Abstract
Viral hemorrhagic fevers (VHFs) are acute infections with high case fatality rates. Important VHF agents are Ebola and Marburg viruses (MBGV/EBOV), Lassa virus (LASV), Crimean-Congo hemorrhagic fever virus (CCHFV), Rift Valley fever virus (RVFV), dengue virus (DENV), and yellow fever virus (YFV). VHFs are clinically difficult to diagnose and to distinguish; a rapid and reliable laboratory diagnosis is required in suspected cases. We have established six one-step, real-time reverse transcription-PCR assays for these pathogens based on the Superscript reverse transcriptase-Platinum Taq polymerase enzyme mixture. Novel primers and/or 5′-nuclease detection probes were designed for RVFV, DENV, YFV, and CCHFV by using the latest DNA database entries. PCR products were detected in real time on a LightCycler instrument by using 5′-nuclease technology (RVFV, DENV, and YFV) or SybrGreen dye intercalation (MBGV/EBOV, LASV, and CCHFV). The inhibitory effect of SybrGreen on reverse transcription was overcome by initial immobilization of the dye in the reaction capillaries. Universal cycling conditions for SybrGreen and 5′-nuclease probe detection were established. Thus, up to three assays could be performed in parallel, facilitating rapid testing for several pathogens. All assays were thoroughly optimized and validated in terms of analytical sensitivity by using in vitro-transcribed RNA. The ≥95% detection limits as determined by probit regression analysis ranged from 1,545 to 2,835 viral genome equivalents/ml of serum (8.6 to 16 RNA copies per assay). The suitability of the assays was exemplified by detection and quantification of viral RNA in serum samples of VHF patients.
Viral hemorrhagic fever (VHF) is a clinical syndrome caused by different viruses. VHF agents belong to the Filoviridae (Marburg virus [MBGV] and Ebola virus [EBOV]), Arenaviridae (Lassa virus [LASV] and Junin, Machupo, Sabia, and Guanarito viruses), Bunyaviridae (Crimean-Congo hemorrhagic fever virus [CCHFV], Rift Valley fever virus [RVFV], and Hanta viruses), and Flaviviridae (yellow fever virus [YFV] and dengue virus [DENV]) (27). The natural reservoirs of these viruses are arthropods, ticks, and rodents; the reservoir of filoviruses is not known. Infections by VHF viruses are associated with a wide spectrum of clinical manifestations such as diarrhea, myalgia, cough, headache, pneumonia, encephalopathy, and hepatitis (4, 18, 19, 25, 33, 37, 40, 41). Hemorrhage is the characteristic manifestation, although nonhemorrhagic infections are also common. VHF is often fatal in spite of modern intensive care. Filoviruses, arenaviruses, and CCHFV are of particular relevance because they can be transmitted from human to human, thus causing epidemics with high mortality rates (3, 13, 24). Nearly all VHF agents are endemic in tropical or subtropical regions. However, infections can also occur outside these regions, for example, in returning travelers (38). Very recently, LASV infections have been imported to Germany (17, 18), The Netherlands (2), and the United Kingdom (1). In 1999 a patient died from imported yellow fever in Germany (41), and there are several earlier reports on the import of VHF infections into Europe, Japan, or North America (19, 42).
In the absence of bleeding or organ manifestation, a VHF is clinically difficult to diagnose, and the various etiologic agents can hardly be distinguished by clinical tests. The range of possible agents can be narrowed down by the travel history. The definitive diagnosis of a VHF relies mainly on laboratory testing. A clinical VHF suspect must be rapidly excluded or the causative virus must be identified to initiate specific treatment, if possible, as well as appropriate case management such as isolation measures or tracking of contact persons.
PCR has been successfully applied in the diagnosis of VHF (7, 8, 22, 28, 30, 31, 36). However, most of the published assays are time-consuming, as they include a separate cDNA synthesis step prior to PCR, agarose gel analysis of PCR products, and in some instances a second round of nested amplification or Southern hybridization. PCRs for different pathogens have to be run assay by assay due to differences in cycling conditions, which complicates broad-range testing in a short period of time. Moreover, post-PCR processing or nested PCR steps included in currently used assays increase the risk of false-positive results due to carryover contamination (26). Very recently, real-time PCR assays for detection of RVFV and DENV and discrimination of EBOV subtypes have been published (5, 14, 15).
To facilitate reliable testing for a wide range of VHF agents in a short period of time, we have established a PCR system comprising six one-step, real-time reverse transcription-PCR (RT-PCR) assays for MBGV and EBOV (MBGV/EBOV), LASV, CCHFV, RVFV, DENV, and YFV. For rapid qualitative testing, all PCRs can be performed in two runs on a LightCycler instrument with two universal cycling conditions. Each assay can be used separately for quantification of viral RNA. The establishment and validation of these PCRs are described, as well as their application for detection and quantification of viral RNA in patient samples.
MATERIALS AND METHODS
Virus propagation.
MBGV/EBOV, LASV, CCHFV, RVFV, DENV, and YFV were grown on Vero cells in the biosafety level 3 and 4 facilities of the Bernhard-Nocht-Institute. Supernatants were taken 3 to 5 days after virus inoculation.
Serum samples.
Serum samples were from one acute and one convalescent case of Ebola hemorrhagic fever (Gulu, Uganda, October 2000 [3]), two cases of imported Lassa fever (Würzburg, Germany, January 2000 [17], and Leiden, The Netherlands, March 2000 [2]), an acute case of Crimean-Congo hemorrhagic fever (Prizren, Kosovo, May 2000), an imported case of yellow fever (Berlin, Germany, August 1999 [41]), and an imported case of acute secondary dengue fever with hemorrhagic diathesis (Hamburg, Germany, August 2001).
