Abstract
Various nucleic acid assays have been developed and implemented for diagnostics and therapeutic monitoring of human immunodeficiency virus type 1 (HIV-1) and hepatitis C virus (HCV) infections. The high-throughput, semiautomated assays described here were developed to provide a method suitable for screening plasma specimens for the presence of HIV-1 and HCV RNAs. Three assays were developed: a multiplex HIV-1/HCV assay for simultaneous detection of HIV-1 and HCV, and discriminatory assays for specific detection of HIV-1 and HCV. The assay systems utilize three proprietary technologies: (i) target capture-based sample preparation, (ii) transcription-mediated amplification (TMA), and (iii) hybridization protection assay (HPA). An internal control is incorporated into each reaction to control for every step of the assay and identify random false-negative reactions. The assays demonstrated a sensitivity of at least 100 copies/ml for each target, and they detected with similar sensitivity all major variants of HCV and HIV-1, including HIV-1 group O strains. Assay sensitivity for one virus was not affected by the presence of the other. The specificity of these TMA-driven assays was ≥99.5% in both normal donor specimens and plasma containing potentially interfering substances or other blood-borne pathogens. Statistical receiver operating characteristic plots of 1 − specificity versus sensitivity data determined very wide analyte cutoff values for each assay at the point at which the assay specificity and sensitivity were both ≥99.5%. The sensitivity, specificity, and throughput capability predict that these assays will be valuable for large-volume plasma screening, either in a blood bank setting or in other diagnostic applications.
Epidemiologic studies have identified human immunodeficiency virus type 1 (HIV-1) as the etiologic agent of AIDS and hepatitis virus C (HCV) as the etiologic agent of most blood-borne non-A, non-B hepatitis infections. Both viruses are transmitted primarily by exposure to infected blood or blood products, by certain body fluids or tissues, or from mother to fetus or child.
As of the end of 1999, 733,374 cases of AIDS had been reported to the Centers for Disease Control and Prevention (CDC), and through June 2000, 438,795 people had died of AIDS in the United States. AIDS prevalence continues to rise, with 320,000 persons living with AIDS in the United States in 1999 (CDC Divisions of HIV/AIDS Prevention Commentary, www.cdc.gov/hiv/stats 12, 2000). The prevalence of HIV-positive blood donations in first-time donors (first-time donors represent 21.1% of all donations) noted by the American Red Cross is 23 per 100,000. Among repeat donors (78.9% of all donations), this prevalence is 3 per 100,000, and the weighted overall prevalence is 7 per 100,000. The window of infectivity before seroconversion is approximately 22 days but may be up to 6 months in some cases. It is estimated that 1.5 HIV-1 infectious units per million donations escape detection with the licensed serological assays currently in place (8, 15).
Currently, detection of HIV-1 infection in the blood bank and clinical settings is based on serologic detection of p24 antigen and antiviral antibodies by enzyme immunoassay. Confirmation of antibody-positive samples is done by supplemental antibody tests such as Western blot or immunofluorescence assays, while confirmation of p24 positivity is done by neutralization. Follow-up of a positive result typically involves antiviral treatment and monitoring of the HIV-1 RNA titer by quantitative HIV-1 assays (2, 3, 15). Although the sensitivity of HIV antibody tests has increased and sensitive tests for p24 antigen have been developed and implemented, a window period still exists between infection and the appearance of detectable serological markers. Addition of the p24 antigen test has reduced the average window period from 22 days to approximately 16 days, since detectable antigenemia generally precedes seroconversion (2, 15). However, p24 antigen tests have a high false-positive rate and are less sensitive than nucleic acid-based amplification testing (NAT) (36). HIV-1 p24 antigen screening of blood donations, implemented in 1996, has only yielded six cases of antigen-positive, HIV-1 antibody-negative donors within more than 24 million donations screened (37). Several studies suggest that use of NAT could reduce the window period by 6 to 11 days, further reducing the risk of acquiring HIV from blood transfusions (7, 6, 9, 15, 19, 30).
At least 40% of chronic liver disease is related to HCV, resulting in an estimated 8,000 to 10,000 deaths each year (CDC data). The overall seroprevalence of HCV in the United States is 3.9 million (1.8%) (CDC National Center for Infectious Diseases Report, www.cdc.gov/ncidod/diseases/hepatitis/c, 2001), and infections can be divided into three groups: resolved infections (15%), such as in patients that are anti-HCV antibody positive and HCV RNA negative and have normal alanine aminotransferase (ALT) levels; persistent infections/death (15%), as in patients that are HCV antibody positive and HCV RNA positive and have elevated ALT levels; and benign persistent disease (70%), as in patients that are HCV antibody positive and HCV RNA positive and have normal or mildly elevated ALT levels.
Data from the Retrovirus Epidemiology Donor Study and recent data from the American Red Cross show that the frequency of HCV-positive U.S. blood donations is 1.12 per 1,000. The American Red Cross rate for first-time donors is 4.25 per 1,000, and for repeat donors it is 0.03 per 1,000. The window of infectivity before seroconversion is calculated to be approximately 70 days (8). With the licensed serological assays, it is estimated that up to 10 HCV infectious units per million donations escape detection and enter the blood supply (8, 15).
Detection of HCV, like that of HIV-1, is also based on serologic screening for antiviral antibodies, using enzyme-linked immunosorbent assays and confirmation with a recombinant strip immunoblot assay. Even though implementation of these tests has significantly reduced the prevalence of posttransfusion HCV infection in the United States and tests with improved sensitivity have allowed earlier detection, the risk of contracting HCV through transfusion still exists. HCV immunoserologic tests are limited by the prolonged, 70-day window of seronegativity after acute infection, the inability to differentiate between active and resolved infections, and false-negative results in rare patients with chronic, antibody-negative infection. Antigen detection tests for HCV are not yet available for blood bank screening. Use of NAT allows the detection of HCV infection about 50 to 60 days earlier than current antibody-based tests, resulting in a window period of approximately 11 days (4, 13, 14, 15, 23, 41).
Detection of viral nucleic acids as markers for HIV-1 and HCV infection and disease progression has become widely accepted as a research tool (3, 7, 34, 37). Nucleic acid tests allow earlier detection of infection because they directly test for the presence of circulating virus rather than measure the individual's antibody response to the virus. In addition, NAT exhibits a very high sensitivity of detection, serves as an independent method for confirming infection in indeterminate serology cases, helps to discriminate between chronic and resolved HCV cases, and confirms infections in HIV- and HCV-seropositive newborns. Several technologies for the detection of HIV and/or HCV nucleic acids are currently available, such as branched DNA signal amplification (10, 12, 18, 39), nucleic acid sequence-based amplification (40, 42), strand displacement amplification (29, 44), the ligase chain reaction (24, 27), transcription-mediated amplification (21, 28), and PCR (1, 25, 31, 38). The main shortcomings of the first-generation NAT technologies, rendering them unsuitable for high-throughput screening, include technical difficulty, significant hands-on time, lengthy incubation times, large sample volume requirements, high cost, relatively high rate of false positives, and inconsistency in detection of genetic variants (11, 17, 33, 34).
To provide a method suitable for high-throughput screening, semiautomated assays were developed for codetection or individual detection of HIV-1 and HCV RNAs. The HIV-1/HCV assay and the corresponding HIV-1 and HCV discriminatory assays utilize three proprietary technologies: (i) target capture-based sample preparation, (ii) transcription-mediated amplification, and (iii) hybridization protection assay. All three assays incorporate an internal control to validate each reaction. A positive result in the HIV-1/HCV multiplex assay does not distinguish between the presence of HIV-1 and HCV in the specimen; therefore, individual HIV-1 and HCV discriminatory assays are subsequently used to identify the virus present. HIV-1 and HCV discriminatory assays could also be used as highly sensitive qualitative assays for diagnostic and epidemiological applications. The assays detect all known HIV-1 and/or HCV subtypes and have sensitivities designed to reduce the window period between infection and detection while maintaining the specificity to discriminate between positive and negative specimens. These assays, described in detail below, are currently used in several blood donation locations under investigational new drug applications and have received a Biologics license from the U.S. Food and Drug Administration.
MATERIALS AND METHODS
Sample preparation.
Sample processing allows the release, stabilization, and capture of viral nucleic acids and the removal of unwanted components of the clinical specimen. The first step of sample processing consists of the addition of target capture reagent to plasma or serum samples. Target capture reagent is a HEPES-buffered detergent solution that contains an internal control RNA, capture oligonucleotides complementary to specific sequences of the HIV-1 and HCV RNAs, and superparamagnetic particles. Target capture reagent causes lysis of the viral particles and inactivates nucleases, thus enhancing viral RNA stability. The capture of the viral RNA is mediated by chimeric oligonucleotides, containing a 3′ homopolymeric dA tail and a 5′ sequence complementary to HIV-1 or HCV RNA sequences. There are five capture oligonucleotides used in the assay. Two are directed against HCV sequences in the 5′ untranslated region, two are directed against both the HIV polymerase region and the Internal Control, and the fifth is directed against the long terminal repeat (LTR) region of HIV.
The 3′ dA sequences of the capture oligonucleotides hybridize to dT oligonucleotides that are covalently bound to the magnetic microparticle solid phase. The purpose of the magnetic particles in sample preparation is to specifically capture and concentrate the HIV-1 and HCV RNA targets from the sample. The superparamagnetic particles are approximately 1 μm in diameter, with a hydrophilic surface that is derivatized with oligo(dT). Under capture conditions, the dT tail binds to the capture oligonucleotides. Target viral RNAs are captured onto magnetic microparticles, readily separated from other plasma components in a magnetic field, and washed twice.
