Abstract
A rapid and reliable method for the identification of five clinically relevant G genotypes (G1 to G4 and G9) of human rotaviruses based on oligonucleotide microarray hybridization has been developed. The genotype-specific oligonucleotides immobilized on the surface of glass slides were selected to bind to the multiple target regions within the VP7 gene that are highly conserved among individual rotavirus genotypes. Rotavirus cDNA was amplified in a PCR with primers common to all group A rotaviruses. A second round of nested PCR amplification was performed in the presence of indodicarbocyanine-dCTP and another pair of degenerate primers also broadly specific for all genotypes. The use of one primer containing 5′-biotin allowed us to prepare fluorescently labeled single-stranded hybridization probe by binding of another strand to magnetic beads. The identification of rotavirus genotype was based on hybridization with several individual genotype-specific oligonucleotides. This approach combines the high sensitivity of PCR with the selectivity of DNA-DNA hybridization. The specificity of oligonucleotide microchip hybridization was evaluated by testing 20 coded rotavirus isolates from different geographic areas for which genotypes were previously determined by conventional methods. Analysis of the coded specimens showed that this microarray-based method is capable of unambiguous identification of all rotavirus strains. Because of the presence of random mutations, each individual virus isolate produced a unique hybridization pattern capable of distinguishing different isolates of the same genotype and, therefore, subgenotype differentiation. This strain information indicates one of several advantages that microarray technology has over conventional PCR techniques.
Rotaviruses are the most important etiological agents of severe diarrhea in infants and young children in both developed and developing countries (15). The rotavirus outer capsid proteins VP4 and VP7 represent neutralization antigens that are the basis for the classification of rotaviruses by P and G serotypes, respectively. VP4- and VP7-specific antibodies provide serotype-specific protection against rotavirus diarrhea (14). Accurate and rapid serological identification is important for the diagnosis of rotavirus infections, host range determination, and global epidemiological surveillance. It is also an important part of molecular epidemiology and can be used for tracing lines of viral transmission, monitoring molecular evolution, identifying new strains, and determining genotype distribution in clinical trials of experimental vaccines. The current classification of rotaviruses is based on neutralization assays with polyclonal or monoclonal antibodies (13). At least 14 G serotypes (VP7 protein) and 13 (not including subtypes) P serotypes (VP4 protein) have been identified by this approach (7, 15).
Nucleotide sequence analysis of the VP7 and VP4 genes allows rotaviruses to be classified into a number of distinct genotypes. The G genotypes have been shown to consistently correlate with the VP7 serotypes (25). The situation is more complicated for P genotypes because genotyping by sequencing has been carried out more extensively than serotyping by neutralization, and therefore correlation between the two methods is incomplete.
A number of assays based on PCR have previously been developed and used for the identification of nucleotide sequences common to different P and G genotypes. The genotypes were identified by a difference in the molecular weights of the DNA fragments synthesized in the presence of genotype-specific primers and separated by gel electrophoresis (8, 9). The sensitivity of PCR amplification tends to be inversely related to its specificity, and as a result, DNA generated in a highly sensitive PCR assay often contains nonspecific products, making it difficult to interpret the resulting band pattern. In addition, genotype-specific PCR primers occasionally fail to amplify specific DNA from a particular isolate if there is a spontaneous mutation(s) in the primer-binding site. However, it may be possible to improve PCR-based genotyping by replacing the gel electrophoresis analysis of the PCR products with DNA-DNA hybridization. The DNA-DNA hybridization enables unambiguous detection of target sequences regardless of the possible presence of nonspecific DNA products and allows the use of PCR primers with broader specificity, permitting a more sensitive and robust amplification of a wider range of organisms.
The hybridization of DNA samples with miniature arrays (microchips) of immobilized gene-specific DNA or oligonucleotide probes has recently become a powerful tool for the quantitative study of gene expression (21, 22), DNA resequencing (11, 17), phylogenetic classification of bacteria (10), mapping of genes (4), and analysis of single-nucleotide polymorphisms (12, 19). This format permits the simultaneous monitoring and analysis of a large number of genetic features in one easy hybridization experiment (20). While cDNA microarrays are suitable for gene expression analysis and identification of known genes and organisms, microchips with short (8 to 30 nucleotides) synthetic oligonucleotides are more sensitive to small genetic differences, such as single nucleotide substitutions. Therefore, they are more appropriate for use in discriminating between microorganisms with minor genetic variations.
There are a number of different ways to make oligonucleotide microchips, including in situ synthesis of oligonucleotides directly on the microchip surface (16) and immobilization of presynthesized oligonucleotides. The former approach produces high-density microarrays but is expensive and requires highly sophisticated industrial equipment. The latter approach can use different methods for immobilization of presynthesized oligonucleotides on a solid support, such as modified or nonmodified glass surfaces (2), gold (24), a gel-covered surface (18), or synthetic membranes (1). All of these methods have both advantages and disadvantages and are currently used in different protocols.
In general, it is not uncommon for viruses of the same species or even serotype to differ by more than 30% at the nucleotide level, making their identification by whole-DNA hybridization inefficient. However, the degree of evolutionary conservation varies widely among different parts of viral genomes and within individual genes. This creates the opportunity for identifying highly conserved regions suitable for efficient and broadly specific PCR amplification of DNA from all members of a group and for finding regions unique for members of a particular genotype of interest. Thus, the use of short genotype-specific oligonucleotides appears to be a promising approach for classifying viruses with variable degrees of relatedness.
In this communication we describe a new approach to G genotyping of human rotaviruses based on hybridization with oligonucleotide microarrays. This includes finding appropriate regions within the VP7 gene that contain multiple sequence targets conserved among viruses of a particular genotype. These regions from the human rotaviruses were easily amplified by PCR with broadly specific primers and labeled with fluorescent dye. We developed and optimized protocols for the fabrication and use of microarrays with aminolink oligonucleotides immobilized on an aldehyde-coated glass surface. Finally, we present the results of a comparison of our new technique with conventional genotyping by PCR amplification.
MATERIALS AND METHODS
Rotavirus strains.
Rotavirus strains grown and plaque-purified three times in MA104 monkey kidney cells were used as the source of genomic RNA. Cells were frozen and thawed three times to maximize virus yield.
RNA extraction and RT-PCR.
