Abstract
PCR-restriction fragment length polymorphism (PCR-RFLP) and PCR-single-strand conformation polymorphism (PCR-SSCP) analyses were carried out on the 1.6-kb groEL gene from 41 strains of 10 different Salmonella serovars. Three HaeIII RFLP profiles were recognized, but no discrimination between the serovars could be achieved by this technique. However, PCR-SSCP analysis of the groEL genes of various Salmonella serovars produced 14 SSCP profiles, indicating the potential of this technique to differentiate different Salmonella serovars (interserovar differentiation). Moreover, PCR-SSCP could differentiate strains within a subset of serovars (intraserovar discrimination), as three SSCP profiles were produced for the 11 Salmonella enterica serovar Enteritidis strains, and two SSCP profiles were generated for the 7 S. enterica serovar Infantis and five S. enterica serovar Newport strains. PCR-SSCP has the potential to complement classical typing methods such as serotyping and phage typing for the typing of Salmonella serovars due to its rapidity, simplicity, and typeability.
Salmonellae are the etiologic agents of different diseases collectively referred to as salmonellosis. Human salmonellosis can be divided into four syndromes: enteric fever (typhoid-like disease), gastroenteritis (food poisoning), bacteremia with or without gastroenteritis, and the asymptomatic carrier state.
On a global scale, the incidence of typhoid fever is decreasing, while that of nontyphoidal salmonellosis is increasing, although both remain major health problems. The World Health Organization has estimated that annually there are close to 17 million cases of typhoid fever, with nearly 600,000 deaths, and 1.3 billion cases of acute gastroenteritis or diarrhea due to nontyphoidal salmonellosis, with 3 million deaths (8, 23, 26).
To curb both typhoidal and nontyphoidal salmonellae, laboratory-based surveillance of human and animal infections is a necessary first step of any prevention strategy. Phenotypic methods play an important role in the identification to the genus level. Serotyping is used for primary typing of strains, while phage typing and antibiogram are used for subdivision of serotypes (33). However, serotyping of Salmonella is laborious due to the large number of recognized serotypes, i.e., over 2,400 (1, 27).
In addition, a number of molecular typing methods have also been used to try to improve the identification of salmonellae and also to differentiate strains below the level of serotypes. These DNA-related techniques include ribotyping (3), pulsed-field gel electrophoresis (PFGE) (18, 20, 32), IS200 fingerprinting (4), PCR-ribotyping (12), ribosomal DNA intergenic spacer amplification and heteroduplex analysis (9), amplified fragment length polymorphism (1, 21), automated 5′ nuclease PCR assay (7), and random amplified polymorphic DNA analysis (30).
In recent times, various molecular techniques that detect base sequence changes in bacteria have been used as tools in epidemiological typing. One of the most widely used techniques for the identification of point mutations, due to its simplicity, sensitivity, and rapidity, is PCR-single-strand conformation polymorphism (PCR-SSCP) (6, 22). SSCP was first designed to detect mutations in oncogenes and allelic variations in the human genome (22). Since then, this technique has played a role in bacterial typing (35) and in Salmonella studies (11, 34). Briefly, amplified double-stranded DNA is denatured to single-stranded DNA and subjected to nondenaturing polyacrylamide gel electrophoresis. The mobility of the single-stranded DNA in the gel is dependent not only on its length but also on its secondary structure, as determined by nucleotide sequence (6).
Here, we investigate the possibility of using PCR-SSCP to differentiate Salmonella strains both at the serovar level (interserovar) and at the intraserovar level, using nucleotide variation in the groEL gene, which encodes a heat shock protein (GroEL) that is a member of the stress response protein (HSP60) family (36).
MATERIALS AND METHODS
Bacterial strains.
Forty-one epidemiologically unrelated strains from 10 different Salmonella serovars were studied (Table 1). These strains were kindly provided by Andre Burnens from the Swiss National Reference Laboratory for Foodborne Diseases, University of Berne, Berne, Switzerland. Five of the serovars (Salmonella enterica serovar Typhimurium, S. enterica serovar Newport, S. enterica serovar Hadar, S. enterica serovar Infantis, and S. enterica serovar Virchow) were the most common serovars isolated in Switzerland at the time of the study. The other five serovars (S. enterica serovar Enteritidis, S. enterica serovar Typhi, S. enterica serovar Arizona, S. enterica serovar Paratyphi A, and S. enterica serovar Paratyphi B) consisted of strains that belonged to the reference collection. All of the strains had been identified, biochemically tested, and serotyped at the institution from which they were obtained.