Amplification with TMA.
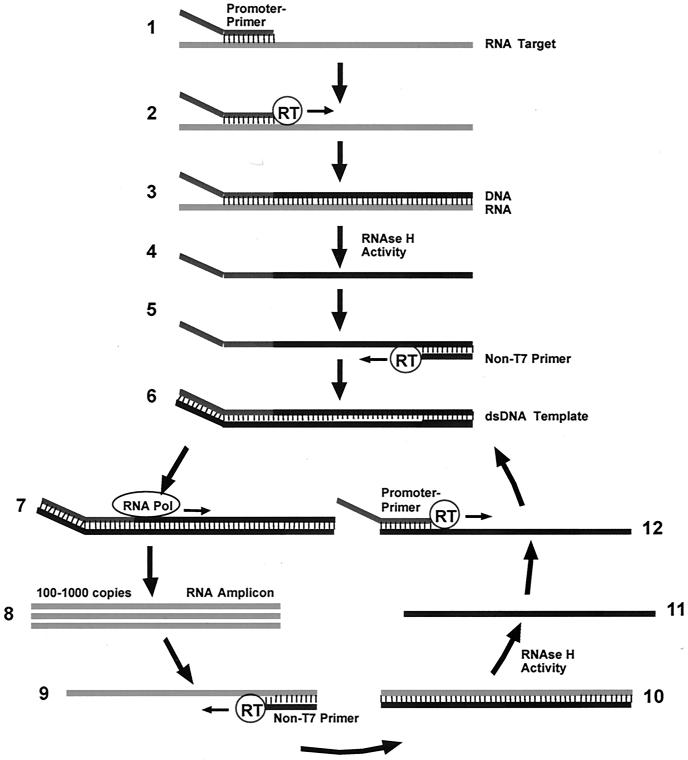
The Gen-Probe Transcription-Mediated Amplification (TMA) (21, 28) technology exponentially amplifies the captured HIV-1 and HCV viral RNAs. TMA is an isothermal transcription-based amplification technique that utilizes a two-enzyme system to produce RNA amplicon via DNA intermediates. TMA requires bacteriophage T7 RNA polymerase, Moloney murine leukemia virus reverse transcriptase (RT), deoxyribo- and ribonucleoside triphosphates, T7 promoter-primers and non-T7 primers, and target nucleic acid RNA. Reverse transcriptase is a recombinant version of the Moloney murine leukemia virus reverse transcriptase that is expressed in Escherichia coli. Moloney murine leukemia virus reverse transcriptase has both RNA-dependent and DNA-dependent DNA polymerase activities as well as RNase H activity. T7 RNA polymerase is a recombinant version of the bacteriophage T7 RNA polymerase that is also expressed in E. coli. There are five T7 promoter-primers and five non-T7 primers used in the assay. Four of the primers are involved in the amplification of HCV sequences, four additional primers are involved in the amplification of HIV polymerase and internal control sequences, and the remaining two are involved in the amplification of HIV LTR sequences. The steps of TMA are outlined in Fig. 1.
FIG. 1.
Cycle of transcription-mediated amplification. TMA includes the following five main activities. (i) A promoter-primer binds to the RNA target and is extended via the DNA polymerase activity of the reverse transcriptase (RT) (steps 1 and 2). The product of this reaction is an RNA/DNA hybrid duplex, with the new DNA strand being complementary to the virion RNA target (step 3). (ii) RNase H activity of the reverse transcriptase specifically digests the RNA strand of the RNA/DNA hybrid, leaving only the cDNA (step 4). (iii) Following the RNA degradation, the non-T7 primer (i.e., the primer without the T7 promoter) base pairs to its complementary sequence on the newly created DNA strand, and the reverse transcriptase catalyzes the synthesis of another new DNA strand using the cDNA as a template (step 5). The product of this reaction is a DNA intermediate that contains the bacteriophage T7 RNA polymerase promoter sequence in double-stranded form (step 6). (iv) The bacteriophage T7 RNA polymerase now recognizes the promoter sequence on the DNA intermediate and transcribes multiple copies of the RNA amplicon (steps 7 and 8). The RNA amplicon strands are opposite in polarity from the original RNA target and contain a region complementary to the probe used for amplicon detection. (v) The RNA strands created in iv are able to base pair to the non-T7 primer (step 9). Subsequently, the reverse transcriptase catalyzes the formation of a cDNA strand by extending the 3′ end of the non-T7 primer, and again an RNA/DNA hybrid duplex results (step 10) as in i, repeating the cycle.
Detection via HPA.
Detection of the amplified viral RNA is accomplished using the Hybridization Protection Assay (HPA) (5). HPA utilizes chemiluminescent acridinium ester-labeled probes in a homogeneous format. The basis of the HPA is the steric protection of the acridinium ester label from alkaline hydrolysis when attached to a hybridized probe. Chemiluminescence of the label on unhybridized probes is destroyed by alkaline hydrolysis during the selection step that follows the hybridization. Hybridized probes maintain their chemiluminescent properties and can thereby be detected in a luminometer. Chemiluminescent detection is achieved by the addition of Auto Detect 1 (0.1% hydrogen peroxide, 1 mM nitric acid) and Auto Detect 2 (1 N NaOH), which results in photon emission by the label on hybridized detection probes. Photons are measured with a luminometer and are reported as relative light units (RLU). The presence of an internal control in each sample tube requires discrimination of the internal control signal from the target signal. Simultaneous discrimination of both signals is accomplished with a variation of the HPA format called the Dual Kinetic Assay (32).
The Dual Kinetic Assay utilizes the kinetic discrimination of chemiluminescent emissions. A probe with rapid emission kinetics, known as a flasher (ortho-fluoroacridinium ester), is used to detect the internal control. Probes with slower emission kinetics, termed glowers (2-methylacridinium ester), are used to detect target viral nucleic acids. At a given pH and temperature, the rate of chemiluminescent emissions from a flasher is more rapid than that from a glower. There are six detection probes used in the assay. Two glower probes are directed against HCV, one glower probe is directed against the LTR region of HIV, two glower probes are directed against the HIV polymerase region, and a flasher probe detects the internal control sequence.
The exponential tail fit algorithm (32) is used to deconvolute the two simultaneous signals from every reaction mixture containing both internal control and target RNA. The shape of the glower signal over time is scalable, and its geometry is constant under any given set of pH and temperature conditions, regardless of the magnitude of the signal. As such, the glower signal at any given time is directly proportional to the total integrated signal/time function. By design, the flasher signal from the hybridized internal control probe is brief and contributes a negligible amount to the magnitude of the glower signal at later time points.
HIV-1 and HCV discriminatory assays.
The discriminatory assays utilize the same three main steps as the multiplex assay (target capture, TMA, and HPA), and the same basic assay procedure is followed. The discriminatory assays are based on chemistry that is identical to the HIV-1/HCV assay except that the labeled probes used and, in the case of the HCV discriminatory probe reagent, the HCV-specific labeled oligonucleotides are present at a higher concentration.
Assay run, calibrators, and controls.
Each TMA assay run can accommodate up to 100 reactions, including nine assay calibrators and up to 91 specimens, and a single operator can manage two assay runs simultaneously. The results obtained from the nine calibrators contained in each TMA assay run are used to determine the validity of the run and to establish the assay cutoffs as described below. The negative calibrator is defibrinated normal human plasma, which is nonreactive for hepatitis B virus surface antigen, antibodies to HIV-1 and HIV-2, and antibodies to human HCV when tested by Food and Drug Administration (FDA)-licensed assays. The HIV-1 positive calibrator consists of heat-inactivated, HIV-1-positive plasma in defibrinated normal human plasma that is nonreactive for hepatitis B virus surface antigen and anti-HCV when tested by FDA-licensed assays. The heat-inactivated HIV-1 virus is present at 500 ± 200 copies/ml. Similarly, the HCV positive calibrator consists of heat-inactivated HCV-positive plasma in defibrinated normal human plasma that is nonreactive for hepatitis B virus surface antigen, anti-HIV-1, and anti-HIV-2 when tested by FDA-licensed assays. The heat-inactivated HCV is present at 560 ± 260 copies/ml.
The internal control serves as a general monitor of assay performance for each specimen tested, as it is added to each test specimen and assay calibrator reaction. The internal control consists of an in vitro-synthesized transcript containing a portion of HIV-1 and a unique sequence targeted by the internal control probe. The internal control signal is distinguished from HIV-1 and HCV analyte signal by the differential kinetics of light emission of the labeled probes used for each target. Prior to carrying out target capture, the internal control is added to a newly opened target capture reagent bottle to ≈250 copies/reaction. As it is added directly to the target capture reagent, the internal control controls for false-negative results that could occur due to problems in target capture, amplification, or detection.
Formula-derived assay cutoff values.
Two cutoffs are determined for the TMA assays, termed the analyte cutoff and internal control cutoff, for the analyte and internal control signals, respectively. These cutoff values are derived from the analyte and internal control RLU signals of the nine assay calibrators that are run at the beginning of each assay run.
Formulas were established to allow determination of a floating analyte cutoff in each assay that falls within the range of RLU values indicated by receiver operating characteristic (ROC) analysis to achieve ≥99.5% sensitivity and specificity. By using a floating cutoff rather than a set value, day-to-day variations due to reagents, operators, and instruments are taken into account. The formulas add a portion of the HIV-1 positive calibrator analyte signal and/or the HCV positive calibrator analyte signal to the negative calibrator analyte signal as defined in Table 1 for the three assays. When a sample demonstrates an analyte signal/analyte cutoff (S/CO) ratio of ≥1, it is considered reactive for the target RNA.
