and T (S) are used as each first character of two of the outer words, respectively, and S (T) is used in one of these two outer words. Note that T and S are exchangeable for each other.
- e.g., sulfoquinovosyl diacylglycerol (SQDG)
- butylated hydroxyanisol (BHA)
- 5) D_FST | DFST
- Both the inner and its outer begin with D. F, S, and T are used as the first three characters in the inner word in this order.
- e.g., 11 beta-hydroxysteroid dehydrogenase (11 beta-HSD)
- 6) FST
- F, S, and T are used as the first three characters in the inner word in this order.
- e.g., IGF-binding protein-3 (IGFBP-3)
- 7) F/pp/S/T | F/S/pp/T‡
- F, S, and T are used as each first character of three of the outer words, respectively, and there is a preposition between F and S (or S and T).
- e.g., National Institutes of Health Stroke Scale (NIHSS)
- 8) F/S | F/T
- F and S (or F and T) are used as each first character of two of the outer words, respectively.
- e.g., cytochrome oxidase (CO)
- 9) F…ST | FS…T | F…S…T
- F is used as the first character of the outer word that has S and T.
- e.g., trifluoroacetic (TFA)
- 4-methylaminoantipyrine (MAA)
- 10) FS | F…S | FT | F…T
- F is used as the first character of the outer word that has S or T.
- e.g., hydroxypyrrole (Hp)
- 11) alF/S/T‡
- Alphabetic characters except F, S, and T are used as the first characters of the outer word that has F. S and T are used as each first character of two of the outer words, respectively. Note that the outer does not begin with F.
- e.g., rt-PA-APSAC patency study (TAPS)
- 12) S/F
- F and S are used as each first character of two of the outer words, respectively. Note that this template is different from template 8 (F/S) since the first outer word does not begin with F but with S.
- e.g., compensatory temperature (TC)
- 13) D'_F'/S' | D'F'/S' (inner)
- Both the inner and its outer begin with D'. F' and S' are used as each first character of two of the inner words, respectively.
- e.g., 5-HIAA (5-hydroxy-indole-acetic acid)
- 14) F'/S' (inner)
- F' and S' are used as each first character of two of the inner words, respectively.
- e.g., CTH (ceramide trihexoside)
- nNOS (nervous NOS)
- 15) D'_F'S' | D'_F'…S' | D'F'S' | D'F'…S' (inner)
- Both the inner and its outer begin with D'. F' is used as the first character of the inner word that has S'.
- e.g., 2 AAF (2-acetylaminofluorene)
- 3MeA (3-methyladenine)
- 16) F'S' | F'…S' (inner)
- F' is used as the first character of the inner word that has S'.
- e.g., NLO (nonlinear optical)
Appendix 5. Steps in evaluating the validity of abbreviation-expansion pairs
[1] When the length of the outer is the same as that of the inner. >> discard
- [2] When the length of the outer is longer than that of the inner,
- - if the outer has neither S nor T. >> discard
- [3] When the length of the inner is longer than that of the outer,
- - if the outer includes a lowercase letter. >> discard
- - if the outer consists of uppercase letters only and the inner includes a word that matches a predefined pattern (not shown). >> discard
[4] When all the alphabetic characters in the short-form are not included in the long-form. >> discard
- [5] When the outer or the inner has a parenthesis,
- - if the inner does not have the same number of left parentheses as the right ones. >> discard
- - if the outer does not have the same number of left parentheses as the right ones. >> discard
- - if the inner begins with a right parenthesis or ends with a left one. >> discard
- - if the outer begins with a right parenthesis or ends with a left one. >> discard
- [6] When the inner is type 3,
- - if the outer includes a digit. >> discard
- [7] When the inner includes digits,
- - if the outer includes the same digits. >>
- - otherwise. >> discard
- [8] When the outer includes digits,
- - if the digits match predefined patterns (not shown). >>

- - otherwise. >> discard
- [9] When the inner matches a predefined pattern (not shown),
- - if the outer includes a word that matches a predefined pattern (not shown). >>

- - otherwise. >> discard
- [10] When the inner consists of lowercase letters only,
- - if all the characters in the inner are included in the outer. >>

- - otherwise. >> discard
[11] When the inner does not consist of lowercase letters only. >>

Appendix 6. An example of the extraction process
Figure 5.
This work was partly supported by a grant from the Grant-in-Aid for Scientific Research in Priority Areas Genome Information Science, Japanese Ministry of Education, Culture, Sports, and Technology. The authors thank the staff of the Department of Computational Biology, University of Tokyo, and the staff of the Basic Research Laboratory, Kanebo Cosmetics, Inc., for their contribution to this study. The authors are also grateful to Yasunori Yamamoto of the Department of Computer Science, University of Tokyo, for editing the manuscript.
Footnotes
In this paper, the term digits refers to one or more digits, and the following terms are used similarly: alphanumeric characters, alphabetic characters, hyphens, spaces, under-bars, periods, primes, commas, uppercase letters, and slashes.
These templates allow T to be null.
References
- 1.Chang JT, Schutze H, Altman RB. Creating an online dictionary of abbreviations from MEDLINE. J Am Med Inform Assoc. 2002;9:612–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Liu H, Aronson AR, Friedman C. A study of abbreviations in MEDLINE abstracts. Proc AMIA Symp. 2002:464–8. [PMC free article] [PubMed]
- 3.Liu H, Friedman C. Mining terminological knowledge in language biomedical corpora. Pacific Symposium on Biocomputing. 2003:415–26. [PubMed]
- 4.Wren JD, Garner HR. Heuristics for identification of acronym-definition patterns within text: towards an automatic construction of comprehensive acronym-definition dictionaries. Methods Inf Med. 2002;41:426–34. [PubMed] [Google Scholar]
- 5.Yu H, Hripcsak G, Friedman C. Mapping abbreviations to full forms in biomedical articles. J Am Med Inform Assoc. 2002;9:262–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Park Y, Byrd RJ. Hybrid text mining for finding abbreviations and their definitions. Conference on Empirical Methods of Natural Language Processing, 2001;126–33.
- 7.Pustejovsky J, Castano J, Cochran B, Kotecki M, Morrell M. Automatic extraction of acronym-meaning pairs from MEDLINE databases. Medinfo. 2001;10:371–5. [PubMed] [Google Scholar]
- 8.Schwartz AS, Hearst MA. A simple algorithm for identifying abbreviation definitions in biomedical text. Pacific Symposium on Biocomputing. 2003:451–62. [PubMed]
- 9.Yeates S. Automatic extraction of acronyms from text. New Zealand Computer Science Research Students' Conference, 1999;117–24.
- 10.Larkey LS, Ogilvie P, Price MA, Tamilio B. Acrophile: an automated acronym extractor and server. Proceedings of the Fifth ACM International Conference on Digital Libraries 2000, 2000;205–14.
- 11.Yoshida M, Fukuda K, Takagi T. PNAD-CSS: a workbench for constructing a protein name abbreviation dictionary. Bioinformatics. 2000;16:169–75. [DOI] [PubMed] [Google Scholar]