Rotavirus double-stranded RNA was extracted from lysates of infected MA104 cells by using Trizol-LS reagent (Life Technologies, Rockville, Md.) according to the manufacturer's protocol with the following modifications to increase the yield of viral RNA. The extraction of double-stranded RNA was repeated twice with Trizol-LS, and the aqueous phases from the first and second extractions were combined. After isopropanol precipitation, the RNA was collected by centrifugation, dissolved, and reprecipitated with 75% ethanol at −20°C overnight in the presence of 0.3 M sodium acetate. RNA was collected by centrifugation, resuspended in RNase-free water, and used for reverse transcription (RT)-PCR.
Full-length cDNA copies of the rotavirus VP7 gene were synthesized by RT-PCR using primers Beg9 and End9 as described previously (9) with the following modifications. The samples were subjected to one cycle of reverse transcription (42°C, 45 min) and 30 cycles of PCR. Each PCR cycle included 1 min at 94°C, 2 min at 50°C, and 3 min at 72°C.
Design of PCR primers and oligonucleotide probes.
More than 150 sequences of the VP7 glycoprotein gene of human rotaviruses were obtained from GenBank and aligned with the ClustalX program (26). This database included only human rotaviruses of all known G genotypes (14). Regions of the rotavirus VP7 gene that were conserved among all the viruses were used to design primers for nested PCR amplification and synthesis of indodicarbocyanine (Cy5)-labeled single-stranded DNA samples for microchip hybridization. Other regions of the VP7 gene, conserved only among individual rotavirus genotypes, were used to design genotype-specific oligonucleotide probes. These probes were then immobilized on a modified glass surface to create the microchip.
The complete list of oligonucleotide primers, probes, and quality control oligonucleotides (see below) is shown in Table 1. Oligonucleotide probes were selected by using the following criteria: lengths of about 20 nucleotides, melting temperatures between 65 and 75°C (calculated by using the nearest-neighbor algorithm) (3), and two or more mismatches with homologous sequences in other genotypes. In some cases, mixtures of nucleotides or inosine were used to cover all genetic variations within the genotype. During the automated oligonucleotide synthesis, the 5′ end of each probe was modified by adding an aminolink group (TFA Aminolink CE reagent; PE Applied Biosystems) to enable covalent immobilizing on the aldehyde-coated glass surface. The synthesized aminolink oligonucleotides were purified by extraction with an equal volume of chloroform and precipitated with 10 volumes of 2% lithium chlorate in acetone (6). This additional purification was used to remove traces of chemicals that could interfere with oligonucleotide immobilization.
TABLE 1.
List of primers and G genotype-specific oligonucleotide probes
| Type and name | Sequencea | Positions | Tm (°C) |
|---|---|---|---|
| Primers for PCR | |||
| LID1 | ATGTATGGTATTGAATATACCA | 48-70 | 66 |
| G-373 | TTAAARTARACIGAHCCHGTIGGCCAICCYTT | 373-404 | 78-82 |
| G-529 | ATATCCATIGGRTTRCAIAICCAYTCRTT | 529-557 | 73-79 |
| G-922 | GTRTARAAIACTTGCCACCA | 922-941 | 66-67 |
| QC oligonucleotides | |||
| Anti-LID1 | TGGTATATTCAATACCATACAT | 48-70 | 66 |
| QCprb | TTGGCAGAAGCTATGAAACGATATGGG | 82 | |
| Cy3-QC | CCCATATCGTTTCATAGCTTCTGCCA | 81 | |
| Random | d(N)21 | ||
| Genotype-specific oligonucleotide probes | |||
| G1-1 | TGATATCARAIAGATTAGAATT | 72-93 | 63-64 |
| G1-2 | ATATAGTTGAGTAGAATRATTG | 92-114 | 64-65 |
| G1-3 | ATRTAGTTGAGTAAAATAACTG | 92-114 | 65 |
| G1-4 | CCATTATTCGRGTCACTGATT | 119-139 | 69-72 |
| G1-5 | CCATTATTCGRGTTACTGATT | 119-139 | 66-70 |
| G1-6 | GTYAAGGCRAATAATGCTAC | 162-191 | 65-70 |
| G1-7 | TTCTGAGCTCTTGTYAAGGC | 184-203 | 72-73 |
| G1-8 | TTCTGAGCTTTAGTYAAGGC | 184-203 | 69-71 |
| G1-9 | AGTACTTGCTTCAGTTGG | 304-321 | 68 |
| G1-10 | ATTTGAGTACTTGCTTCAGT | 308-326 | 69 |
| G2-1 | ARATATCAAAATGGTCAGAATT | 72-93 | 67 |
| G2-2 | GTCCATYGTATTAGTTATAGTTT | 119-141 | 67 |
| G2-3 | CTYACAAATGGTGAYATCAGA | 172-194 | 67-68 |
| G2-4 | AATTTGTRTATACAGCGTCTA | 136-156 | 68 |
| G2-5 | AATTCGTATATRCAGCGTCTA | 136-156 | 69-72 |
| G2-6 | TAGAAATGIYTCTCCACTAGT | 259-279 | 68-70 |
| G2-7 | TCATTTTTAGCTTCTGYTGGATA | 301-323 | 73-74 |
| G2-8 | AATTGTGATAGAGTATTTTCC | 342-362 | 66 |
| G2-9 | ATGTAGTAATRTCATTGTAGT | 407-428 | 63-64 |
| G3-1 | TCAAAAAGGTTAAAACTGTGG | 68-88 | 69 |
| G3-2 | TCAGAAAGGTYAGAACTGTGG | 68-88 | 71-72 |
| G3-3 | CTAGTTAAIGATTTGAGYAYGTA | 109-131 | 67-72 |
| G3-4 | CATTATTCTASGTTAAIGATTTGAG | 115-137 | 68-70 |
| G3-5 | GCATTAAGGARTGGTGACA | 179-197 | 68-69 |
| G3-6 | TTTIGTGCATTAAGGARTGG | 184-203 | 67-68 |
| G3-7 | TTCTGTGCATTAAGGARTGG | 184-203 | 70-71 |
| G3-8 | TTGATCCAGTAATCGGAAG | 217-235 | 69 |
| G3-9 | TTGCAGTGTAGCGTCGTAYT | 476-495 | 75 |
| G4-1 | TATCAARTAAAATAGAACTGTG | 59-90 | 65-66 |
| G4-2 | TAACTCACAAGAACGAACGATATCAA | 85-111 | 77 |
| G4-3 | GTTTTYARAATRTAACTCACAAG | 100-122 | 64-67 |
| G4-4 | ACTACAATYAYAAATGTTATTCTATA | 151-175 | 66-67 |
| G4-5 | CAACAATTATAAATGTTACTCT | 153-174 | 65 |
| G4-6 | GCATTCGATAATACTGATAATAC | 175-197 | 69 |
| G4-7 | GCATTAGACAGTACTGATAATA | 165-197 | 68 |
| G4-8 | GTAATTGGCAAATTTATTCCATA | 205-227 | 68 |
| G4-9 | TCACTRATTTGAGTTGGAGCYTC | 310-332 | 73-76 |
| G9-1 | TTATAAAGTCCATYGCACTAG | 128-150 | 69 |
| G9-2 | TCCATGGAGCCAGTGATC | 222-239 | 71 |
| G9-3 | TTCTTGCTGTGATGAATTTG | 251-270 | 69 |
| G9-4 | GTATCTAGCTGTGATGAGTTTG | 251-272 | 72 |
| G9-5 | ATCTCCAATTTGAGTTGATGC | 313-333 | 71 |
| G9-6 | TTCCATTCYGTRTCYCCAATTTGA | 321-344 | 70-74 |
| G9-7 | CATTCYGTATCTCCAATTTGA | 321-341 | 68-70 |
| G9-8 | TGAAGCGATRTCRGTGTATTC | 406-426 | 69-74 |
| G9-9 | TGAAGCGATGTCAGTATATTC | 406-426 | 71 |
| G9-10 | TRTCTAGCTCTAACGTTGAAT | 482-502 | 70-71 |
Nucleotide abbreviations are according to the IUPAC system of nomenclature: R, A+G; Y, C+T; H, A+C+T; S, C+G; N, A+C+G+T; I, inosine.