TABLE 1.
Analyte cutoff formulas
| Assay | Analyte cutoff |
|---|---|
| HIV-1/HCV | (negative calibrator analyte signal average) + (0.02 × HIV positive calibrator analyte signal average) + (0.04 × HCV positive calibrator analyte signal average) |
| HIV-1 discriminatory | (negative calibrator analyte signal average) + (0.04 × HIV positive calibrator analyte signal average) |
| HCV discriminatory | (negative calibrator analyte signal average) + (0.04 × HCV positive calibrator analyte signal average) |
The internal control cutoff is also established for each individual assay run, with the cutoff calculation positioned to minimize the number of invalid results while at the same time preventing the occurrence of false-negative results. A cutoff calculated as 50% of the negative calibrator internal control signal was found to meet these conditions for the HIV-1/HCV assay, the HIV-1 discriminatory assay, and the HCV discriminatory assay. Like the analyte S/CO, when a sample demonstrates an internal control signal/internal control cutoff (internal control S/CO) ratio of ≥1, it is considered reactive, and an internal control S/CO ratio of <1.0 indicates a nonreactive result. In negative samples, a positive internal control result validates the reaction. When samples are reactive for analyte signal as determined from the analyte S/CO, the internal control result is not used to validate the reaction.
Instrumentation.
The testing system is composed of the following components: TECAN GENESIS series RSP model 150/8 Instrument pipettor, Gen-Probe target capture system revision B, Gen-Probe LEADER HC+ luminometer, and dedicated personal computer with TMA blood testing data reduction software (Chiron Procleix 2.0), circulating waterbaths for incubation steps, and multitube vortexers for mixing steps. Data reported here were transferred from the LEADER HC+ instruments to a structured query language server database, from which the data were retrieved and analyzed with Microsoft Excel 97.
Specificity testing.
Specificity data were generated by testing frozen EDTA plasma samples from individual normal donors obtained from Gary Tegtmeier of the Community Blood Center of Greater Kansas City (Kansas City, Mo.). Approximately 1,500 samples were tested with three lots of reagents.
Analytical sensitivity evaluation.
Analytical sensitivity panels comprised of HIV-1 type B and HCV subtype 1a at concentrations of 300, 100, 30, 10, 3, 1, and 0 viral copies/ml were used to evaluate assay sensitivity. The HIV-1 panel members were prepared by serial dilution of an HIV-1 type B isolate from a tissue culture supernatant. HCV panel members were made by serial dilution of patient plasma samples containing HCV subtype 1a. The viral stocks used to make the panel were quantified using in-house HIV-1 or HCV quantitative assays that were calibrated according to the Virology Quality Assurance laboratory of the AIDS Clinical Trials Group or World Health Organization (WHO) standards, respectively. The analytical sensitivity panel members were frozen in 5.6-ml single-use aliquots and thawed at room temperature immediately before they were tested in the TMA assays. Approximately 80 replicates of each were tested with each of three reagent lots in each of the three test assays. To prepare the coinfected panels used in this study, 33 ml of HIV-1-infected plasma and 33 ml of HCV-infected plasma were combined at the appropriate concentration. Limits of detection of HIV-1 and HCV were also determined using NAT standards obtained from the Center for Biologics Evaluation and Research (CBER, FDA), the WHO, and Bioclinical Partners, Inc. (Franklin, Mass.).
Effect of donor and donation factors on sensitivity and specificity.
The specificity and sensitivity of the TMA HIV-1/HCV assay, the HIV-1 discriminatory assay, and the HCV discriminatory assay were evaluated with respect to the effects of donors and donation factors in various types of specimens. Conditions, factors, and sample characteristics that were evaluated included the following: (i) infections other than HIV-1 or HCV (herpes simplex virus types 1 and 2, human T-cell lymphotrophic virus types I [n = 8] and II [n = 8], hepatitis A virus, hepatitis B virus, HIV-2, cytomegalovirus, Epstein-Barr virus, rubella virus, and parvovirus B19) and vaccination status (influenza and hepatitis B virus [n = 10 each]) (ProMedDx, LLC, Norton, Mass.; BioClinical Partners, Inc., Franklin, Mass.; and Boston Biomedica, Inc., West Bridgewater, Mass.); (ii) specimens contaminated with bacterial, yeast, or fungal pathogens (Staphylococcus epidermidis, Staphylococcus aureus, Micrococcus luteus, Corynebacterium diphtheriae, Propionibacterium acnes, Candida albicans, and Pneumocystis carinii) (normal blood donations from the Community Blood Center of Greater Kansas City spiked to 105 CFU/ml); (iii) plasma samples from donors with autoimmune and other diseases (rheumatoid arthritis, rheumatoid factor, antinuclear antibody, lupus, multiple sclerosis, multiple myeloma, hypergammaglobulinemia [immunoglobulin G {IgG} and IgM], alcoholic cirrhosis, and elevated alanine aminotransferase levels) (ProMedDx, LLC; BioClinical Partners, Inc.); (iv) specimens containing potentially interfering substances (hemolyzed, icteric, and lipemic specimens) (ProMedDx, LLC) and an analytical panel prepared in-house containing bilirubin (200 mg/liter), lipids (2,752 mg/dl), hemoglobin (5,000 mg/liter), or protein (225 g/liter); (v) serum samples and plasma specimens collected in various anticoagulants (acid citrate dextrose, plasma preparation tube with dipotassium EDTA as the anticoagulant, dipotassium EDTA and tripotassium EDTA, sodium citrate, and sodium heparin) (ProMedDx, LLC).
Overall, approximately 830 HIV-1 and HCV antibody-negative specimens were tested in the three assays for specificity analysis. Aliquots of each specimen were also spiked with HIV-1 or HCV target (200 copies/ml) for analysis of assay sensitivity. For comparison to the assorted conditions investigated, control specimens from healthy blood donors were used as HIV-1 and HCV antibody-negative specimens for specificity analysis, and healthy donor specimens were spiked with HIV-1 or HCV for sensitivity analysis (Community Blood Center of Greater Kansas City, Kansas City, Mo.).
Assay reproducibility.
The reproducibility of the HIV-1/HCV assay was evaluated with respect to variations introduced by operator, reagent lot, LEADER HC+ luminometer system, and assay run. Three operators carried out the assay using three luminometers and three lots of reagents. Fifty-four runs were completed for the study and included in the analysis. The HIV-1 and HCV samples evaluated were panels containing HIV-1 type B, HIV-1 group O, or HCV 1a at 100 copies/ml or HCV 2b at 300 copies/ml. The HIV-1-containing panels were constructed by dilution of cultured virus in HIV-1/HCV-negative serum, and gentamicin sulfate was included at 12.5 μg/ml. Similarly, the HCV-containing panels were made by diluting HIV-1-negative serum containing either HCV genotype 1a or HCV genotype 2b into HIV-1/HCV-negative serum, with gentamicin sulfate included at 12.5 μg/ml.
Statistical determination of assay cutoff values.
ROC curves were used to assess the diagnostic performance of the assay in terms of specificity and sensitivity for each possible cutoff value of the test. Briefly, the sensitivity and specificity data were combined and sorted according to ascending RLU result. With each specific RLU result, or k, the number of reactive and nonreactive decisions that would occur with a cutoff >k but less than the following data point was calculated. The sensitivity (number of positive decisions/true-positive samples) and specificity (number of negative decisions/true-negative samples) for the cutoff point was determined for each cutoff point implemented. The use of actual data points to determine the cutoff values used, rather than using constant small intervals, or bins, results in variable bin width but provides a more optimal resolution of the acceptable cutoff values. These calculations were carried out using a Visual Basic program in Microsoft Excel 97, and the program was verified through comparison to results obtained with SAS software (SAS Institute, Cary, N.C.).
The parameters of sensitivity and 1 − specificity were then plotted for each cutoff value as an ROC curve, where the upper left-hand corner of the curve identifies the specificity and sensitivity limits of the assay. From these analyses, a range of cutoff values was determined that allows ≥99.5% sensitivity and ≥99.5% specificity in accordance with the assay requirements. The point at which the sensitivity and the specificity were mutually maximized was also determined from the ROC curve analysis.
Samples containing at least 100 copies of HIV-1 subtype B/ml or 300 copies of other HIV-1 subtypes/ml were included in the ROC analysis of the HIV-1/HCV assay and the HIV-1 discriminatory assay. Samples containing at least 100 copies of HCV subtype 1a/ml or 300 copies of other HIV-1 subtypes/ml were included in the ROC analysis of the HIV-1/HCV assay and the HCV discriminatory assay. Panel samples containing 0 copies/ml and the specimens from healthy blood donors were included in the ROC analyses as specificity samples. The specificity and sensitivity samples tested under various donor and donation factor conditions were also included in the ROC analyses as they met these described qualifications as well.
RESULTS
Assay sensitivity.
To assess the analytical sensitivity of the HIV-1/HCV assay, the HIV-1 discriminatory assay, and the HCV discriminatory assay, samples containing known quantities of HIV-1 type B or HCV subtype 1a were diluted to 300, 100, 30, 10, 3, 1, and 0 copies/ml. Eighty replicates of each dilution were tested with three lots of reagents, for a total of 240 data points at each copy level for each assay. At 100 copies/ml, detection of 100% (confidence interval [CI] = 98.5 to 100%) was obtained for HIV-1 type B and HCV subtype 1a. Probit analysis of the sensitivity samples was used to predict the virus copy levels at which the detection probability is 50 or 95% as described in Table 2. The assays demonstrated 95% detection rates for HIV-1 type B and HCV subtype 1a, well below the 100 copies/ml design objective of the assays. HIV-1 type B has a 95% detection probability of 11 and 13 copies/ml when tested in the HIV-1/HCV assay and HIV-1 discriminatory assay, respectively. Similarly, the probit analysis indicated that HCV subtype 1a has a 95% detection probability at 29 and 34 copies/ml in the HIV-1/HCV assay and HCV discriminatory assay, respectively.