TABLE 1.
Salmonella serovars used in the SSCP study
| Strain no. | Isolate designation | Salmonella serovar | Source |
|---|---|---|---|
| 1 | 1604-97 | Typhimurium | Human feces |
| 2 | 3917-97 | Typhimurium | Rabbit feces |
| 3 | SARB 65 | Typhimurium | Human |
| 4 | SARB 66 | Typhimurium | Parrot |
| 5 | SARB 67 | Typhimurium | |
| 6 | SARB 16 | Enteritidis | |
| 7 | SARB 17 | Enteritidis | |
| 8 | SARB 18 | Enteritidis | |
| 9 | SARB 20 | Enteritidis | |
| 10 | REF 337 | Enteritidis | |
| 11 | REF 340 | Enteritidis | |
| 12 | REF 344 | Enteritidis | |
| 13 | REF 345 | Enteritidis | |
| 14 | REF 347 | Enteritidis | |
| 15 | REF 352 | Enteritidis | |
| 16 | REF 356 | Enteritidis | |
| 17 | 782-95 | Infantis | Human feces |
| 18 | 1523-95 | Infantis | Meat |
| 19 | 1763-95 | Infantis | Beef |
| 20 | 1766-95 | Infantis | Beef |
| 21 | 1792-95 | Infantis | Beef |
| 22 | SARB 27 | Infantis | |
| 23 | SARB 26 | Infantis | Human |
| 24 | 1021-98 | Newport | Human feces |
| 25 | 1069-98 | Newport | Human feces |
| 26 | SARB 38 | Newport | Snake |
| 27 | SARB 36 | Newport | Human |
| 28 | SARB 37 | Newport | Human |
| 29 | 1588-98 | Hadar | Human feces |
| 30 | 1957-98 | Hadar | Human feces |
| 31 | 2102-98 | Hadar | Human feces |
| 32 | 2128-98 | Hadar | Human feces |
| 33 | 2273-98 | Hadar | Human feces |
| 34 | 32-95 | Virchow | Human feces |
| 35 | 326-95 | Virchow | Bird |
| 36 | 372-95 | Virchow | Human feces |
| 37 | MM177 | Paratyphi A | Human |
| 38 | MM168 | Paratyphi B | Human |
| 39 | PNG1 | Typhi | Human feces |
| 40 | PNG27 | Typhi | Human feces |
| 41 | MM260 | Arizona |
The bacteria were maintained on Luria-Bertani (LB) agar plates. Repeated subculturing of isolates was avoided, and for long term maintenance, all the isolates were kept in LB broth with 20% glycerol at −70°C (2).
PCR-restriction fragment length polymorphism (PCR-RFLP) and PCR-SSCP analyses of Salmonella serovars. (i) Genomic DNA isolation.
Genomic DNAs of the Salmonella isolates were prepared by a modification of the method of Saito and Miura (28). A single colony of Salmonella was grown overnight at 37°C in 5 ml of LB broth. The cells were harvested by centrifugation at 4,500 × g for 15 min at room temperature. The cell pellet was resuspended in 50 μl of solution I (0.15 M NaCl, 0.1 M EDTA [pH 8]) by repeated pipetting, and 10 mg of lysozyme (20 mg/ml) was added. The cell suspension was then incubated at 37°C with gentle shaking for 30 min in a shaking water bath. Forty microliters of solution II (1% sodium dodecyl sulfate, 0.1 M NaCl, 0.1 M Tris-HCl [pH 8]) was then added, followed by 10 min of incubation at 60°C. The solution was cooled to room temperature, and 100 μl of 5 M sodium perchlorate was added to help in the dissociation of proteins from nucleic acids. The suspension was mixed thoroughly but gently. Four hundred fifty microliters of TE (10 mM Tris, 10 mM EDTA [pH 7.4])-saturated phenol-chloroform-isoamyl alcohol (25:24:1) was mixed with the suspension and shaken gently for 20 min. The aqueous phase was recovered by centrifugation at 15,000 × g for 10 to 15 min at room temperature and transferred to a new tube. Two volumes of ice-cold 100% ethanol were added. The tube was vortexed and left at −20°C for at least 2 h.