Synthesis of Cy5-labeled samples of rotavirus cDNA.
Fluorescently labeled samples for hybridization were generated by nested PCR with primers complementary to highly conserved target regions of the VP7 gene (see Table 1). Briefly, 25 μl of a reaction mixture containing 1× AmpliTaq PCR buffer with 1.5 mM MgCl2, 300 nM each primer, 20 μM Cy5-dCTP, 20 μM dCTP, 100 μM each dATP, dGTP, and dTTP, 1 U of AmpliTaq polymerase (Applied Biosystems), and 0.1 μl of PCR product from the first round of RT-PCR (see above) was subjected to 35 cycles of PCR. Each PCR cycle included 30 s at 94°C, 30 s at 50°C, and 60 s at 72°C. The resulting double-stranded DNA containing one biotinylated chain was immobilized on streptavidin-coated GenoPrep magnetic beads (GenoVision Inc.) according to the manufacturer's protocol. The beads were washed twice with 1× TE (Tris-EDTA), and the single-stranded fluorescent probe was eluted by 50 μl of 0.1 M NaOH at room temperature. The fluorescent probe was desalted by spin centrifugation through CentriSep columns (Princeton Separations) and concentrated by either ethanol precipitation or by vacuum drying. Finally, the fluorescent probe was resuspended in a small volume (5 μl) of water; the probe concentration was measured with a UV spectrophotometer (Ultrospec 3100pro; Amersham Pharmacia Biotech, Inc.).
Synthesis of Cy3-QC oligonucleotide.
The indocarbocyanine (Cy3)-labeled quality control (Cy3-QC) oligonucleotide was prepared by coupling the 5′-aminolink QC oligonucleotide with Cy3 monofunctional dye using a FluoroLink Cy3 dye kit (Amersham Pharmacia Biotech, Inc.) according to the manufacturer's protocol. The labeled product was separated from unincorporated fluorescent reagent by gel filtration through a Sephadex G-25 column in water. The efficiency of dye incorporation was determined by the UV-visible spectrum in the 220- to 700-nm range.
Microchip fabrication.
Microchips were printed on silylated (aldehyde-coated) glass slides (3 in. by 1 in. [ca. 8 cm by 2.5 cm]; Cell Associate, Inc.) using a contact microspotting robot (Cartesian Technologies, Inc.) and a ChipMaker microspotting device with a single CMP-7 pin delivering approximately 2 to 3 nl of a spotting mixture per spot (TeleChem International, Inc.). To control the spotting procedure, each oligonucleotide probe was spiked with QCprb oligonucleotide of nonviral origin (see Table 1). The final spotting mixture contained 50 μM genotype-specific oligonucleotide probe and 10 μM QCprb in 0.25 M acetic acid.
A range of 70 to 75% humidity was maintained in an environmental chamber to prevent rapid evaporation of spotted drops and efficient coupling of amino-modified oligonucleotides with the aldehyde groups on the glass surface. Ten identical rotavirus arrays were printed on each slide, allowing us to simultaneously analyze 10 rotavirus samples on one glass slide. The size of spots did not exceed 250 μm in diameter, and the distance between individual spots in arrays was 400 μm. To prevent rapid evaporation of the oligonucleotide solution at lower humidity (40 to 60%), the spotting procedure can be performed in 50% dimethyl sulfoxide-water solution instead of acetic acid. Printed slides were dried for 20 min at 75°C. To make the bonds between oligonucleotides and the glass surface irreversible, the slides were incubated for 5 min in a freshly prepared 0.25% aqueous solution of NaBH4, followed by washing once with a 0.2% aqueous solution of sodium dodecyl sulfate (SDS) for 1 min and twice with distilled water (1 min) to remove unbound oligonucleotides. Microchips prepared according to this protocol can be stored for 2 months at room temperature.
Microarray hybridization.
Immediately before hybridization, single-stranded Cy5-labeled rotavirus sample and Cy3-QC oligonucleotide were mixed with an equal volume of 2× hybridization buffer, containing 12× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate), 10× Denhardt's solution, and 0.2% Tween 20 and denatured for 1 min at 95°C, followed by chilling on ice for 1 min. The final concentration of each fluorescent probe in the hybridization solution was usually above 0.1 μM. This concentration ensures rapid and efficient hybridization of a fluorescent sample with immobilized probes. Aliquots (2 to 3 μl) from each sample were applied to the microarray area and covered with a plastic coverslip (4 by 7 mm) to prevent evaporation of the sample during hybridization in the incubation chamber (ArrayIt). Hybridization was performed for 30 min at 45°C. Slides were then washed once with 6× SSC containing 0.2% Tween 20 for 1 min at room temperature, once with 6× SSC for 1 min, and once with 2× SSC and dried by an air stream to completely remove any remaining solution.