TABLE 2.
Probit analysis of the HIV-1/HCV assay, the HIV-1 discriminatory assay, and the HCV discriminatory assay
| Target | Assay | Detection probabilities (copies/ml)
|
|
|---|---|---|---|
| 50% (95% fiducial limits) | 95% (95% fiducial limits) | ||
| HIV-1 type B | HIV-1/HCV | 4.8 (4.4 to 5.2) | 10.8 (10.0 to 11.7) |
| HIV-1 discriminatory | 5.0 (4.5 to 5.5) | 13.1 (12.1 to 14.4) | |
| HCV subtype 1a | HIV-1/HCV | 12.1 (11.1 to 13.2) | 29.3 (27.1 to 32.0) |
| HCV discriminatory | 13.2 (12.2 to 14.3) | 34.4 (33.4 to 35.5) | |
The limits of detection were also tested using currently available HIV-1 and HCV NAT standards. The results confirmed the results obtained with the in-house analytical sensitivity panels in which ≥95% detection at 100 copies/ml for HIV-1 type B or HCV type 1 was shown. Detection was seen down to 2.7 copies/ml for HIV-1 (Bioclinical Partners HIV-1 Reference Panel) and to 0.1 IU/ml for HCV type 1 (WHO HCV 96/790). Detection of HIV-1 genotype samples (CBER HIV-1 Genotype Panel) was also demonstrated at concentrations below 100 copies/ml (data not shown) (35; P. N. Lelie et al., unpublished data).
Assay sensitivity under coinfection conditions.
The analytical sensitivities of the HIV-1/HCV assay as well as the HIV-1 discriminatory assay and HCV discriminatory assay were evaluated under conditions of HIV-1 and HCV coinfection. The coinfection panels (Table 3) essentially consisted of the following combinations: one virus present at 107 copies/ml and the other at 30 copies/ml (panels A and B), and one virus at 30 copies/ml and the other virus at 0 copies/ml (panels C and D). Upon testing of 10 replicates of panels A to D in the HIV-1/HCV assay, 100% positivity was achieved regardless of the combination of viruses included (Table 3). The signal-to-cutoff ratios observed in the multiplex assay ranged from 9.66 for panel C, containing only HCV at 30 copies/ml, to 34.12 for panel A, containing both viruses. All panel members containing HIV-1 were 100% reactive in the HIV-1 discriminatory assay, including panel B, which contained 107 copies of HCV/ml and only 30 copies of HIV-1/ml.
TABLE 3.
Detection of HIV-1 and HCV in coinfected samples
| Panel | HIV-1 (copies/ml) | HCV (copies/ml) | Statistic | HIV-1/HCV assay (n = 10) | HIV-1 discriminatory (n = 10) | HCV discriminatory (n = 10) |
|---|---|---|---|---|---|---|
| A | 107 | 30 | Mean S/CO | 34.12 | 19.69 | 20.77 |
| CV (%) | 1.29 | 1.52 | 14.03 | |||
| % Positive | 100 | 100 | 100 | |||
| B | 30 | 107 | Mean S/CO | 18.95 | 7.46 | 22.28 |
| CV (%) | 6.61 | 12.40 | 2.05 | |||
| % Positive | 100 | 100 | 100 | |||
| C | 0 | 30 | Mean S/CO | 9.66 | 0.16 | 22.36 |
| CV (%) | 2.28 | 34.64 | 1.91 | |||
| % Positive | 100 | 0 | 100 | |||
| D | 30 | 0 | Mean S/CO | 18.46 | 12.25 | 0.19 |
| CV (%) | 29.42 | 37.91 | 148.08 | |||
| % Positive | 100 | 100 | 0 |
HCV caused no significant interference in the HIV-1 discriminatory assay, as determined from the positivity rates and from the S/CO ratios of 7.46 and 12.25 for testing in the presence and absence of HCV, respectively. Similarly, all panels containing HCV were 100% reactive in the HCV discriminatory assay, including panel A, which contained 107 copies of HIV-1/ml and only 30 copies of HCV/ml. HIV-1 had no significant effects on the analyte signal in the HCV discriminatory assay, with S/CO values of 20.77 and 22.36 observed in the presence and absence of HIV-1, respectively. Panel C, containing only HCV RNA, was nonreactive in the HIV-1 discriminatory assay, and panel D, containing only HIV-1, was nonreactive in the HCV discriminatory assay.
Assay specificity in healthy blood donor specimens.
In nonreactive samples, the internal control S/CO ratio indicates whether the reaction was complete in each individual reaction by controlling for the steps of target capture, amplification, and detection. If the internal control S/CO ratio is ≥1.0, the reaction is considered valid, while any result less than 1.0 is interpreted as an internal control failure or invalid result. However, if the analyte result is positive, the result is treated as valid regardless of the internal control signal. In testing the 1,500 healthy blood donor specimens, there were incidences of initial internal control failure in each of the TMA assays (Table 4), which generally indicates insufficient amplification due to nonspecific inhibition or other assay protocol deficiency in one or more of the steps of target capture, amplification, or detection. In all cases, retesting of the samples with initially invalid internal control signals resulted in valid internal control amplification, indicating that the sample itself did not inhibit the assay reaction.
TABLE 4.
Specificities of the HIV-1/HCV assay, the HIV-1 discriminatory assay, and the HCV discriminatory assay in samples from healthy donors
| Parametera | Value for assay
|
||
|---|---|---|---|
| HIV-1/HCV | HIV-1 discriminatory | HCV discriminatory | |
| No. tested | 1,500 | 1,500 | 1,500 |
| Initial reactive | 1 | 0 | 2 |
| Initial reactive rate (%) | 0.06 | 0.00 | 0.13 |
| Repetitive reactive rate | 0.00 | 0.00 | 0.00 |
| Mean analyte S/CO (SD) | 0.20 (0.06) | 0.17 (0.05) | 0.12 (0.07) |
| Initial IC failures | 13 | 12 | 11 |
| Initial IC failure rate (%) | 0.87 | 0.80 | 0.73 |
| Repetitive IC failure rate | 0.00 | 0.00 | 0.00 |
| Mean IC S/CO (SD) | 2.04 (0.13) | 2.03 (0.14) | 2.03 (0.15) |
IC, internal control.
Specimens with an analyte S/CO ratio of ≥1.0 are considered reactive, or positive for HIV-1 and/or HCV RNA, while those with an analyte S/CO ratio of <1.0 are considered nonreactive, or negative for HIV-1 and HCV RNAs. Of 1,500 specimens from donors with negative serology tested in the HIV-1/HCV assay, only one was initially reactive. This sample was not repeatedly reactive upon retesting (Table 4). The initial reactive result could be the result of amplicon or target contamination of the sample or incomplete selection during the HPA step. When these specimens were tested in the discriminatory assays, there were no initial reactive samples in the HIV-1 discriminatory assay, and only 2 of the 1,500 normal specimens tested in the HCV discriminatory assay were initially reactive with nonreactive results upon retest (Table 4).
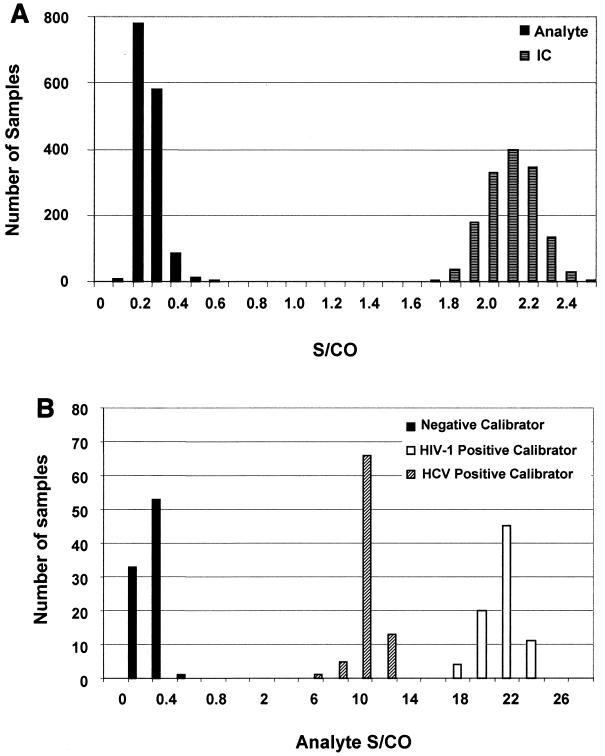
The distribution of internal control S/CO ratios in the HIV-1/HCV assay is summarized graphically in Fig. 2A; the average internal control S/CO value was 2.04 with a standard deviation (SD) of 0.13. This demonstrates clear separation of 7.6 SD from the cutoff of 1.0. The distribution of internal control S/CO ratios for the discriminatory assays is similar to that shown in Fig. 2A for the HIV-1/HCV assay, with mean ratios ± SD of 2.03 ± 0.14 and 2.03 ± 0.15 for the HIV-1 and HCV discriminatory assays, respectively. Likewise, the distribution of analyte S/CO ratios for the negative specimens is also summarized graphically in Fig. 2A. The mean analyte S/CO value of this population was 0.20 with an SD of 0.06 and demonstrates a clear separation of 13.0 standard deviations from the cutoff of 1.0. The distribution of analyte S/CO ratios for the discriminatory assays is similar to that shown in Fig. 2A for the HIV-1/HCV assay, with mean S/CO ratios ± SD of 0.17 ± 0.05 and 0.12 ± 0.07 for the HIV-1 and HCV discriminatory assays, respectively.