RNA preparation.
Human sera and cell culture supernatants were cleared by centrifugation in a table-top centrifuge at 10,000 × g for 10 min. Viral RNA was prepared from 140 μl of the cell-free fluid by using the Qiamp viral RNA kit (Qiagen, Hilden, Germany) without modification. RNA was eluted in 50 μl.
Oligonucleotide design.
All sequences available from the EMBL, GenBank, and DDBJ databases (accessed autumn 2000) were included in the search for possible primer binding regions. Sequence alignments to identify conserved regions were done using the BLAST algorithm at the National Center for Biotechnology Information website. Primers and corresponding 5′-nuclease detection probes were designed with Primer Express software (Applied Biosystems, Weiterstadt, Germany). Several possible primers and probes were synthesized (Tib Molbiol, Berlin, Germany). All 5′-nuclease probes were labeled with 6-carboxyfluorescein at the 5′ end and with 6-carboxy-N,N,N′,N′-tetramethylrhodamine at the 3′ end. The 3′ end of each probe was phosphorylated to prevent elongation during PCR. The final oligonucleotide sequences are given in Table 1.
TABLE 1.
Primers and 5′-nuclease probes
| Virus (GenBank accession no.) | Sense primer, antisense primer, probe (sequence [position]) | Genomic target region | Amplicon length (bp) | Reference |
|---|---|---|---|---|
| EBOV (AF086833)/MBGV (MRVMBGL) | Filo A (ATCGGAATTTTTCTTTCTCATT [13213-13234]), Filo B (ATGTG GTGGGTTATAATAATCACTGACATG [13631-13601]) | L gene | 419 | 36 |
| LASV (J04324) | 36E2 (ACCGGGGATCCTAGGCATTT [5-24]), 80F2 (ATATAATGATGA CTGTTGTTCTTTGTGCA [339-311]) | GPC gene | 334 | 7 |
| CCHFV (U88413) | CCS (ATGCAGGAACCATTAARTCTTGGGA [351-375]), CCAs (CTAAT CATATCTGACAACATTTC plus CTAATCATGTCTGACAGCATCTC, 1:1 [579-557]) | NP gene | 228 | This article |
| RVFV (AF134508) | RVS (AAAGGAACAATGGACTCTGGTCA [349-371]), RVAs (CACTTCT TACTACCATGTCCTCCAAT [443-417]), RVP (AAAGCTTTGATATCT CTCAGTGCCCCAA [388-416]) | G2 gene | 94 | This article |
| YFV (NC_002031) | YFS (AATCGAGTTGCTAGGCAATAAACAC [29-64]), YFAs (TCCCTGA GCTTTACGACCAGA [141-122]), YFP (ATCGTTCGTTGAGCGATTAG CAG [81-103]) | 5′ noncoding region | 112 | This article |
| DENV (M87512) | DenS (GGATAGACCAGAGATCCTGCTGT [10615-10636]), DenAs (CAT TCCATTTTCTGGCGTTC plus CAATCCATCTTGCGGCGCTC, 1:1 [10694-10675]), DenP (CAGCATCATTCCAGGCACAG [10656-10675]) | 3′ noncoding region | 79 | This article |
RT-PCR conditions.
The one-step RT-PCR system combining Superscript reverse transcriptase with Platinum Taq-polymerase (Life Technologies, Karlsruhe, Germany) was used in 5′-nuclease and SybrGreen assays. The 20-μl assay contained 10 μl of reaction mix provided with the kit (including the basic level of MgSO4), 40 ng of bovine serum albumin (Sigma, Munich, Germany) per μl, and 2 μl of RNA. The reaction capillaries for MBGV/EBOV, LASV, and CCHFV RT-PCR were prepared as follows: 1 μl of SybrGreen stock solution (Roche Molecular Biochemicals, Mannheim, Germany) diluted 1:5,000 in dimethyl sulfoxide was centrifuged to the bottom of the LightCycler capillary and lyophilized for 40 min without prior freezing. The concentrations of additional MgSO4, sense and antisense primers, and 5′-nuclease probe (only for RVFV, DENV, and YFV) are listed for each virus in Table 2.
TABLE 2.
RT-PCR conditions
| Reaction component (concn unit) | Concn for:
|
|||||
|---|---|---|---|---|---|---|
| MBGV/ EBOV | LASV | CCHFV | RVFV | DENV | YFV | |
| Additional MgSO4 (mM)a | 0.75 | 2.5 | 3.25 | 2.5 | 3.25 | 2.5 |
| Sense primer (μM) | 1 | 0.3 | 0.25 | 1 | 0.15 | 0.25 |
| Antisense primer (μM) | 1 | 0.2 | 0.5b | 1 | 0.3b | 0.25 |
| Probe (μM) | 0.2 | 0.2 | 0.2 | |||
The reaction mix contains an MgSO4 basis concentration of 1.2 mM which is not included.
Consists of a 1:1 molar ratio of two primers.
Universal cycling profiles for qualitative RT-PCR.
RT-PCR of MBGV/EBOV, LASV, and CCHFV with SybrGreen detection involved the following steps: reverse transcription at 50°C for 20 min; initial denaturation at 95°C for 5 min; 10 precycles with 95°C for 5 s, 60°C for 5 s with a temperature decrease of 1°C per cycle, and 72°C for 25 s; and 40 cycles with 95°C for 5 s, 56°C for 10 s, and 72°C for 30 s. Since SybrGreen intercalates nonspecifically into PCR products (32), a melting curve analysis was performed following PCR to identify the correct product by its specific melting temperature. Melting curve analysis included 95°C for 5 s, 65°C for 15 s, and heating to 95°C at a rate of 0.1°C/s with continuous reading of fluorescence. RT-PCR of RVFV, DENV, and YFV with 5′-nuclease probe detection (20, 29) involved reverse transcription at 45°C for 30 min, initial denaturation at 95°C for 5 min, and 45 cycles with 95°C for 5 s and 57°C for 35 s. Fluorescence was read at the combined annealing-extension step at 57°C.