Microchip scanning and data analysis.
Microchip images were taken using a confocal fluorescent scanner ScanArray 5000 (GSI Lumonics) with green and red HeNe lasers (543 nm for excitation of Cy3 and 632 nm for Cy5). Fluorescent images obtained at 570 nm (Cy3) and 694 nm (Cy5) were analyzed using QuantArray software (Packard BioScience). Absolute values of Cy5 fluorescence signals obtained from each oligonucleotide probe were normalized to the Cy3 signal from the QC probe of the same spot to compensate for possible spot size variations. The final results are presented as the percentage of normalized signal from each probe to the total signal from all array probes.
Sequencing of rotavirus strains.
To confirm the genotype of rotaviruses used in this study, the partial sequence of the VP7 gene of each strain was determined with an ABI Prism rhodamine terminator cycle sequencing reaction kit (PE Applied Biosystems) and an automated ABI Prism 310 genetic analyzer (PE Applied Biosystems).
RESULTS
Design of PCR primers for G typing of rotaviruses.
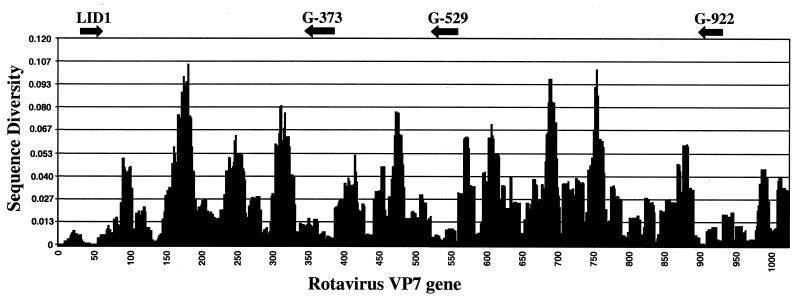
To identify regions of the VP7 gene that are appropriate for G-type discrimination, more than 150 sequences of human rotavirus VP7 genes available in GenBank were aligned using ClustalX software and analyzed by using custom-made OligoScan software to identify regions appropriate for both the selection of PCR primers and genotype-specific oligonucleotides. The most conserved regions of the VP7 gene, suitable for selecting broadly specific PCR primers, are shown on the diversity profile (Fig. 1). In addition to previously published highly conserved segments of the VP7 gene (9), we found other regions that could be used to design primers for the synthesis of fluorescently labeled DNA samples. One such region, LID1, was located at the 5′ end of VP7 mRNA (nucleotides 48 to 70) and was used for forward PCR priming (see Table 1).
FIG. 1.
Diversity profile of rotavirus VP7 gene based on analysis of nucleotide sequences of 150 individual strains. The diversity score is calculated by counting the number of mismatches between all combinations of the sequences at each position in the array of aligned sequences. Matching nucleotides are those that can bind the same complementary base. Arrows show the positions of optimized primers for synthesis of fluorescent samples for this microarray assay.
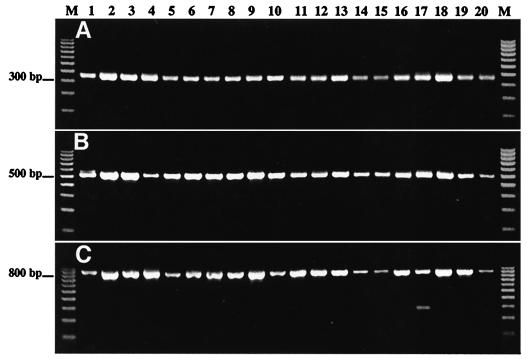
Three other conserved regions, 373 to 404, 529 to 557, and 922 to 941, were used to make reverse primers G-373, G-529, and G-922, respectively. The efficiency of each selected primer was tested by nested PCR amplification using 20 coded templates representing different rotavirus G genotypes (Table 2). The first round of RT-PCR was conducted in the presence of the conventional universal primers Beg9 and End9 (9). Small aliquots of the first-round PCR amplification products were used as templates for nested PCR with different combinations of both newly designed forward and reverse primers. The results of amplification of the samples are presented in Fig. 2. The forward primer LID1 was found to be efficient in combination with either reverse primer.
TABLE 2.
Results of genotyping of coded samples
| Coded sample | Rotavirus strain | Genotype
|
Serotype (neutralization test) | |
|---|---|---|---|---|
| Microarray assay | Sequencing | |||
| 1 | Hochi | G4 | G4 | G4 |
| 2 | KU | G1 | G1 | G1 |
| 3 | M37 | G1 | G1 | G1 |
| 4 | R143 | G9 | G9 | G9 |
| 5 | Hosokawa | G4 | G4 | G4 |
| 6 | McN | G3 | G3 | G3 |
| 7 | McN | G3 | G3 | G3 |
| 8 | 1076 | G2 | G2 | G2 |
| 9 | Nemoto | G3 | G3 | G3 |
| 10 | AJ | G2 | G2 | G2 |
| 11 | AU32 | G9 | G9 | G9 |
| 12 | 116E | G9 | G9 | G9 |
| 13 | S2 | G2 | G2 | G2 |
| 14 | VA-70 | G4 | G4 | G4 |
| 15 | 57M | G4 | G4 | G4 |
| 16 | WI61 | G9 | G9 | G9 |
| 17 | DS-1 | G2 | G2 | G2 |
| 18 | Wa | G1 | G1 | G1 |
| 19 | P | G3 | G3 | G3 |
| 20 | ST3 | G4 | G4 | G4 |
FIG. 2.
PCR amplification of 20 coded rotavirus templates with different pairs of designed primers: LID1 and G-373 (A), LID1 and G-529 (B), and LID1 and G-922 (C). Lane M, 100-bp molecular size markers.
After testing the newly designed primers, we selected LID1 and G-529 for the microarray assay. This primer pair ensured a high efficiency of amplification, and the resulting DNA segment (507 bp) contained a number of loci that can be used for genotyping all five clinically important rotavirus G types (G1 to G4 and G9). Two other reverse primers, G-373 and G-922, that we identified in this study can also be used for microarray assay when primer G-529 does not work efficiently.
Design of genotype-specific oligonucleotides.