FIG. 2.
Distribution of S/CO ratios of normal blood bank specimens and calibrators tested with the TMA HIV-1/HCV assay. Approximately 1,500 specimens from healthy donors were analyzed in the HIV-1/HCV assay, and their resulting internal control (IC) and analyte S/CO ratios are plotted as black and gray bars, respectively (A). The cutoff values used for the S/CO calculation were obtained from the formulas listed in Table 1. (B) Distribution of analyte S/CO ratios for the calibrators included in the specificity analysis assay runs. Bars represent the number of reactions at each S/CO, with black, white, and hatched bars representative of the negative calibrator, HIV-1 positive calibrator, and HCV positive calibrator, respectively.
The distribution of analyte signal was also evaluated for the negative calibrators, HIV-1 positive calibrators, and HCV positive calibrators tested in each run with the specificity samples. From Fig. 2B, a relationship between calibrator type and analyte S/CO is apparent. The negative calibrators have the lowest analyte S/CO ratios, as expected, with a mean ± SD of 0.2 ± 0.1. The positive calibrators, while both having much larger analyte RLU values, are clearly distinguishable, with mean ± SD values of 20.5 ± 1.6 and 9.1 ± 0.9 for the HIV-1 and HCV positive calibrators, respectively. The difference in the mean of the HIV-1 versus the HCV S/CO reflects the quantity of labeled probes present in the probe reagent, as the amount of HIV-1 probe included in the reactions is greater than that of the HCV probe.
Donor and donation effects on assay sensitivity and specificity.
The specificities and sensitivities of the HIV-1/HCV assay, the HIV-1 discriminatory assay, and the HCV discriminatory assay were evaluated with two reagent lots with respect to the effect of donor and donation factors. Conditions and characteristics evaluated included infections other than HIV-1 and HCV, specimens contaminated by bacterial, yeast, or fungal pathogens, problematic plasma samples from donors with autoimmune and other diseases, specimens containing potentially interfering substances (hemolyzed, icteric, and lipemic specimens and samples containing high levels of bilirubin, lipids, albumin, and hemoglobin), and plasma specimens collected in various anticoagulants and serum specimens. Approximately 10 individual specimens for each category were tested, for a total of ≈830 specimens. These specimens, which were HIV-1 and HCV antibody negative, were tested to evaluate assay specificity.
Aliquots of each specimen were also spiked with approximately 200 copies of HIV-1 or HCV/ml, and assay sensitivity was assessed. Control specimens for these studies consisted of 91 samples from healthy blood donors for specificity evaluation. Additional specimens from healthy blood donors were also spiked with HIV-1-infected plasma (targeted input of 200 copies/ml, n = 91) or HCV-infected plasma (targeted input of 200 copies/ml, n = 91) for assessment of assay sensitivity.
Table 5 shows the positivity rates obtained with samples representing various donor and donation factors tested in the HIV-1/HCV assay. All HIV-1- and HCV-seronegative specimens, regardless of specimen type, displayed positivity rates of 0%, as expected for antibody-negative samples. Furthermore, the positivity rate of 0% and confidence intervals of 0 to 0.4% were not statistically different from those determined with the control specimens (0% [CI = 0 to 4%]). Similar results were obtained in the HIV-1 and HCV discriminatory assays, with 0% reactivity in both assays and confidence intervals of 0 to 0.4% and 0 to 0.5%, respectively (data not shown). There was no particular group of donors or type of donation factor that resulted in nonspecific reactivity or false-positive results.
TABLE 5.
Effect of donor and donation factors on sensitivity and specificity
| Condition | No. reactive/no. tested (% positivity)
|
||
|---|---|---|---|
| HIV-1 and HCV antibody negative | HIV-1 positive | HCV positive | |
| Infections (not HIV-1 or HCV)a | 0/232 (0) | 232/232 (100) | 230/232 (99.1) |
| Microbe contaminationb | 0/140 (0) | 140/140 (100) | 140/140 (100) |
| Autoimmune and other diseasesc | 0/192 (0) | 192/192 (100) | 192/192 (100) |
| Potentially interfering substancesd | 0/125 (0) | 131/131 (100) | 132/132 (100) |
| Various anticoagulantse | 0/139 (0) | 140/140 (100) | 140/140 (100) |
| Overall % positivityf | 0 (0-0.4) | 100 (99.6-100) | 99.8 (99.1-100) |
| Controlf (n = 91) | 0 (0-4) | 100 (96-100) | 100 (96-100) |
The agents tested are listed in the text.
Staphylococcus aureus, Streptococcus epidermidis, Candida albicans, P. carinii, M. luteus, P. acnes, C. diptheriae (10 donors per microbe).
Lupus, antinuclear antibody, rheumatoid arthritis, rheumatoid factor, multiple myeloma, elevated ALT, alcoholic cirrhosis, multiple sclerosis, hyperglobulinemia IgG (n = 11), hyperglobulinemia IgM (n = 6) (10 donors per type, unless noted).
Lipids, hemoglobin, bilirubin, albumin (12 replicates per type); icteric, lipemic, and hemolyzed (five donors per type).
Dipotassium and tripotassium EDTA, plasma preparation tube, sodium citrate, sodium heparin, acid citrate dextrose, serum (10 donors per type).
Values in parentheses are CIs.
When aliquots of the specimens spiked with HIV-1 were tested in the TMA HIV-1/HCV assay, the HIV-1-positive specimens achieved 100% positivity in each group, with a CI of 99.6 to 100%, the same as that found in HIV-1-spiked controls (100% [CI= 96 to 100%]). Among the HCV-positive specimens, two were nonreactive in the assay, resulting in an overall positivity rate of 99.8% (CI = 99.1 to 100%), which was not statistically different than that observed in the HCV-spiked controls (100% [CI = 96 to 100%]) (Table 5). Similar overall results were obtained in the HIV-1 and HCV discriminatory assays, with positivity rates of 100% (CI = 99.6 to 100) and 99.8% (CI = 99.1 to 100%), respectively (data not shown). There was no particular group of donors or type of donation factor that resulted in interference or false-negative reactions.
Overall, the specificity and sensitivity of the three assays demonstrated less than 0.5% repetitive reactive rates with negative specimens and greater than 95% detection at higher than 100 copies of HIV-1 or HCV/ml under all conditions tested.
Reproducibility of the HIV-1/HCV assay.
A total of 2,151 valid reaction results were analyzed for assay reproducibility in the HIV-1/HCV assay to determine the influences of operator, reagent lot, luminometer, and run on assay variability. Table 6 shows the positivity rates and coefficients of variance (CV) obtained with in-house HIV-1- and HCV-infected panels. Overall sensitivity results for this study indicated that the HIV-1 type B and HCV 1a panels containing 100 copies/ml both achieved high (99.6% [CI = 98.7 to 100%]) detection rates. Testing of the HIV-1 group O panel containing 100 copies/ml and the HCV 2b panel containing 300 copies/ml resulted in 97.4% (CI = 95.7 to 98.6%) and 95% (CI = 92.8 to 96.7%) positivity rates, respectively.
TABLE 6.
Evaluation of HIV-1/HCV assay reproducibility
| Panel | No. of valid reactions | % Positive (95% CI) | CVa (%) of RLU value
|
||||
|---|---|---|---|---|---|---|---|
| Interoperator | Interlot | Interleader | Interrun | Intrarun | |||
| HIV-1 type B | 540 | 99.6 (98.7-100) | 2.1 | 6.7 | 1.5 | 7.8 | 24.9 |
| HIV-1 group O | 537 | 97.4 (95.7-98.6) | 7.5 | 8.7 | 3.1 | 11.4 | 28.8 |
| HCV 1a | 537 | 99.6 (98.7-100) | 4.4 | 4.1 | 1.5 | 4.8 | 11.0 |
| HCV 2b | 537 | 95.0 (92.8-96.7) | 19.8 | 14.4 | 4.8 | 10.9 | 29.6 |
Nonreactive results were not included for the CV analyses; three operators, three reagent lots, three Leader HC+ luminometers, and 54 runs were compared for the analyses.
The RLU values obtained with reactive samples were compared with respect to operator (three), reagent lot (three), LEADER HC+ luminometer system (three), and across runs (n = 54), and the effects on CV of these potential variability-inducing factors were determined. The mean RLU for HIV-1 type B was 794,000, with CV values ranging only from 1.5 to 7.8% for this target under all conditions tested. The variance was similarly unaffected by these factors when specimens containing HCV 1a were tested, as the mean RLU was 455,510 with low CV values ranging from 1.5 to 4.8%. The subtypes HIV-1 group O and HCV 2b were slightly more influenced by the variability factor assessed, as the CV ranged from 3.1 to 11.4% (highest observed across runs) and 4.3 to 19.8% (highest observed among operators). However, overall the CV was not dramatically affected by any of the factors analyzed, indicating minimal effect on variability due to operator, lot, leader, and run. The RLU values obtained in reactive samples were also compared with respect to the results obtained within each run. The variance within the run, also known as random error, introduced more variability than the other factors assessed, with CV values ranging from 11.0 to 29.6%.