Cycling profiles for quantification of viral RNA.
For quantification of MBGV/EBOV, LASV, and CCHFV RNAs, PCR was done essentially as described for qualitative detection, except that precycling was followed by 40 cycles with 95°C for 5 s, 56°C for 10 s, 72°C for 25 s, and a fluorescence read step at 79°C (MBGV/EBOV and CCHFV) or at 83°C (LASV) for 15 s. Quantification by 5′-nuclease probe detection (DENV, YFV, and RVFV) was done as described for qualitative detection.
In vitro transcription.
The diagnostic target regions were amplified by RT-PCR using viral RNA prepared from cell culture supernatant and cloned into T7 polymerase expression vector pT-Adv (Clontech, Heidelberg, Germany). The complete inserts including the T7 promoter were amplified with vector-specific primers. The PCR product was purified and then in vitro transcribed and DNase digested using the MegaScript T7 in vitro transcription kit (Ambion, Austin, Tex.). The RNA was purified with RNeasy columns (Qiagen) and quantified spectrophotometrically.
Statistical analysis.
The ≥95% detection limits of the RT-PCRs were determined by probit analysis (12) using the Statgraphics plus 5.0 software package (Statistical Graphics Inc.). The experimental input data for this nonlinear regression model were the different test concentrations of RNA (0 to 8,000 RNA copies/ml) and the corresponding proportions of positive results after replicate (n = 6) PCRs.
RESULTS
Design of primers and 5′-nuclease detection probes.
Real-time detection of PCR products by 5′-nuclease technology is highly specific. A fluorescent dye-labeled detection probe hybridizes with the specific PCR product and is cleaved during PCR by the 5′-exonuclease activity of Taq polymerase, which activates the dye (20, 29). Primer-probe combinations for this technology were designed for YFV, DENV, and RVFV (Table 1). Conserved sequences were used as target sites for computer-aided design of several possible sense and antisense primers as well as hybridization probes for 5′-nuclease detection. Novel primers were also designed for CCHFV, while no conserved binding site for a 5′-nuclease probe could be found. The genomic target regions and the optimal primer-probe combinations as determined experimentally (see below) are shown in Fig. 1. None of the primer sequences showed homologies above the statistical threshold to other database sequences by BLAST searching. Novel primers were not designed for MBGV/EBOV and LASV, because these viruses were found to be too variable for design of 5′-nuclease probes and published primers (7, 36) largely matched the above-described criteria.
FIG.1.
Target regions of newly designed primers and 5′-nuclease probes. All sequences of the target regions available at GenBank were aligned. Each type of sequence variation within this region is shown once, with its frequency on the right. GenBank accession numbers of representative sequences for each type of variation are indicated on the left. Primer and probe sequences as well as their designations are given above the alignment. The antisense primer sequences are reverse complement (rc). If two primers were chosen for one binding site, both primer variants are shown, with variable positions underlined. Boldface nucleotides in the DENV alignment indicate positions which are shared by the binding site of the sense strand probe and the antisense primer. An asterisk indicates the end of a GenBank entry, and a slash indicates a gap in the alignment.
Establishment of one-step RT-PCR with real-time SybrGreen detection.
Since probes could not be designed for MBGV/EBOV, LASV, and CCHFV, the suitability of the DNA-intercalating dye SybrGreen I for real-time detection of PCR products was tested. A SybrGreen concentration of 0.001% (vol/vol) of the stock solution (the manufacturer does not indicate the concentration of the stock solution), as required for detection of PCR products in the LightCycler, did not interfere with the activity of Taq DNA polymerase. However, this concentration inhibited all one-step RT-PCR assays tested (Brilliant RT-PCR system [Stratagene], Thermoscript and Superscript RT-PCR systems [Life Technologies], and a combination of Superscript reverse transcriptase [Life Technologies] with AmpliTaqGold DNA polymerase [Applied Biosystems]). Further experiments revealed that only the reverse transcription step was inhibited. To overcome this inhibitory effect, we wondered whether it might be possible to initially separate the dye from the reaction but to enable its release during PCR. To this end, 1 μl of SybrGreen stock solution diluted 1:5,000 in dimethyl sulfoxide was centrifuged to the bottom of the LightCycler reaction capillaries and lyophilized. One-step RT-PCRs running with this simple modification were as sensitive as those running without dye (Fig. 2), whereas real-time detection of the PCR product was possible. Prepared capillaries were stored for up to 1 week at room temperature without affecting RT-PCR performance.
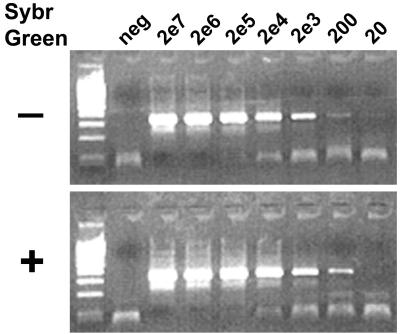
FIG. 2.
Sensitivity of one-step RT-PCR in the absence (−) and presence (+) of SybrGreen. The dye was lyophilized at the bottom of the capillary. In vitro-transcribed LASV RNA was amplified in a LightCycler as described in Materials and Methods. The PCR products were visualized in ethidium bromide-stained agarose gel. Input RNA copy numbers are indicated above the lanes. The negative control (neg) contained RNA extracted from negative plasma.