The length of the oligonucleotide probes was selected so that the melting temperature of the oligonucleotides was between 65 and 75°C. Only probes that differed at two or more nucleotides with homologous sequences in other genotypes were chosen. The genotype-specific probes are listed in Table 1. All selected oligonucleotides are located within the most variable regions of the VP7 gene as previously identified (15). Therefore, each rotavirus genotype was identified not by a single specific probe, but by an average of nine different ones. This redundancy is a major advantage over the conventional PCR genotyping technique, since it makes the assay more reliable and less vulnerable to spontaneous mutations that often occur in clinical isolates. G genotyping of viruses based on hybridization with multiple independent probes not only increases confidence in the results but also allows detection of small genetic differences between strains belonging to the same genotype (see below).
Design of rotavirus microarray.
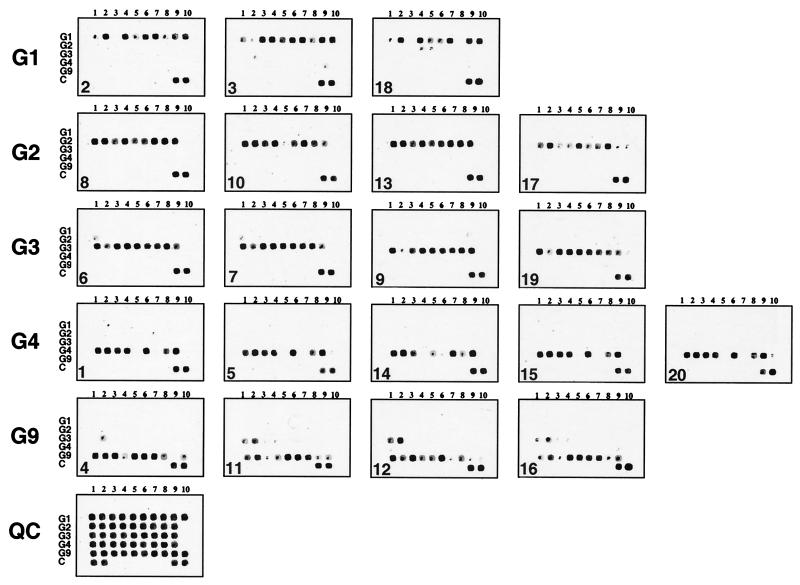
To simplify the analysis and facilitate the interpretation of the microarray data, all 1 to 10 oligonucleotide probes specific for the G1, G2, G3, G4, and G9 genotypes were arrayed in separate rows (Fig. 3). The only exception occurred at the first two G3-specific probes, which were common for both the G3 and G9 genotypes, which were placed in the third row. Thus, the G9-derived fluorescent samples were expected to produce positive signals with the first two G3 probes in addition to the G9-specific probes placed in the fifth row. Quality control oligonucleotides were placed in the last row.
FIG. 3.
Microarray hybridization patterns of 20 rotavirus samples under code. Microarray images (Cy5 fluorescence signal) are sorted into separate rows based on their G genotype. The sample code is shown in the left bottom corner of each image. The typical Cy3 image (QC) of the array is shown at the bottom of the figure, showing the layout of oligonucleotides in the microarray. Random 21-base oligonucleotides are spotted into the first two positions of row C, whereas QCprb oligonucleotide is spotted into positions 9 and 10.
Design and fabrication of oligonucleotide microchips.
There are a number of alternative protocols for the immobilization of oligonucleotides on a solid surface. We chose the method based on coupling oligonucleotides containing an amino group at the 5′ end with an aldehyde-coated glass surface. The Schiff base formation between the amino groups of oligonucleotides and aldehyde residues on the glass surface is transient and leads to reversible complexes that can be stabilized by reduction with NaBH4.
To achieve efficient immobilization of aminolink-modified oligonucleotides with an aldehyde-coated glass surface, it was necessary to optimize the conditions for the binding reaction. Oligonucleotides synthesized with an automated DNA synthesizer contain traces of chemicals, most likely ammonia, which interfere with coupling. We found that an additional purification by chloroform extraction followed by acetone precipitation of the oligonucleotides resulted in an improvement of immobilization and spot morphology. We also found that adjustment of the oligonucleotide concentration to 50 μM provided a robust, uniform, and specific florescent signal with low background.
The composition of the spotting solution and reaction conditions were found to be very important for ensuring the uniform size and good quality of the oligonucleotide spots. The low pH (3.0 to 4.0) of the spotting solution seems to be optimal for catalysis of Schiff base formation (23). At 70 to 75% humidity inside the spot arrayer's environmental chamber, the oligonucleotide spots dry completely in about 10 to 15 s and undergo efficient immobilization (data not shown). Alternatively, a 50% dimethyl sulfoxide-water solution was used for spotting instead of acetic acid. This allowed us to perform spotting at much lower humidity (40 to 60%) and to repeatedly use the same source plate containing oligonucleotides without any visible change in their concentration. The main disadvantage, however, of both methods is the difficulty of visual control of the spotting procedure, since the tiny drops of oligonucleotides become invisible after drying. Therefore, we have developed the quality control system described below, which allows us to monitor not only for spotting uniformity but also for detection of potential irregularities during the hybridization step.
Quality control of microchip fabrication and hybridization process.
Quality control is an important aspect of the microarray procedure, since the large number of microarray elements makes it impossible to inspect each microchip manually. To address this concern, we used a three-tier system of control. First, prior to spotting the oligonucleotides onto the glass surface, we spiked them with the reference oligonucleotide QCprb. The hybridization reactions always contained a mixture of rotavirus-specific sample and the oligonucleotide complementary to QCprb labeled with fluorescent dye (Cy3-QC). Scanning of the microchip at two wavelengths produced information about distributions of both specific and reference oligonucleotides (Fig. 3). Uniformity of the latter ensures that the fabrication of the chip and hybridization do not result in irregularities. Since the amount of reference oligonucleotide should be identical in all the spots, the pattern of hybridization can be used for normalization of experimental values by expressing them as ratios of a specific signal to the reference signal (Fig. 4).