Statistical analysis of assay specificity and sensitivity.
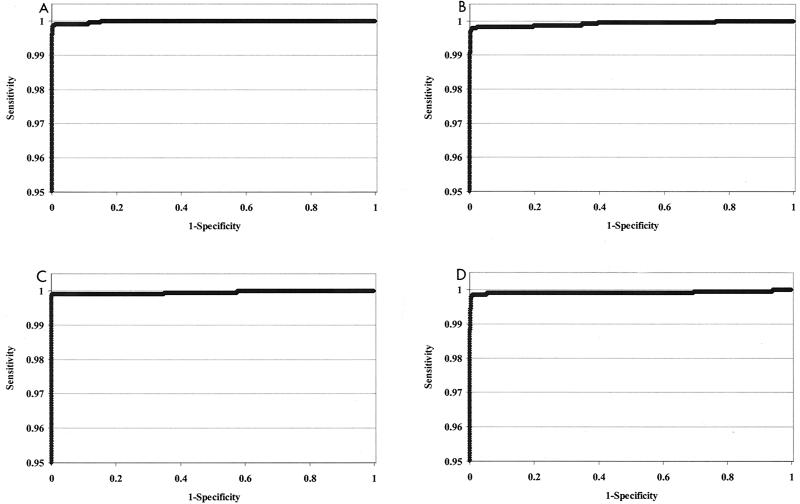
The specificity specimens and sensitivity samples containing ≥100 copies of HIV-1, HCV, or both per ml were statistically analyzed by ROC as described in Materials and Methods. Conventional ROC plots were constructed for data obtained with the HIV-1/HCV assay, the HIV-1 discriminatory assay, and the HCV discriminatory assay, as demonstrated in Fig. 3. There are two plots presented for the HIV-1/HCV assay, with a separate ROC analysis for each of the targets, HIV-1 and HCV. This analysis clearly shows the very high specificity and sensitivity of the three assays. In all cases, cutoff ranges, which allow for ≥99.5% sensitivity and ≥99.5% specificity, were determined from the analyses (Table 7). Results obtained from the ROC analysis of analyte RLUs of the HIV-1/HCV assay indicated that cutoff values between 37,777 and 284,560 RLU achieve greater than 99.5% specificity and sensitivity for the detection of HIV-1. Likewise, cutoff values between 37,777 and 121,856 RLU achieve the same specificity and sensitivity in the HIV-1/HCV assay for the detection of HCV. Analysis of the HIV-1 discriminatory assay determined a cutoff range of 29,026 to 152,941 RLU, and the cutoff range for the HCV discriminatory assay was 48,860 to 132,007 RLU. The cutoff values that maximized the sensitivity and specificity were quite similar between assays, ranging from 38,198 to 62,880 RLU (Table 7).
FIG. 3.
ROC plots of the specificity and sensitivity of the TMA HIV-1/HCV assay, the HIV-1 discriminatory assay, and the HCV discriminatory assay. Specificity and sensitivity data were assessed for each assay by ROC analysis as described in Materials and Methods. The results represent the plots of the TMA HIV-1/HCV assay detection of HIV-1 (A), the TMA HIV-1/HCV assay detection of HCV (B), the HIV-1 discriminatory assay (C), and the HCV discriminatory assay (D). The scale of the y axis is limited to better show the sensitivity and specificity limits.
TABLE 7.
Statistical results of ROC curve analysis
| Assay | Target | Cutoff (RLU)a | Sensitivity (%) | Specificity (%) |
|---|---|---|---|---|
| HIV-1/HCV | HIV-1 | 37,777 | 99.9 | 99.5 |
| 44,718 | 99.9 | 99.6 | ||
| 284,560 | 99.5 | 99.9 | ||
| HIV-1/HCV | HCV | 37,777 | 99.7 | 99.5 |
| 58,972 | 99.7 | 99.7 | ||
| 121,856 | 99.5 | 99.8 | ||
| HIV-1 discriminatory | HIV-1 | 29,026 | 99.9 | 99.5 |
| 38,198 | 99.9 | 99.8 | ||
| 152,941 | 99.5 | 100.0 | ||
| HCV discriminatory | HCV | 48,860 | 99.8 | 99.5 |
| 62,880 | 99.8 | 99.6 | ||
| 132,007 | 99.5 | 99.7 |
Listed are the lower limit cutoff, the cutoff with maximized sensitivity and specificity, and the upper limit cutoff.
Formulas were derived that calculated a floating cutoff value designed to fall within the range of analyte RLUs defined by ROC analysis to achieve ≥99.5% sensitivity and specificity. The cutoff formula for each assay averages the analyte signal from the negative calibrators and then, depending on the assay, adds a fraction of the analyte signal values obtained for the HIV-1 and/or HCV positive calibrators (Table 1).
Table 8 demonstrates the calculated cutoff determined from the 245 runs of the HIV-1/HCV assay that were analyzed by ROC in Fig. 3. An average calculated cutoff of 51,076 RLU was indicated, falling within the ranges of 37,777 to 284,560 RLU and 37,777 to 121,856 RLU determined by ROC analysis to achieve ≥99.5% sensitivity and specificity in the detection of HIV-1 and HCV, respectively (Table 7). The highest and lowest analyte cutoff calculated for these 245 runs also fell within the ranges defined in Table 7. A total of 133 runs were analyzed in the HIV-1 discriminatory assay and were found to have an average calculated cutoff of 50,139 RLU. This cutoff falls within the range of 29,026 to 152,941 RLU defined for the same runs by ROC analysis to achieve ≥99.5% sensitivity and specificity. Even the highest and lowest calculated cutoffs within these runs (Table 8) fall within the defined range.
TABLE 8.
Experimental cutoff RLU values calculated in TMA assay runsa
| Assay | No. of runs | Calculated highest cutoff | Calculated average cutoff | Calculated lowest cutoff |
|---|---|---|---|---|
| HIV-1/HCV | 245 | 74,750 | 51,076 | 37,825 |
| HIV-1 discriminatory | 133 | 75,866 | 50,139 | 34,750 |
| HCV discriminatory | 133 | 83,700 | 63,449 | 48,631 |
Cutoffs were calculated using the formulas defined in Table 1 for each individual run. The average cutoff is the mean of all cutoffs, while the lowest and highest cutoffs are the extremes observed within the run set.
Similarly, the average cutoff of 63,449 RLU determined for the 133 runs carried out with the HCV discriminatory assay falls within the range defined by the ROC plot of 48,860 to 132,007. The highest calculated cutoff of 83,700 RLU lies within this range, and the lowest calculated cutoff of 48,631 RLU lies just below the lower limit defined by the ROC analysis. To maximize the sensitivity of the assay and reduce the chance of false-negative results, the formulas were designed to achieve cutoffs in the lower end of the range defined by ROC plots.
DISCUSSION
The Gen-Probe TMA HIV-/HCV assay, the HIV-1 discriminatory assay, and the HCV discriminatory assay demonstrated specificity of 99.5% and sensitivity of detection of ≥99.5% for samples containing HIV-1 or HCV RNA at greater than or equal to 100 copies/ml. This was achieved with specimens of varied characteristics and was confirmed under conditions of coinfection of the viruses. The sensitivity of the assay is much greater than the established assay goal of 100 copies/ml, as probit analysis predicts 95% detection rates for HIV-1 type B and HCV subtype 1a at 13 and 34 copies/ml, respectively (Table 2). Results from testing NAT standards corroborates these results (P. N. Lelie, A. A. van Drimmelen, H. T. M. Cuypers, S. Best, E. M. Dax, S. L. Stramer, C. Hyland, J. P. Allain, P. Moncharmont, C. Defer, M. Nubling, A. Glauser, M. da Silva Cardoso, J. F. Viret, M. Lankinen, L. Grillner, U. Wirthmuller, J. Coste, V. Schottstedt, and B. Masecar, unpublished data). These limits of detection are lower than those reported previously for HIV-1 or HCV assays using reverse transcription-PCR or branched DNA technology (≈50 to 100 copies/ml) (10, 12). Furthermore, the assay detects the most divergent HIV-1 and HCV subtypes, including HIV-1 group O and HCV 2b, with similar sensitivities (Table 6) (16, 26).
The statistical relationship of assay specificity and sensitivity obtained by ROC plots (Fig. 3; Table 7) demonstrated that all three assays show excellent discrimination between positive and negative samples and a broad cutoff range within which the specificity and sensitivity are greater than or equal to 99.5%, consistent with the proposed claims of the assay. The wide separation of average S/CO and the high degree of sensitivity and specificity of the assay will help ensure accurate detection of HIV-1 or HCV in the blood bank setting or in other diagnostic applications. Furthermore, the use of floating analyte cutoffs reduces the effects of day-to-day variations and allows the calculation of cutoff values which consistently fall within a range of RLU analyte cutoff values defined by ROC analysis which allows the assay to achieve ≥99.5% specificity and sensitivity at 100 copies/ml. Overall, the ROC analysis indicates that in the hands of various users, the assay will consistently determine an analyte cutoff value that allows reproducible sensitivity and specificity of detection of HIV-1 and/or HCV RNA.
The overall specificity and sensitivity goals of 100 copies/ml and ≥99.5% were reached even when potentially interfering donor factors were present in specimens, such as additional viruses, pathogens, or potentially interfering substances, or when samples from patients with autoimmune or other diseases were tested (Table 5). This robustness of the HIV-1/HCV assay, even when testing nontypical specimens, was observed for all three assays. The nature of the target capture technology minimizes interferences, as potentially inhibitory substances are removed in the first step of the assay. In addition, donation factors such as the chosen anticoagulant for sample collection had no effect on the specificity or sensitivity of the assays. This is in contrast to some reports of target recovery issues with certain anticoagulants with other technologies (20) and implies that the TMA assays could readily be applied to additional targets and specimen types in a variety of applications.