SybrGreen intercalates independent of the sequence into double-stranded DNA (32). To prevent detection of nonspecific primer-dimer artifacts, a separate fluorescence read step was added at the end of each cycle. The temperature at this reading step exceeded the melting point of short primer-dimers (i.e., they are single stranded and thus are not a target for SybrGreen) but not that of the specific PCR product. By using this reading procedure, the nonspecific signal that was initially observed in the negative controls (data not shown) could be completely eliminated (see negative controls in Fig. 4, left panels). The optimum reading temperatures as determined for each assay are stated in Materials and Methods.
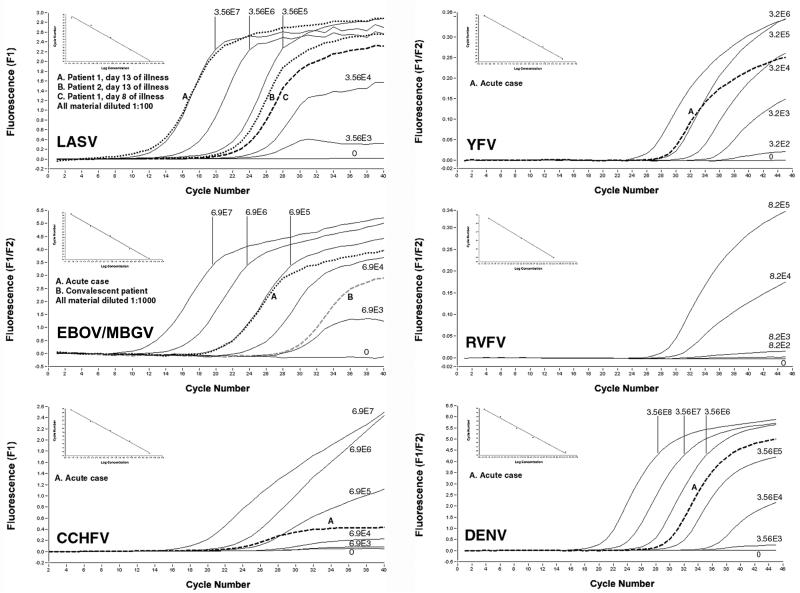
FIG. 4.
Detection and quantification of VHF pathogens in clinical specimens. RNA was prepared from serum samples of VHF patients (see Materials and Methods for details on the patients). Sera from patients with Ebola fever were diluted 1:1,000 to overcome inhibition, and those from Lassa fever patients were diluted 1:100 to save serum. A log10 dilution series of in vitro-transcribed RNA was amplified as a standard; the copy number per milliliter of plasma is shown at the curves. The graphs show the real-time detection of the specific PCR products by fluorescence (y axis) (F1 detection wavelength or F1/F2 ratio) dependent on the PCR cycle number (x axis). Despite the low plateau level in some reactions with low template copy numbers, the signals are clearly positive in both melting curve analysis and agarose gel analysis. The insert in the upper left corner of each graph depicts the standard curve (x axis, RNA concentration; y axis, cycle number at first detection of PCR product). For RVFV, no clinical sample was available (the graph is shown to illustrate the technical performance of the PCR). The specimens from the patients with Ebola hemorrhagic fever were diluted 1:1,000 to reverse RT-PCR inhibition, which probably resulted from extensive hemolysis.
Optimization of reaction and cycling conditions.
In pilot experiments, the Superscript II RT/Platinum Taq polymerase one-step RT-PCR kit (Life Technologies) was found to be more sensitive and robust than other enzyme combinations tested. It was used as a basis for all six PCR assays. Designed primers and probes were tested in all possible combinations, and the most sensitive pair of primers was chosen for further optimization. PCR products generated with these primers were cloned and in vitro transcribed. About 103 transcript copies were used as a template in subsequent titration experiments. In order to most accurately determine the optimal reaction conditions, various concentrations (≥4 concentrations) of one component were simultaneously tested versus various concentrations (≥4 concentrations) of another component (4 × 4 = 16 reactions) (16). The following titrations were performed for each PCR: sense versus antisense primer concentration, 5′-nuclease probe versus total primer concentration, and Mg2+ versus total primer concentration. Criteria for optimal conditions were both the intensity of fluorescence at the end of the reaction (as high as possible) and the number of cycles needed before a signal became detectable (as low as possible). The optimum concentrations of these four parameters are summarized in Table 2.
The one-step RT-PCR SybrGreen I kit (Roche Molecular Biochemicals), a preassembled reaction mix containing Tth polymerase and SybrGreen, underwent a similar optimization process with LASV RNA as a template. However, even after extensive optimization the sensitivity was lower than that with the Superscript II/Platinum kit and immobilized dye.
In order to shorten the total running time if several assays have to be performed in parallel, two universal RT-PCR profiles were established (see Materials and Methods). One profile was optimized for simultaneous processing of YFV, RVFV, and DENV PCRs with 5′-nuclease probe detection, and the other profile was optimized for MBGV/EBOV, LASV, and CCHFV PCRs with qualitative SybrGreen detection.
Analytical sensitivity and specificity.