A pellet was obtained by centrifugation for 5 min at 15,000 × g. The pellet was washed with 1 ml of cold 70% ethanol. The ethanol was discarded, and the pellet was dried under vacuum for 5 min. The pellet was resuspended in 200 μl of 1× TE. The DNA solution was treated with 5 μl of RNase (20 μg/ml) at 37°C for 30 min. The DNA concentration was determined spectrophotometrically at wavelengths of 260 and 280 nm (Novaspec II; Pharmacia LKB).
(ii) Amplification of the groEL gene.
Two oligonucleotide PCR primers which amplify the 1.6-kb groEL gene encoding a heat shock protein (HSP60) of S. enterica serovar Typhi were used (15). The sequences of the primers were as follows: primer H1, 5′-GAT CCA TAT GGC AGC TAA AGA CGT AAA ATT CGG-3′; and primer H2, 5′-CTA GGT CGA CTT ACA TCA TGC GGC CCA TGC CAC-3′.
PCRs were performed in a volume of 25 μl containing 50 mM Tris-HCl (pH 9.0); 50 mM KCl; 2.5 mM MgCl2; 400 μM (each) dCTP, dGTP, dATP. and dTTP (Promega, Madison, Wis.); 0.2 μM (each) primers; and 1 U of Taq DNA polymerase (Promega). The substrate was 100 ng of genomic DNA from the Salmonella serovar isolates. Amplification was performed in a 480 DNA thermal cycler (Perkin-Elmer, Norwalk, Conn.) programmed as follows: initial denaturation of the DNA templates at 95°C for 3 min, followed by 35 cycles comprising consecutive denaturation (1 min, 93°C), annealing (2 min, 55°C), and elongation (1.5 min, 72°C).
After PCR, 10 μl of the amplified product was subjected to electrophoresis at 100 V on a 1.5% agarose gel (type II medium electrophoresis grade; Sigma, St. Louis, Mo.) in a 0.5× TBE (45 mM Tris, 45 mM boric acid, 10 mM EDTA [pH 8.0]) buffer system. Following electrophoresis, the gel was stained in ethidium bromide (1 μg/ml) and photographed under UV light.
(iii) Restriction of the groEL gene.
The PCR-amplified groEL gene was digested with 10 U of HaeIII (Promega) at 37°C for 2 h according to the manufacturer's instructions. Restriction DNA fragments were separated by electrophoresis at 70 V in horizontal gel containing 1.5% agarose (type II medium electroendosmosis grade; Sigma) in a 0.5× TBE buffer system for 1 to 2 h. Following electrophoresis the gel was stained in ethidium bromide (1 μg/ml) and photographed under UV light.
(iv) SSCP electrophoresis.
Five microliters of digested DNA (groEL gene) was mixed with 5 μl of denaturation buffer (5 mM EDTA, 0.05% bromophenol blue, and 0.05% xylene cyanole in formamide) and incubated at 95°C for 7 min. The denatured DNA was then placed directly in ice for 10 min before being loaded onto a nondenaturing 10% polyacrylamide gel (CleanGel 36S; Pharmacia Biotech, Uppsala, Sweden). Samples (2 to 4 μl) were separated with DNA Disk buffer (Pharmacia Biotech, Uppsala, Sweden) on a 2117 Multiphor II Electrophoresis Unit (Pharmacia Biotech) at 4°C, using 100 V for 30 min followed by 400 V for 180 min. Two to four microliters of undenatured digested DNA (gene) was run as a control.
After electrophoresis, the 10% polyacrylamide gels were silver stained. Briefly, the silver staining procedure was as follows. The gels were first fixed in 10% acetic acid for approximately 30 min at room temperature and washed with deionized water three times for 2 min each time. Color impregnation lasted for 20 min at room temperature with silver nitrate solution (0.1% [wt/vol] AgNO3 and 0.036% [wt/vol] formaldehyde in distilled water). The gels were then washed for 5 to 10 s with deionized water, followed by color development for 3 to 10 min with a color development solution (2.5% [wt/vol] Na2CO3, 0.036% [wt/vol] formaldehyde, and 0.002% [wt/vol] sodium thiosulfate in distilled water). The color reaction was stopped with a stop solution (1.46% [wt/vol] EDTA in distilled water). The bands were fixed with a fixing solution (28.8% [vol/vol] 96% ethanol and 3.91% [vol/vol] 85% glycerol in distilled water). The stained gels were air dried for a approximately 2 h. SSCP profiles were interpreted visually.