The signals of the internal control and calibrators have been optimized to result in clear separation of the internal control from the cutoff (≈8 SD) and distinct analyte S/CO ranges for the three calibrators (Fig. 2). Similar to the information provided by calibrator S/CO distribution, the analyte S/CO values in the HIV-1/HCV assay allow preliminary discrimination between HIV-1 signal and HCV signal (Table 3). Figure 3D, containing HIV-1 RNA only, has an S/CO ratio of 18.46, similar to that of the HIV-1 positive calibrator. Likewise, the S/CO ratios for Fig. 3C, containing HCV RNA only, is 9.66 and essentially the same as that of the HCV positive calibrator signal. In addition, the panels containing both HIV-1 and HCV show the general trend of having an S/CO ratio that is additive to that which is observed for the individual targets. While this does not eliminate the need for using the discriminatory assays to determine whether the sample is infected with HIV-1 or HCV or both, it does allow the user to have some indication of the reactive target.
The multiplex format of the HIV-1/HCV assay has advantages with respect to time and cost compared to assays designed with a single-target format. The time needed to complete initial screening is cut in half due to the ability to amplify multiple targets simultaneously. Also, the batch sample processing method of this assay is highly effective and allows the processing of 200 specimens (either single donor unit or plasma pool sample) and calibrators by a single operator in less than 2 h and completion of 182 test results for each target in 6 h. This is in contrast to another recently presented methodology that describes assay batch sizes that are approximately 10 times smaller (45).
The necessity for NAT of blood donations in addition to or in lieu of some of the currently licensed serological techniques is clear due to the ability of these assays to significantly reduce the window period between infection and detection (3, 15). A negative result by serological screening may be a false-negative result if the donor was recently infected. Due to the additional sensitivity and specificity provided by NAT, genomic detection could readily replace screening for HIV-1 p24 antigen, which has not yielded more than six instances of antigen-positive, antibody-negative donations in over 24 million specimens tested (8, 37). The added sensitivity of NAT would likely have detected these six infected donations and may have identified additional infected specimens. The technology of the HIV-1/HCV assay could also be used instead of techniques such as the Western blot and recombinant strip immunoblot assay, assays currently implemented for confirmation of HIV-1 and HCV antibody-positive specimens. Detailed studies with commercially available seroconversion panels further demonstrate the utility of implementing NAT to replace these HIV-1 and HCV confirmation techniques (22, 43).
The test system discussed in this report is currently used in various blood donation locations under investigational new drug applications approved by the FDA. As of August 2000, approximately 11 million blood donor units had been tested using the HIV-1/HCV and discriminatory assays at the American Red Cross, America's blood centers, and America's independent blood centers. Within this study set, 43 HCV NAT-reactive, seronegative donations and three HIV-1 NAT-reactive, p24 antigen-negative and seronegative donations were identified (37).
In addition to earlier detection of HIV-1 and HCV in blood donations and prevention of infection by transfusion, the use of the HIV-1/HCV assay technology in diagnostic applications would provide increased sensitivity for monitoring viral RNA status. Implementation of the HIV-1 and HCV discriminatory assay technologies would improve the monitoring of patients undergoing treatment by providing lower detection limits than the ones provided by current quantitative assays, aiding in the evaluation of therapeutic endpoints. The ability to detect multiple subtypes of HIV-1 and HCV further supports the worldwide utility of the TMA assays. Furthermore, the technology is readily adaptable to include other targets such as hepatitis A virus, hepatitis B virus, and parvovirus as the need for blood bank screening and diagnostic monitoring of these and/or additional infectious agents evolves.
Acknowledgments
We thank Joseph Baroldi, Amy Broulik, David Cox, Michael Gilker, Kim Greenbaum, Edlyn Halili, Stephanie Hotaling, Alexis Long, Eric Peterson, Alanna Umali, Alyshia Vaughn, and Barbara Weinbaum for expert technical assistance. We also appreciate the assistance of Songbai Wang, who carried out the statistical analysis of the reproducibility specimens, and Tony Eistetter (Chiron Corporation, Emeryville, Calif.), who oversaw the assay specificity testing in normal samples.
This project has been funded in part with federal funds from the National Heart, Lung and Blood Institute under contract no. NHLBI-HB-67130.
REFERENCES
- 1.Abravaya, K., C. Esping, R. Hoenle, J. Gorzowski, R. Perry, P. Kroeger, J. Robinson, and R. Flanders. 2000. Performance of a multiplex qualitative PCR LCx assay for detection of human immunodeficiency virus type 1 (HIV-1) group M subtypes, group O, and HIV-2. J. Clin. Microbiol. 38:716-723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Allain, J. P. 2000. Genomic screening for blood-borne viruses in transfusion settings. Clin. Lab. Haematol. 22:1-10. [DOI] [PubMed] [Google Scholar]
- 3.Allain, J. P. 2000. Will genome detection replace serology in blood screening for microbial agents? Baillieres Best Pract. Res. Clin. Haematol. 13:615-629. [DOI] [PubMed] [Google Scholar]
- 4.Alter, H. J., R. H. Purcell, J. W. Shih, J. C. Melpolder, M. Houghton, Q. L. Choo, and G. Kuo. 1989. Detection of antibody to hepatitis C virus in prospectively followed transfusion recipients with acute and chronic non-A, non-B hepatitis. N. Engl. J. Med. 321:1494-1500. [DOI] [PubMed] [Google Scholar]
- 5.Arnold, L., P. W. Hammond, W. A. Wiese, and N. C. Nelson. 1989. Assay formats involving acridinium-ester-labeled DNA probes. Clin. Chem. 35:1588-1594. [PubMed] [Google Scholar]
- 6.Busch, M. P. 1994. HIV and blood transfusions: focus on seroconversion. Vox Sang. 67(Suppl. 3):13-18. [DOI] [PubMed] [Google Scholar]
- 7.Busch, M. P. 1995. Testing blood donors for HIV: current controversies. Immunol. Investig. 24:147-154. [DOI] [PubMed] [Google Scholar]
- 8.Busch, M. P., and S. H. Kleinman. 2000. Nucleic acid amplification testing of blood donors for transfusion-transmitted infectious diseases: report of the Interorganizational Task Force on Nucleic Acid Amplification Testing of Blood Donors. Transfusion 40:143-159. [DOI] [PubMed] [Google Scholar]
- 9.Chuansumrit, A., W. Varavithya, P. Isarangkura, S. Sirinavin, P. Chiewsilp, S. Tanprasert, and P. Hathirat. 1996. Transfusion-transmitted AIDS with blood negative for anti-HIV and HIV-antigen. Vox Sang. 71:64-65. [DOI] [PubMed] [Google Scholar]
- 10.Collins, M. L., B. Irvine, D. Tyner, E. Fine, C. Zayati, C. Chang, T. Horn, D. Ahle, J. Detmer, L. P. Shen, J. Kolberg, S. Bushnell, M. S. Urdea, and D. D. Ho. 1997. A branched DNA signal amplification assay for quantification of nucleic acid targets below 100 molecules/ml. Nucleic Acids Res. 25:2979-2984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dunne, A. L., and S. M. Crowe. 1997. Comparison of branched DNA and reverse transcriptase polymerase chain reaction for quantifying six different HIV-1 subtypes in plasma. AIDS 11:126-127. [PubMed] [Google Scholar]
- 12.Erice, A., D. Brambilla, J. Bremer, J. B. Jackson, R. Kokka, B. Yen-Lieberman, and R. W. Coombs. 2000. Performance characteristics of the QUANTIPLEX HIV-1 RNA 3.0 assay for detection and quantitation of human immunodeficiency virus type 1 RNA in plasma. J. Clin. Microbiol. 38:2837-2845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Esteban, J. I., R. Esteban, L. Viladomiu, J. C. Lopez-Talavera, A. Gonzalez, J. M. Hernandez, M. Roget, V. Vargas, J. Genesca, and M. Buti. 1989.. Hepatitis C virus antibodies among risk groups in Spain. Lancet ii:294-297. [DOI] [PubMed]
- 14.Fried, M. W., J. O. Draguesku, M. Shindo, L. H. Simpson, S. M. Banks, J. H. Hoofnagle, and A. M. Di Bisceglie. 1993. Clinical and serological differentiation of autoimmune and hepatitis C virus-related chronic hepatitis. Dig. Dis. Sci. 38:631-636. [DOI] [PubMed] [Google Scholar]
- 15.Gallarda, J. L., and E. Dragon. 2000. Blood screening by nucleic acid amplification technology: current issues, future challenges. Mol. Diagn. 5:11-22. [DOI] [PubMed] [Google Scholar]
- 16.Giachetti, C., D. Kolk, J. Dockter, J. Knowlton, R. Wang, H. Hotaling, and S. McDonough. 1998. High throughput assay for sensitive detection of HIV-1 RNA of diverse origins, including type O strains, p. 151-155. In 12th World AIDS Conference. Monduzzi Editore, Bologna, Italy.