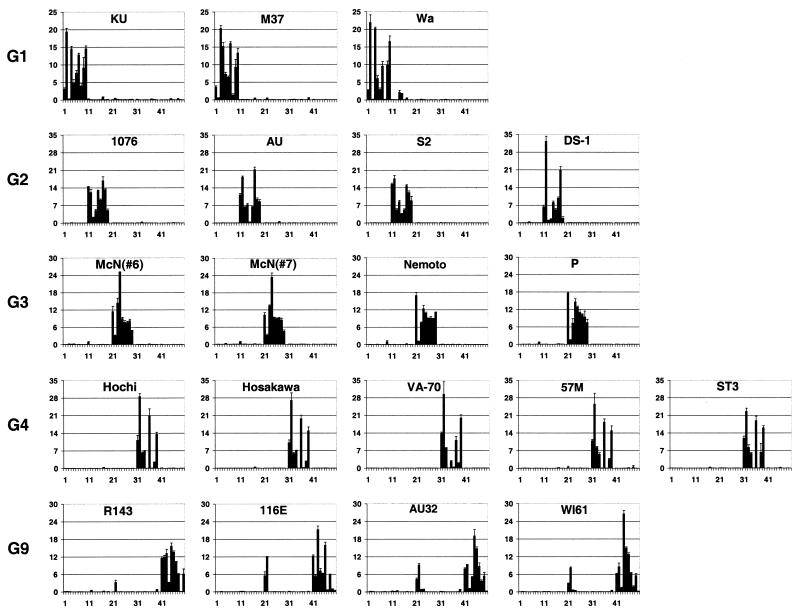
FIG. 4.
Quantitative fluorescence (Cy5) profiles of rotavirus microarrays. Normalized fluorescent signals from each oligonucleotide are shown on the y axis. Numbered locations of probes are shown on the x axis. All G1 genotype-specific oligonucleotides have numbers from 1 to 10, G2 genotype from 11 to 19, G3 genotype from 21 to 29, G4 genotype from 31 to 39, and G4 genotype from 41 to 50. All fluorescence data were obtained from two or three independent hybridizations on different microchips.
The second aspect of quality control included additional spots of oligonucleotides complementary to one of the primers used for PCR amplification (the last two spots in array row QC, Fig. 3). These sequences must be present in the fluorescent sample prepared from any rotavirus genotype and provide a positive control for sample preparation and hybridization conditions. The absence of a positive signal from these spots indicates problems with either the fluorescent sample synthesis or hybridization conditions. Finally, the third quality control was a 21-nucleotide-long random heteropolymer containing all four bases (the first two spots in array row C, Fig. 3). This control was used to measure the background caused by nonspecific sample binding of a fluorescent probe to the oligonucleotide template.
Analysis of coded samples.
The ultimate validation of any diagnostic procedure involves testing of samples of unknown identity. The 20 samples used in this study were prepared in a different laboratory and submitted for microchip analysis in a coded form, and their identity was revealed to the operator only after results were finalized. All fluorescent samples for analysis were prepared as described in Materials and Methods. Use of a single-stranded rather than a double-stranded DNA sample resulted in a higher reproducibility of hybridization results and a lower background and, in addition, did not indicate a dependence of the fluorescent signal on the location of the probe in the sample DNA molecule, as observed previously (5).
To identify the rotavirus genotype, the fluorescent samples were hybridized to microarrays in 6× SSC-5× Denhardt's solution with 0.1% Tween 20 at 42°C for 30 min. Longer incubation times did not appear to affect the efficiency of hybridization. The microarrays were hybridized with single-stranded DNA prepared as described in Materials and Methods, and the fluorescent images of microarrays were taken with a confocal laser scanner at 633 and 543 nm (Fig. 3). Since the thermodynamic parameters of oligonucleotide probes binding to sample DNA are slightly different, the fluorescent signals are not identical at any given temperature. Nonetheless, in all cases the fluorescence was observed in only one row of oligonucleotides corresponding to a certain rotavirus G genotype. All genotype-specific probes were highly discriminating, and there was very little cross-reactivity with samples of heterologous genotypes. Only a few exceptions were observed, e.g., the positive signals from the first two G3 probes (G3-1 and G3-2) due to virtually identical sequences between the G3 and G9 genotype sequences in the region of recognition by those probes. The G genotype of all 20 rotavirus strains used in this study was confirmed by direct sequencing of PCR fragments generated with the LID1 and G-529 primers. The results of this comparison are summarized in Table 2. The assay correctly identified 100% of the samples.
In additional experiments with other rotaviruses, we successfully identified the genotype of strains that could not be determined by PCR-based methods (data not shown), possibly because of spontaneous mutations in the primer-binding regions. Since the primers that we used for sample preparation bind to more conserved parts of the rotavirus genome than the primers for genotype-specific PCR, amplification and fluorescent sample preparation steps were broadly specific and more robust than those of conventional PCR.
Subgenotype differentiation.
As mentioned above, the intensity of signals from individual oligonucleotides specific to the same genotype differ, resulting in a varying pattern or profile of intensities. These profiles can potentially contain additional information about the presence of random mutations in individual viral isolates. To examine this possibility, we analyzed the microarray results quantitatively with QuantArray image analysis software. The results presented in Fig. 4 show that closely related rotavirus strains always had very similar patterns. For instance, the Ku and Wa strains (G1 genotype) had very similar profiles, with strong reactivity with oligonucleotide G1-2 and weak binding to G1-3. Another G1 strain, M37, reacted strongly with G1-3, while there was almost no binding to G1-2. As expected, the G3 genotype samples 6 and 7, representing the duplication of strain McN, showed virtually identical profiles, while the Nemoto and P strains differed from them but were very similar to each other. The Hochi, Hosakawa, 57 M, and ST3 strains of genotype G4 are highly homologous and had identical profiles, while strain VA-70 was somewhat different.
In some cases minor cross-reactivity between genotypes can be used as a signature inherent to a particular isolate. For example, strain Wa (G1) showed low but consistent cross-hybridization with two probes of the G2 set. This is a characteristic feature only of strain Wa and distinguishes it from two other G1 strains, Ku and M37. This observation is consistent with the results of sequence analysis of strain Wa at the site of recognition by oligonucleotides G2-4 and G2-5. The Wa strain had only two mismatches, which were located close to the ends of those probes and diminished their discriminating ability.
DISCUSSION
Identification and differentiation of microorganisms have been and together remain an important task with numerous applications in clinical microbiology, epidemiology, biotechnology, forensics, etc. Over the past few decades, the focus of such methods has gradually shifted from analysis of whole microorganisms to the identification of their components, including proteins (immunological methods) and nucleic acids. Progress in molecular methodologies has led to an enormous increase in the sensitivity and specificity of such assays. The current state of the art in nucleic acid identification is represented by a variety of PCR-based methods, which are highly sensitive and can potentially amplify single molecules of DNA. PCR with primers unique to a species of interest results in selective DNA amplification, and the determination is made by the match between the size of the PCR-amplified DNA (usually determined by gel electrophoresis) and the size of the expected or reference DNA products. However, as the sensitivity of PCRs increases, nonspecific products tend to accumulate, complicating analysis and leading to ambiguities and misidentification.