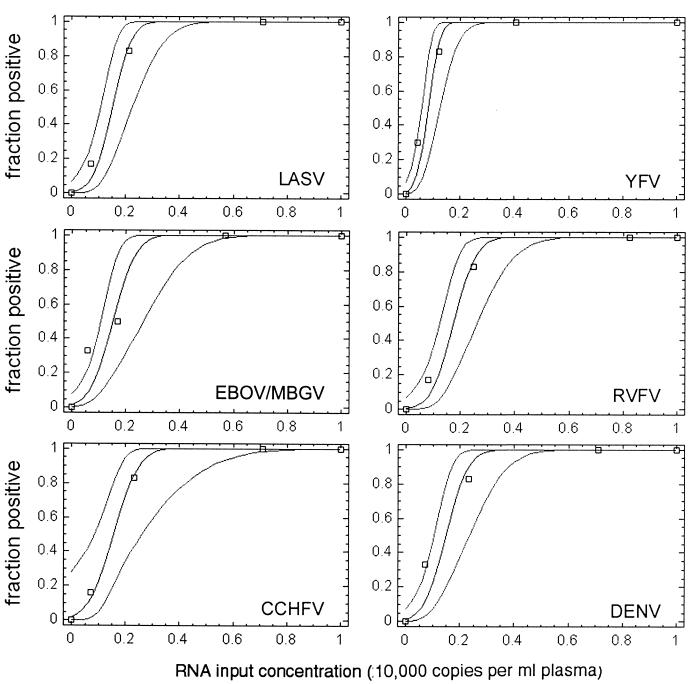
For statistically precise determination of the detection limit, four different concentrations of RNA transcript were spiked into human plasma prior to RNA preparation and tested in six replicates (24 test reactions per PCR assay). The numbers of positive and negative reactions obtained with each of the four RNA concentrations were subjected to probit regression analysis (12) to calculate the probability of achieving a positive result at any RNA concentration within the range of 0 to 10,000 copies per ml of plasma (Fig. 3). Virus genome equivalents (geq) per milliliter of plasma which could be detected with ≥95% probability were as follows: LASV, 2,445 geq/ml (95% confidence interval, 1,848 to 3,903); MBGV/EBOV, 2,647 geq/ml (1,887 to 4,964); CCHFV, 2,779 geq/ml (2,021 to 6,017); YFV, 1,545 geq/ml (1,003 to 2,207); RVFV, 2,835 geq/ml (2,143 to 4,525); and DENV, 2,550 geq/ml (1,871 to 4,212). These detection limits corresponded to 8.6 to 16 geq per reaction, taking into account that RNA was prepared from 140 μl of plasma and 1/25 of the RNA preparation was used as the template and assuming 100% efficiency in RNA preparation.
FIG. 3.
Determination of RT-PCR detection limits by probit regression analysis. Negative human plasma was spiked with defined amounts of in vitro-transcribed RNA and prepared three times in parallel. Each RNA preparation was amplified two times in parallel, resulting in six replicate RT-PCRs per RNA test concentration. The experimentally determined fraction of positive reactions (npositive/ntested) (y axis) at the corresponding RNA test concentration (copies per milliliter of plasma) (x axis) is shown by squares. The calculated regression curves (middle curve) indicate the probability (y axis) of obtaining a positive result at any RNA concentration. The 95% confidence intervals for this probability are shown by curves at the left and right of the middle curve. The RNA concentration at which a positive result is achieved with a probability of 95% is given in the text.
Specificity of the VHF assays was evaluated by testing samples containing heterologous nucleic acid. None of the assays amplified human immunodeficiency virus type 1, hepatitis C virus, hepatitis B virus, herpes simplex virus type 1, cytomegalovirus, Modoc virus, Mycobacterium tuberculosis, Mycobacterium leprae, Borrelia spp., Leptospira spp., Neisseria spp., Plasmodium spp., or Leishmania spp.
Suitability in diagnostics and quantification of virus in VHF patient samples.
The PCR set was used for about 30 suspected cases of VHF. As an inhibition control, in vitro-transcribed EBOV RNA was spiked into an aliquot of each RNA preparation and amplified in parallel with the test samples. Purification of RNA and performance of all six assays in two LightCycler runs was usually accomplished in less than 6 h. We have not encountered any false-positive RT-PCR results due to contamination.
To exemplify the suitability of the assays and to obtain first data on virus RNA levels in VHF patients, viral RNA concentrations in serum samples from patients with Ebola hemorrhagic fever, Lassa fever, Crimean-Congo hemorrhagic fever, yellow fever, and dengue fever were measured (Fig. 4). The samples were amplified in parallel with in vitro-transcribed RNA of the corresponding virus as a quantification standard. All assays showed an excellent correlation (r ≥ 0.9) between cycle number and RNA concentration over a broad dynamic range (Fig. 4, standard curves). The patients were found to have high virus RNA concentrations in serum: one patient with Lassa fever had 7 ×106 and 4 ×109 geq/ml at days 4 and 13 of illness, respectively, and the other had 9 × 106 geq/ml on day 13 of illness; the patient with acute Ebola fever had 6.9 × 108 geq/ml, and the convalescent Ebola fever patient had 7 × 106 geq/ml; the patient with Crimean-Congo fever had 7.7 × 105 geq/ml; the patient with yellow fever had 4 × 105 geq/ml; and the patient with dengue fever had 8 × 105 geq/ml (Fig. 4). Thus, virus concentrations in patients in the acute phase of VHF were orders of magnitudes above the 95% detection limits of the assays.
DISCUSSION
This report describes the establishment of a PCR system, comprising six real-time, one-step RT-PCR assays, for rapid diagnosis of important VHF agents. Both specific 5′-nuclease probes and a newly developed SybrGreen method were used for real-time detection. The assays were thoroughly validated with respect to their analytical sensitivity by using in vitro-transcribed RNA. Their suitability was exemplified by detection and quantification of VHF agents in human serum samples.
A major problem in designing oligonucleotides for diagnostic PCRs of RNA viruses is the considerable genetic variability of these viruses. In order to reduce the risk of PCR failure due to mismatches in primer binding sites, as many sequences as possible, covering known genotypes or phylogenetic lineages, must be included in primer design. This strategy was followed in designing novel oligonucleotides for the assays described in this study. The primers are based on 30 to 90 sequences of a conserved region in each virus. Therefore, it is assumed that these novel primers and probes will bind with high likelihood to unknown virus isolates. They may be further optimized when novel sequences become available in the future.