- 17.Gretch, D. R. 1997. Diagnostic tests for hepatitis C. Hepatology 26:43S-47S. [DOI] [PubMed]
- 18.Gretch, D. R., C. dela Rosa, R. L. Carithers, Jr., R. A. Willson, B. Williams, and L. Corey. 1995. Assessment of hepatitis C viremia using molecular amplification technologies: correlations and clinical implications. Ann. Intern. Med. 123:321-329. [DOI] [PubMed] [Google Scholar]
- 19.Henrard, D. R., E. Daar, H. Farzadegan, S. J. Clark, J. Phillips, G. M. Shaw, and M. P. Busch. 1995. Virologic and immunologic characterization of symptomatic and asymptomatic primary HIV-1 infection. J. Acquir. Immune Defic. Syndr. Hum. Retrovirol. 9:305-310. [PubMed] [Google Scholar]
- 20.Holodniy, M., L. Mole, and B. Yen-Lieberman. 1995. Comparative stabilities of quantitative human immunodeficiency virus RNA in plasma from samples collected in VACUTAINER CPT, VACUTAINER PPT, and standard VACUTAINER tubes. J. Clin. Microbiol. 33:1562-1566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kacian, D. L., and T. J. Fultz. March1995. Nucleic acid sequence amplification methods. U.S. patent 5,399,491.
- 22.Kolk, D. P., J. Dockter, J. Linnen, M. Ho-Sing-Loy, K. Gillotte-Taylor, S. H. McDonough, L. Mimms, and C. Giachetti. 2002. Significant closure of the HIV-1 and HCV preseroconversion detection windows with a TMA-driven HIV-1/HCV assay. J. Clin. Microbiol. 40:1761-1766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kuo, G., Q. L. Choo, H. J. Alter, G. L. Gitnick, A. G. Redeker, R. H. Purcell, T. Miyamura, J. L. Dienstag, M. J. Alter, and C. E. Stevens. 1989. An assay for circulating antibodies to a major etiologic virus of human non-A, non-B hepatitis. Science 244:362-364. [DOI] [PubMed] [Google Scholar]
- 24.Laffler, T. G., J. J. Carrino, and R. L. Marshall. 1993. The ligase chain reaction in DNA-based diagnosis. Ann. Biol. Clin. (Paris) 51:821-826. [PubMed] [Google Scholar]
- 25.Lee, S. C., A. Antony, N. Lee, J. Leibow, J. Q. Yang, S. Soviero, K. Gutekunst, and M. Rosenstraus. 2000. Improved version 2.0 qualitative and quantitative AMPLICOR reverse transcription-PCR tests for hepatitis C virus RNA: calibration to international units, enhanced genotype reactivity, and performance characteristics. J. Clin. Microbiol. 38:4171-4179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Linnen, J. M., J. M. Gilker, A. Menez, A. Vaughn, A. Broulik, J. Dockter, K. Gillotte-Taylor, K. Greenbaum, D. P. Kolk, L. T. Mimms, and C. Giachetti. 2002. Sensitive detection of genetic variants of HIV-1 and HCV with an HIV-1/HCV assay based on Transcription-Mediated Amplification. J. Virol. Methods 102:139-155. [DOI] [PubMed] [Google Scholar]
- 27.Marshall, R. L., T. G. Laffler, M. B. Cerney, J. C. Sustachek, J. D. Kratochvil, and R. L. Morgan. 1994. Detection of HCV RNA by the asymmetric gap ligase chain reaction. PCR Methods Appl. 4:80-84. [DOI] [PubMed] [Google Scholar]
- 28.McDonough, S. H., C. Giachetti, Y. Yang, D. P. Kolk, E. Billyard, and L. Mimms. 1998. High throughput assay for the simultaneous or separate detection of human immunodeficiency virus (HIV) and hepatitis type C virus (HCV). Infusionsther. Transfusionsmed. 25:164-169. [Google Scholar]
- 29.Mehrpouyan, M., J. E. Bishop, N. Ostrerova, M. Van Cleve, and K. L. Lohman. 1997. A rapid and sensitive method for non-isotopic quantitation of HIV-1 RNA using thermophilic strand displacement amplification and flow cytometry. Mol. Cell. Probes 11:337-347. [DOI] [PubMed] [Google Scholar]
- 30.Morandi, P. A., G. A. Schockmel, S. Yerly, P. Burgisser, P. Erb, L. Matter, R. Sitavanc, and L. Perrin. 1998. Detection of human immunodeficiency virus type 1 (HIV-1) RNA in pools of sera negative for antibodies to HIV-1 and HIV-2. J. Clin. Microbiol. 36:1534-1538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mulder, J., N. McKinney, C. Christopherson, J. Sninsky, L. Greenfield, and S. Kwok. 1994. Rapid and simple PCR assay for quantitation of human immunodeficiency virus type 1 RNA in plasma: application to acute retroviral infection. J. Clin. Microbiol. 32:292-300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nelson, N. C., A. BenCheikh, E. Matsuda, and M. Becker. 1996. Simultaneous detection of multiple nucleic acid targets in a homogeneous format. Biochemistry 35:8429-8438. [DOI] [PubMed] [Google Scholar]
- 33.Nolte, F. S., J. Boysza, C. Thurmond, W. S. Clark, and J. L. Lennox. 1998. Clinical comparison of an enhanced-sensitivity branched-DNA assay and reverse transcription-PCR for quantitation of human immunodeficiency virus type 1 RNA in plasma. J. Clin. Microbiol. 36:716-720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Reichelderfer, P. S., and R. W. Coombs. 1995. Virologic parameters as surrogate markers for clinical outcome in HIV-1 disease: verification, variation, and validation. J. Acquir. Immune Defic. Syndr. Hum. Retrovirol. 10(Suppl. 2):S19-S24. [PubMed]
- 35.Saldanha, J., A. Heath, N. Lelie, G. Pisani, M. Nubling, and M. Yu. 2000. Calibration of HCV working reagents for NAT assays against the HCV international standard. The Collaborative Study Group. Vox Sang. 78:217-224. [DOI] [PubMed] [Google Scholar]
- 36.Sayre, K. R., R. Y. Dodd, G. Tegtmeier, L. Layug, S. S. Alexander, and M. P. Busch. 1996. False-positive human immunodeficiency virus type 1 western blot tests in noninfected blood donors. Transfusion 36:45-52. [DOI] [PubMed] [Google Scholar]
- 37.Stramer, S. L., S. Caglioti, and D. M. Strong. 2000. NAT of the United States and Canadian blood supply. Transfusion 40:1165-1168. [DOI] [PubMed] [Google Scholar]
- 38.Sun, R., W. Schilling, H. Jayakar, J. Ku, J. Wang, M. Rosensraus, and J. Spadoro. 1999. Simultaneous extraction of hepatitis C virus (HCV), hepatitis B virus, and HIV-1 from plasma and detection of HCV RNA by a reverse transcriptase-polymerase chain reaction assay designed for screening pooled units of donated blood. Transfusion 39:1111-1119. [DOI] [PubMed] [Google Scholar]
- 39.Todd, J., C. Pachl, R. White, T. Yeghiazarian, P. Johnson, B. Taylor, M. Holodniy, D. Kern, S. Hamren, and D. Chernoff. 1995.. Performance characteristics for the quantitation of plasma HIV-1 RNA using branched DNA signal amplification technology. J. Acquir. Immune. Defic. Syndr. Hum. Retrovirol. 10(Suppl. 2):S35-S44. [PubMed]
- 40.Vandamme, A. M., J. C. Schmit, S. Van Dooren, K. Van Laethem, E. Gobbers, W. Kok, P. Goubau, M. Witvrouw, W. Peetermans, E. De Clercq, and J. Desmyter. 1996. Quantification of HIV-1 RNA in plasma: comparable results with the nucleic acid sequence-based amplification HIV-1 RNA QT and the AMPLICOR HIV monitor test. J. Acquir. Immune Defic. Syndr. Hum. Retrovirol. 13:127-139. [DOI] [PubMed] [Google Scholar]
- 41.van der Poel, C. L., H. W. Reesink, P. N. Lelie, A. Leentvaar-Kuypers, Q. L. Choo, G. Kuo, and M. Houghton. 1989.. Anti-hepatitis C antibodies and non-A, non-B post-transfusion hepatitis in The Netherlands. Lancet ii:297-298. [DOI] [PubMed]
- 42.van Gemen, B., T. Kievits, P. Nara, H. G. Huisman, S. Jurriaans, J. Goudsmit, and P. Lens. 1993.. Qualitative and quantitative detection of HIV-1 RNA by nucleic acid sequence-based amplification. AIDS 7(Suppl. 2):S107-S110. [DOI] [PubMed]
- 43.Vargo, J. M., K. Smith, C. Knott, S. Wang, C. Fang, S. McDonough, C. Giachetti, S. Caglioti, R. Gammon, D. Gilbert, J. B. Jackson, W. Richards, S. Stramer, and L. Mimms. Clinical specificity and sensitivity of a blood screening assay for detection of HIV-1 and HCV RNA. Transfusion, in press. [DOI] [PubMed]
- 44.Walker, G. T., M. S. Fraiser, J. L. Schram, M. C. Little, J. G. Nadeau, and D. P. Malinowski. 1992. Strand displacement amplification—an isothermal, in vitro DNA amplification technique. Nucleic Acids Res. 20:1691-1696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yu, M. L., W. L. Chuang, C. Y. Dai, S. C. Chen, Z. Y. Lin, M. Y. Hsieh, L. Y. Wang, and W. Y. Chang. 2000. Clinical evaluation of the automated COBAS AMPLICOR HCV MONITOR test version 2.0 for quantifying serum hepatitis C virus RNA and comparison to the quantiplex HCV version 2.0 test. J. Clin. Microbiol. 38:2933-2939. [DOI] [PMC free article] [PubMed] [Google Scholar]