Recently, a novel PCR-based method was introduced which takes advantage of a third oligonucleotide to confirm the identity of PCR products (TaqMan technology). However, this approach is limited to only one specific marker per reaction. Nucleic acid hybridization is highly specific and can be controlled by various reaction conditions. Combining the sensitivity of PCR and specificity of hybridization can create a new powerful identification tool. Since the invention of the Northern blot procedure, use of probes immobilized on a solid support has become a popular format for nucleic acid hybridizations. Over the years it has evolved into microarray hybridization, which allows simultaneous reactions with hundreds and thousands of individual probes immobilized on a solid surface or embedded in a three-dimensional gel matrix. This format is widely used for a variety of applications, including quantitative gene expression analysis, detection of single-nucleotide polymorphisms, gene mapping, and pathogen detection.
A widely used variant of microarray hybridization involves the immobilization of DNA fragments representing the different genes (or their parts) of a target organism and the subsequent hybridization of the microarray with samples under study. While this approach is very efficient for gene identification and quantification of mRNA profiles in cells and tissues, it is not suitable for the detection of minor genetic differences (low genomic divergence or single point mutations) between closely related species or for the identification of unknown pathogens. The immobilization of shorter DNA probes (oligonucleotides with lengths of 15 to 50 nucleotides) that bind to genomic regions, conserved among members of a certain group of interest, can provide a way to achieve the desirable balance between specificity and sensitivity. This approach could be used for rapid, high-throughput screening and highly specific genotyping of a variety of viral and bacterial pathogens characterized by substantial degrees of genetic diversity.
Recently, we have successfully used this approach for detection of virulence factors in Escherichia coli and other enteric bacteria (5), for genotyping of influenza B virus segments, herpesviruses, and orthopoxviruses, for the quantitative analysis of mutants in live mumps virus vaccine, and for the physical mapping of poliovirus recombinant genomes (unpublished data). This approach appears to be highly robust and informative and can be adapted to analyze a broad variety of microorganisms.
Here we optimized all steps in the protocol and found that commercially produced silylated slides can be used for efficient immobilization of aminolink-modified oligonucleotides to produce high-density microarrays. The use of acetic acid solution ensured optimal conditions needed for Schiff base formation and did not leave solid residues distorting the shape of the oligonucleotide spots. Hybridization with a fluorescent single-stranded DNA sample (instead of double-stranded DNA) increased the intensity of the resulting signals and improved reproducibility and specificity. The biotin-modified primer used for PCR amplification was used for strand separation and allowed us to obtain high-quality single-stranded DNA samples.
One of the major advantages of this method is that it shortens the time needed for completion of the assay. PCR amplification takes about 2 h, followed by strand separation, which can be completed in 20 min. The hybridization step is performed for another 20 to 30 min. The entire assay can be completed in less than 4 h (time required for the first-round RT-PCR is not included). More than 10 independent assays can be done on one glass slide, making this method a high-throughput tool suitable for large-scale genotyping.
Rotaviruses are double-stranded RNA viruses that cause disease in humans and animals. They are an important cause of morbidity among infants and young children in developed countries and a major cause of morbidity and mortality in developing countries in the same age group. The rotavirus genome consists of 11 segments of double-stranded RNA that code for viral proteins involved in RNA replication and viral capsid formation. Two of the outer capsid proteins, VP4 and VP7, are immunogenic, and antibodies to these proteins play major roles in antiviral immunity. A number of distinct serotypes exist for each of these antigens. It appears that serotype-specific protection is important, thus complicating vaccine development. Rotaviruses are capable of highly efficient reassortment by exchanging individual genomic segments, further increasing their diversity. Analysis of the epidemiology and molecular evolution of human rotaviruses is an important part of the strategy for containment of the disease and requires methods that enable rapid and accurate genotyping of field isolates. In addition, rapid genotyping methods could facilitate the creation of novel reassorted attenuated strains that will aid in the analysis of samples obtained during clinical trials of experimental vaccines.
In the present study we combined the hybridization of fluorescently labeled single-stranded DNA prepared by broadly specific PCR amplification with microarrays of immobilized oligonucleotides. By comparing a large number of VP7 gene sequences of rotaviruses isolated from different mammalian species, we found that the LID1 and G-529 primers can amplify most rotavirus genotypes, potentially making this approach an important addition to assays used for the classification of rotaviruses. Multiple oligonucleotides conserved in individual rotavirus genotypes were selected for the G1, G2, G3, G4, and G9 genotypes. Our analysis also showed that it might be possible to design sets of oligonucleotide probes that would discriminate among all known mammalian rotavirus G and P genotypes.
We have demonstrated that the genotype-specific oligonucleotides unambiguously identified different genotypes of human rotaviruses. The use of these oligonucleotides for solid-phase hybridization in a microarray format, with fluorescently labeled DNA amplified by PCR with highly conserved and thus broadly specific primers, proved to be an efficient tool for rapid genotyping. Several features make this approach very efficient. The use of multiple probes ensures a high redundancy of results, thereby increasing confidence in the genotype determinations. This redundancy is especially critical with viruses that mutate rapidly and therefore lose some of their diagnostic markers. The use of highly conserved primers for PCR amplification and for the preparation of fluorescent hybridization samples allows us to develop an amplification protocol resulting in the highest sensitivity and specificity without producing nonspecific DNA. In addition to unambiguous discrimination among rotavirus genotypes, the approach described in this communication can become a versatile tool for genotyping other viruses and bacteria.
Acknowledgments
This work was supported in part by a grant from the Defense Advanced Research Project Agency (DAPRPA).