When PCR is to be used in a diagnostic setting, a thorough validation of the analytical performance, especially sensitivity, is mandatory. Standardized virus stocks with defined numbers or units of genomes per milliliter are the test material of choice. They are available for widely used PCR assays detecting prevalent viruses, such as hepatitis C virus, hepatitis B virus, or human immunodeficiency virus type 1 (9, 21, 35), but not for VHF pathogens. For less common viruses, PCR sensitivity has been determined by comparison with classical virological quantification methods (22). However, the ratio between infectious units and viral RNA copies depends on the fraction of noninfectious RNA-containing virus such as defective interfering particles and will predictably differ between various viruses or stocks of the same virus. Therefore, PFU do not provide precise information on the concentration of RNA-containing particles in a virus stock. We have circumvented this problem by using quantified, in vitro-transcribed RNA spiked into plasma to determine the analytical sensitivity. Although replicate testing of defined RNA concentrations followed by statistical probit regression analysis is laborious, it is one of the most accurate methods to determine the analytical sensitivity of PCR (9, 12, 39). The calculation that 9 to 16 RNA copies per reaction can be detected with a probability of 95% indicates that all RT-PCRs are well optimized. These copy numbers are close to the detection limits of commercial PCR assays for hepatitis C virus or human immunodeficiency virus (43).
There are presently no data available on viral RNA concentrations in patients with VHF. Using the established assays, we have analyzed a few sera of patients with VHF. The RNA concentrations in all patients were orders of magnitudes above the detection limits, suggesting that the assays are sufficiently sensitive to diagnose VHF during the acute febrile phase. To define the clinical sensitivity of the assays more precisely would require testing of well-characterized serum panels from patients with different courses of VHF and in different stages of the disease. However, such panels are not available. The possibility of quantifying viral RNA may prove to be useful in therapy monitoring and to estimate the prognosis.
The specificity of the assays was evaluated by testing specimens containing nucleic acids of a variety of common pathogens. However, the negative results with these samples do not completely exclude the possibility of nonspecific amplification in other samples. While in the 5′-nuclease only the specific PCR product is detected due to the sequence-specific hybridization of the probe, the melting point in the SybrGreen assays is much less specific. Therefore, even if the melting point of a PCR product corresponds to that of the positive control, the presence of the specific product needs to be verified by agarose gel analysis and sequencing. In this regard, it should be mentioned that the assays were validated only for serum, which contains a low background level of heterologous nucleic acid.
Especially when the detection of several possible pathogens is needed in short periods of time, as is usually the case in VHF diagnostics, it is necessary to perform tests in parallel rather than sequentially. The simultaneous detection of various pathogens has often been addressed by multiplex PCR. Up to six different sequences were detected (23). However, the more sequences are targeted in one PCR, the more difficult it becomes to achieve optimal reaction conditions for each individual pair of primers. Nonspecific interference of oligonucleotides is generally thought to be an additional limitation of multiplex PCR (11). Low sensitivity has been observed in several multiplex PCRs and was overcome by nested PCR (6, 10, 34). This, however, increases the risk of contamination and prolongs the processing time. For these reasons we have chosen to detect the pathogens in assays which are optimized separately with regard to reaction components but which share common thermal cycling conditions. This strategy facilitates running of different assays in parallel but only marginally increases the hands-on time compared with that in multiplex PCR. Furthermore, the range of agents tested for can easily be adapted according to the travel history of the patient. MBGV/EBOV, LASV, and CCHFV, which can cause epidemics by human-to-human transmission, are tested together in the first LightCycler run. Thus, the first results on these most important viruses are available within 3 h after arrival of the samples in the laboratory. However, definitive diagnosis or exclusion of a VHF should not be based on a single PCR result. The described set of PCRs must be embedded in a diagnostic procedure involving additional tests, such as virus isolation in cell culture, antigen enzyme-linked immunosorbent assay, confirmatory PCRs, or detection of virus-specific immunoglobulin M and immunoglobulin G by enzyme-linked immunosorbent assay or immunofluorescence.
Acknowledgments
We thank Nadine Peters for excellent technical assistance and Matthias Grade, Clinical Department of the Bernhard-Nocht-Institute, for supplying serum samples from Ugandan patients with Ebola hemorrhagic fever.
This work was supported by grants E/B31E/M0171/M5916 and E/B41G/1G309/1A403 from the Bundesamt für Wehrtechnik und Beschaffung. The Bernhard-Nocht-Institute is supported by the Bundesministerium für Gesundheit and the Freie und Hansestadt Hamburg.
REFERENCES
- 1.Anonymous. 2000. Lassa fever imported to England. Commun. Dis. Rep. CDR Wkly. 10:99.. [PubMed] [Google Scholar]
- 2.Anonymous. 2000. Lassa fever, imported case, Netherlands. Wkly. Epidemiol. Rec. 75:265.. [PubMed] [Google Scholar]
- 3.Anonymous. 2001. Outbreak of Ebola haemorrhagic fever, Uganda, August 2000-January 2001. Wkly. Epidemiol. Rec. 76:41-46. [PubMed] [Google Scholar]
- 4.Bwaka, M. A., M. J. Bonnet, P. Calain, R. Colebunders, A. De Roo, Y. Guimard, K. R. Katwiki, K. Kibadi, M. A. Kipasa, K. J. Kuvula, B. B. Mapanda, M. Massamba, K. D. Mupapa, J. J. Muyembe-Tamfum, E. Ndaberey, C. J. Peters, P. E. Rollin, and E. Van den Enden. 1999.. Ebola hemorrhagic fever in Kikwit, Democratic Republic of the Congo: clinical observations in 103 patients. J. Infect. Dis. 179(Suppl. 1):S1-S7. [DOI] [PubMed]
- 5.Callahan, J. D., S. J. Wu, A. Dion-Schultz, B. E. Mangold, L. F. Peruski, D. M. Watts, K. R. Porter, G. R. Murphy, W. Suharyono, C. C. King, C. G. Hayes, and J. J. Temenak. 2001. Development and evaluation of serotype- and group-specific fluorogenic reverse transcriptase PCR (TaqMan) assays for dengue virus. J. Clin. Microbiol. 39:4119-4124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cassinotti, P., and G. Siegl. 1998. A nested-PCR assay for the simultaneous amplification of HSV-1, HSV-2, and HCMV genomes in patients with presumed herpetic CNS infections. J. Virol. Methods 71:105-114. [DOI] [PubMed] [Google Scholar]
- 7.Demby, A. H., J. Chamberlain, D. W. Brown, and C. Clegg. 1994. Early diagnosis of Lassa fever by reverse transcription PCR. J. Clin. Microbiol. 32:2898-2903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Deubel, V., M. Huerre, G. Cathomas, M. T. Drouet, N. Wuscher, B. Le Guenno, and A. F. Widmer. 1997. Molecular detection and characterization of yellow fever virus in blood and liver specimens of a non-vaccinated fatal human case. J. Med. Virol. 53:212-217. [PubMed] [Google Scholar]
- 9.Drosten, C., M. Weber, E. Seifried, and W. K. Roth. 2000. Evaluation of a new PCR assay with competitive internal control sequence for blood donor screening. Transfusion 40:718-724. [DOI] [PubMed] [Google Scholar]
- 10.Ellis, J. S., D. M. Fleming, and M. C. Zambon. 1997. Multiplex reverse transcription-PCR for surveillance of influenza A and B viruses in England and Wales in 1995 and 1996. J. Clin. Microbiol. 35:2076-2082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Elnifro, E. M., A. M. Ashshi, R. J. Cooper, and P. E. Klapper. 2000. Multiplex PCR: optimization and application in diagnostic virology. Clin. Microbiol. Rev. 13:559-570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Finney, D. J. 1971. Probit analysis, 3rd ed. Cambridge University Press, Cambridge, United Kingdom.
- 13.Fisher-Hoch, S. P., O. Tomori, A. Nasidi, G. I. Perez-Oronoz, Y. Fakile, L. Hutwagner, and J. B. McCormick. 1995. Review of cases of nosocomial Lassa fever in Nigeria: the high price of poor medical practice. Br. Med. J. 311:857-859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Garcia, S., J. M. Crance, A. Billecocq, A. Peinnequin, A. Jouan, M. Bouloy, and D. Garin. 2001. Quantitative real-time PCR detection of Rift Valley fever virus and its application to evaluation of antiviral compounds. J. Clin. Microbiol. 39:4456-4461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gibb, T. R., D. A. Norwood, Jr., N. Woollen, and E. A. Henchal. 2001. Development and evaluation of a fluorogenic 5′ nuclease assay to detect and differentiate between Ebola virus subtypes Zaire and Sudan. J. Clin. Microbiol. 39:4125-4130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Grunwald, H. W., F. Rosner, and Y. Hirshaut. 1978. Two dimensional view of combination chemotherapy in vitro. Am. J. Hematol. 4:35-46. [DOI] [PubMed] [Google Scholar]
- 17.Günther, S., P. Emmerich, T. Laue, O. Kühle, M. Asper, A. Jung, T. Grewing, J. ter Meulen, and H. Schmitz. 2000. Imported Lassa fever in Germany: molecular characterization of a new Lassa virus strain. Emerg. Infect. Dis. 6:466-476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Günther, S., B. Weisner, A. Roth, T. Grewing, M. Asper, C. Drosten, P. Emmerich, J. Petersen, M. Wilczek, and H. Schmitz. 2001. Lassa fever encephalopathy: Lassa virus in cerebrospinal fluid but not in serum. J. Infect. Dis. 184:345-349. [DOI] [PubMed] [Google Scholar]
- 19.Hirabayashi, Y., S. Oka, H. Goto, K. Shimada, T. Kurata, S. P. Fisher-Hoch, and J. B. McCormick. 1988. An imported case of Lassa fever with late appearance of polyserositis. J. Infect. Dis. 158:872-875. [DOI] [PubMed] [Google Scholar]
- 20.Holland, P. M., R. D. Abramson, R. Watson, and D. H. Gelfand. 1991. Detection of specific polymerase chain reaction product by utilizing the 5′-3′ exonuclease activity of Thermus aquaticus DNA polymerase. Proc. Natl. Acad. Sci. USA 88:7276-7280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Holmes, H., C. Davis, A. Heath, I. Hewlett, and N. Lelie. 2001. An international collaborative study to establish the 1st international standard for HIV-1 RNA for use in nucleic acid-based techniques. J. Virol. Methods 92:141-150. [DOI] [PubMed] [Google Scholar]