REFERENCES
- 1.Baldwin, D., V. Crane, and D. Rice. 1999. A comparison of gel-based, nylon filter and microarray techniques to detect differential RNA expression in plants. Curr. Opin. Plant Biol. 2:96-103. [DOI] [PubMed] [Google Scholar]
- 2.Beier, M., and J. D. Hoheisel. 1999. Versatile derivatisation of solid support media for covalent bonding on DNA-microchips. Nucleic Acids Res. 27:1970-1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Breslauer, K. J., R. Frank, H. Blocker, and L. A. Marky. 1986. Predicting DNA duplex stability from the base sequence. Proc. Natl. Acad. Sci. USA 83:3746-3750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cheung, V. G., J. P. Gregg, K. J. Gogolin-Ewens, J. Bandong, C. A. Stanley, L. Baker, M. J. Higgins, N. J. Nowak, T. B. Shows, W. J. Ewens, S. F. Nelson, and R. S. Spielman. 1998. Linkage-disequilibrium mapping without genotyping. Nat. Genet. 18:225-230. [DOI] [PubMed] [Google Scholar]
- 5.Chizhikov, V., A. Rasooly, K. Chumakov, and D. D. Levy. 2001. Microarray analysis of microbial virulence factors. Appl. Environ. Microbiol. 67:3258-3263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Daniliuk, N. K., S. I. Iastrebov, T. P. Artamonova, and S. G. Popov. 1986. A simplified variant of the Maxam-Gilbert method for determining the primary structure of oligonucleotides and DNA fragments. Bioorg. Khim. 12:1185-1188. (In Russian.) [PubMed] [Google Scholar]
- 7.Estes, M. K. 2001. Rotaviruses and their replication, p. 1747-1785. In D. M. Knipe and P. M. Howley (ed.), Fields virology, 4th ed. Lippincott, Williams & Wilkins, Philadelphia, Pa.
- 8.Gentsch, J. R., R. I. Glass, P. Woods, V. Gouvea, M. Gorziglia, J. Flores, B. K. Das, and M. K. Bhan. 1992. Identification of group A rotavirus gene 4 types by polymerase chain reaction. J. Clin. Microbiol. 30:1365-1373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gouvea, V., R. I. Glass, P. Woods, K. Taniguchi, H. F. Clark, B. Forrester, and Z. Y. Fang. 1990. Polymerase chain reaction amplification and typing of rotavirus nucleic acid from stool specimens. J. Clin. Microbiol. 28:276-282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Guschin, D. Y., B. K. Mobarry, D. Proudnikov, D. A. Stahl, B. E. Rittmann, and A. D. Mirzabekov. 1997. Oligonucleotide microchips as genosensors for determinative and environmental studies in microbiology. Appl. Environ. Microbiol. 63:2397-2402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hacia, J. G. 1999. Resequencing and mutational analysis using oligonucleotide microarrays. Nat. Genet. 21:42-47. [DOI] [PubMed] [Google Scholar]
- 12.Hacia, J. G., J. B. Fan, O. Ryder, L. Jin, K. Edgemon, G. Ghandour, R. A. Mayer, B. Sun, L. Hsie, C. M. Robbins, L. C. Brody, D. Wang, E. S. Lander, R. Lipshutz, S. P. Fodor, and F. S. Collins. 1999. Determination of ancestral alleles for human single-nucleotide polymorphisms using high-density oligonucleotide arrays. Nat. Genet. 22:164-167. [DOI] [PubMed] [Google Scholar]
- 13.Hoshino, Y., and A. Z. Kapikian. 1996. Classification of rotavirus VP4 and VP7 serotypes. Arch. Virol. Suppl. 12:99-111. [DOI] [PubMed] [Google Scholar]
- 14.Hoshino, Y., and A. Z. Kapikian. 2000. Rotavirus serotypes: classification and importance in epidemiology, immunity, and vaccine development. J. Health Popul. Nutr. 18:5-14. [PubMed] [Google Scholar]
- 15.Kapikian, A. Z., and R. M. Chanock. 2001. Rotaviruses, p. 1787-1833. In D. M. Knipe and P. M. Howley (ed.), Fields virology, 4th ed. Lippincott, Williams & Wilkins, Philadelphia, Pa.
- 16.Lipshutz, R. J., S. P. Fodor, T. R. Gingeras, and D. J. Lockhart. 1999. High density synthetic oligonucleotide arrays. Nat. Genet. 21:20-24. [DOI] [PubMed] [Google Scholar]
- 17.Parinov, S., V. Barsky, G. Yershov, E. Kirillov, E. Timofeev, A. Belgovskiy, and A. Mirzabekov. 1996. DNA sequencing by hybridization to microchip octa- and decanucleotides extended by stacked pentanucleotides. Nucleic Acids Res. 24:2998-3004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Proudnikov, D., E. Timofeev, and A. Mirzabekov. 1998. Immobilization of DNA in polyacrylamide gel for the manufacture of DNA and DNA-oligonucleotide microchips. Anal. Biochem. 259:34-41. [DOI] [PubMed] [Google Scholar]
- 19.Raitio, M., K. Lindroos, M. Laukkanen, T. Pastinen, P. Sistonen, A. Sajantila, and A. C. Syvanen. 2001. Y-chromosomal SNPs in Finno-Ugric-speaking populations analyzed by minisequencing on microarrays. Genome Res. 11:471-482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ramsay, G. 1998. DNA chips: state-of-the art. Nat. Biotechnol. 16:40-44. [DOI] [PubMed] [Google Scholar]
- 21.Schena, M., D. Shalon, R. W. Davis, and P. O. Brown. 1995. Quantitative monitoring of gene expression patterns with a complementary DNA microarray. Science 270:467-470. [DOI] [PubMed] [Google Scholar]
- 22.Schena, M., D. Shalon, R. Heller, A. Chai, P. O. Brown, and R. W. Davis. 1996. Parallel human genome analysis: microarray-based expression monitoring of 1000 genes. Proc. Natl. Acad. Sci. USA 93:10614-10619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sollenberger, P. Y., and R. B. Martin. 1968. Carbon-nitrogen and nitrogen-nitrogen double bond condensation reactions, p. 349-406. In S. Patai (ed.), The chemistry of amino group. Interscience Publishers, New York, N.Y.
- 24.Steel, A. B., R. L. Levicky, T. M. Herne, and M. J. Tarlov. 2000. Immobilization of nucleic acids at solid surfaces: effect of oligonucleotide length on layer assembly. Biophys. J. 79:975-981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Taniguchi, K., F. Wakasugi, Y. Pongsuwanna, T. Urasawa, S. Ukae, S. Chiba, and S. Urasawa. 1992. Identification of human and bovine rotavirus serotypes by polymerase chain reaction. Epidemiol. Infect. 109:303-312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]