- 22.Ibrahim, M. S., M. J. Turell, F. K. Knauert, and R. S. Lofts. 1997. Detection of Rift Valley fever virus in mosquitoes by RT-PCR. Mol. Cell. Probes 11:49-53. [DOI] [PubMed] [Google Scholar]
- 23.Kehl, S. C., K. J. Henrickson, W. Hua, and J. Fan. 2001. Evaluation of the Hexaplex assay for detection of respiratory viruses in children. J. Clin. Microbiol. 39:1696-1701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Khan, A. S., G. O. Maupin, P. E. Rollin, A. M. Noor, H. H. Shurie, A. G. Shalabi, S. Wasef, Y. M. Haddad, R. Sadek, K. Ijaz, C. J. Peters, and T. G. Ksiazek. 1997. An outbreak of Crimean-Congo hemorrhagic fever in the United Arab Emirates, 1994-1995. Am. J. Trop. Med. Hyg. 57:519-525. [DOI] [PubMed] [Google Scholar]
- 25.Knobloch, J., J. B. McCormick, P. A. Webb, M. Dietrich, H. H. Schumacher, and E. Dennis. 1980. Clinical observations in 42 patients with Lassa fever. Tropenmed. Parasitol. 31:389-398. [PubMed] [Google Scholar]
- 26.Kwok, S., and R. Higuchi. 1989. Avoiding false positives with PCR. Nature 339:237-238. [DOI] [PubMed] [Google Scholar]
- 27.Le Guenno, B. 1995. Emerging viruses. Sci. Am. 273:56-64. [DOI] [PubMed] [Google Scholar]
- 28.Leroy, E. M., S. Baize, C. Y. Lu, J. B. McCormick, A. J. Georges, M. C. Georges-Courbot, J. Lansoud-Soukate, and S. P. Fisher-Hoch. 2000. Diagnosis of Ebola haemorrhagic fever by RT-PCR in an epidemic setting. J. Med. Virol. 60:463-467. [PubMed] [Google Scholar]
- 29.Livak, K. J., S. J. Flood, J. Marmaro, W. Giusti, and K. Deetz. 1995. Oligonucleotides with fluorescent dyes at opposite ends provide a quenched probe system useful for detecting PCR product and nucleic acid hybridization. PCR Methods Appl. 4:357-362. [DOI] [PubMed] [Google Scholar]
- 30.Lunkenheimer, K., F. T. Hufert, and H. Schmitz. 1990. Detection of Lassa virus RNA in specimens from patients with Lassa fever by using the polymerase chain reaction. J. Clin. Microbiol. 28:2689-2692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Morita, K., T. Mariko, and I. Akika. 1991. Rapid Identification of dengue virus serotypes by using polymerase chain reaction. J. Clin. Microbiol. 29:2107-2110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Morrison, T. B., J. J. Weis, and C. T. Wittwer. 1998. Quantification of low-copy transcripts by continuous SYBR Green I monitoring during amplification. BioTechniques 24:954-958. [PubMed] [Google Scholar]
- 33.Peters, C. J., C. T. Liu, G. W. Anderson, Jr., J. C. Morrill, and P. B. Jahrling. 1989.. Pathogenesis of viral hemorrhagic fevers: Rift Valley fever and Lassa fever contrasted. Rev. Infect. Dis. 11(Suppl. 4):S743-S749. [DOI] [PubMed]
- 34.Read, S. J., J. L. Mitchell, and C. G. Fink. 2001. Lightcycler multiplex PCR for the laboratory diagnosis of common viral infections of the central nervous system. J. Clin. Microbiol. 39:3056-3059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Saldanha, J., N. Lelie, and A. Heath. 1999. Establishment of the first international standard for nucleic acid amplification technology (NAT) assays for HCV RNA. Vox Sang. 76:149-158. [DOI] [PubMed] [Google Scholar]
- 36.Sanchez, A., T. G. Ksiazek, P. E. Rollin, M. E. Miranda, S. G. Trappier, A. S. Khan, C. J. Peters, and S. T. Nichol. 1999.. Detection and molecular characterization of Ebola viruses causing disease in human and nonhuman primates. J. Infect. Dis. 179(Suppl. 1):S164-S169. [DOI] [PubMed]
- 37.Schmitz, H. 2000. Virological and epidemiological aspects of Dengue fever. Nova Acta Leopoldina 80:153-161. [Google Scholar]
- 38.Schmitz, H., P. Emmerich, and J. ter Meulen. 1996. Imported tropical virus infections in Germany. Arch Virol. Suppl. 11:67-74. [DOI] [PubMed] [Google Scholar]
- 39.Smieja, M., J. B. Mahony, C. H. Goldsmith, S. Chong, A. Petrich, and M. Chernesky. 2001. Replicate PCR testing and probit analysis for detection and quantitation of Chlamydia pneumoniae in clinical specimens. J. Clin. Microbiol. 39:1796-1801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Suleiman, M. N., J. M. Muscat-Baron, J. R. Harries, A. G. Satti, G. S. Platt, E. T. Bowen, and D. I. Simpson. 1980.. Congo/Crimean haemorrhagic fever in Dubai. An outbreak at the Rashid Hospital. Lancet ii:939-941. [PubMed] [Google Scholar]
- 41.Teichmann, D., M. P. Grobusch, H. Wesselmann, B. Temmesfeld-Wollbruck, T. Breuer, M. Dietel, P. Emmerich, H. Schmitz, and N. Suttorp. 1999. A haemorrhagic fever from the Cote d'Ivoire. Lancet 354:1608.. [DOI] [PubMed] [Google Scholar]
- 42.Woodruff, A. W., T. P. Monath, A. A. Mahmoud, A. K. Pain, and C. A. Morris. 1973. Lassa fever in Britain: an imported case. Br. Med. J. 3:616-617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yang, Y., M. H. Lamendola, M. Mendoza, D. Xu, M. Nguyen, S. Yeh, Y. Wu, J. Ku, M. Rosenstraus, and R. Sun. 2001. Performance characteristics of the COBAS AmpliScreen HIV-1 test, version 1.5, an assay designed for screening plasma mini-pools. Transfusion 41:643-651. [DOI] [PubMed] [Google Scholar]