RESULTS
PCR-RFLP and PCR-SSCP analyses were carried out with 41 strains belonging to 10 different Salmonella serovars (Table 1). The antigenic properties of the O antigen have formed the basis of the serological classification of Salmonella (14). The 10 different Salmonella serovars investigated represented five different serogroups (Table 2).
TABLE 2.
PCR-RFLP and PCR-SSCP analysis of Salmonella serovars
| Strain no. | Salmonella serovar | Sero- group | No. of groEL amplicons (1.6 kb) | No. of PCR-RFLP profiles (groEL [HaeIII]) | No. of PCR-SSCP profiles groEL [HaeIII] |
|---|---|---|---|---|---|
| 1 | Typhimurium | B | 1 | 1 | 1 |
| 2 | Typhimurium | B | 1 | 1 | 1 |
| 3 | Typhimurium | B | 1 | 1 | 1 |
| 4 | Typhimurium | B | 1 | 1 | 1 |
| 5 | Typhimurium | B | 1 | 1 | 1 |
| 6 | Enteritidis | D1 | 1 | 2 | 2 |
| 7 | Enteritidis | D1 | 1 | 2 | 3 |
| 8 | Enteritidis | D1 | 1 | 2 | 4 |
| 9 | Enteritidis | D1 | 1 | 2 | 2 |
| 10 | Enteritidis | D1 | 1 | 2 | 2 |
| 11 | Enteritidis | D1 | 1 | 2 | 2 |
| 12 | Enteritidis | D1 | 1 | 2 | 2 |
| 13 | Enteritidis | D1 | 1 | 2 | 2 |
| 14 | Enteritidis | D1 | 1 | 2 | 2 |
| 15 | Enteritidis | D1 | 1 | 2 | 2 |
| 16 | Enteritidis | D1 | 1 | 2 | 2 |
| 17 | Infantis | C1 | 1 | 1 | 11 |
| 18 | Infantis | C1 | 1 | 1 | 11 |
| 19 | Infantis | C1 | 1 | 1 | 11 |
| 20 | Infantis | C1 | 1 | 1 | 11 |
| 21 | Infantis | C1 | 1 | 1 | 11 |
| 22 | Infantis | C1 | 1 | 2 | 7 |
| 23 | Infantis | C1 | 1 | 1 | 11 |
| 24 | Newport | C2 | 1 | 2 | 5 |
| 25 | Newport | C2 | 1 | 1 | 12 |
| 26 | Newport | C2 | 1 | 2 | 5 |
| 27 | Newport | C2 | 1 | 1 | 12 |
| 28 | Newport | C2 | 1 | 2 | 5 |
| 29 | Hadar | C2 | 1 | 2 | 6 |
| 30 | Hadar | C2 | 1 | 2 | 6 |
| 31 | Hadar | C2 | 1 | 2 | 6 |
| 32 | Hadar | C2 | 1 | 2 | 6 |
| 33 | Hadar | C2 | 1 | 2 | 6 |
| 34 | Virchow | C1 | 1 | 1 | 13 |
| 35 | Virchow | C1 | 1 | 1 | 13 |
| 36 | Virchow | C1 | 1 | 1 | 13 |
| 37 | Paratyphi A | A | 1 | 2 | 8 |
| 38 | Paratyphi B | B | 1 | 2 | 9 |
| 39 | Typhi | D1 | 1 | 2 | 10 |
| 40 | Typhi | D1 | 1 | 2 | 10 |
| 41 | Arizona | C1 | 1 | 3 | 14 |
Amplification of the 1.6-kb groEL gene produced an identical single profile for all of the 41 Salmonella strains (Table 2). The published groEL gene sequence was analyzed using the DNASIS software program (Hitachi, Brisbane, Calif.) to search for useful restriction enzyme cleavage sites. Based on this program, HaeIII was used to restrict the groEL gene for PCR-RFLP analysis.
The sensitivity of PCR-SSCP tends to decrease with increasing fragment length. SSCP is reported to be capable of detecting 99% of point mutations in DNA molecules of 100 to 300 bp in length and 89% of mutations in molecules of 300 to 450 bp in length (6, 19, 22). Therefore, the 1.6-kb groEL amplicon was digested with HaeIII to generate shorter fragments before analysis by SSCP.
Undenatured, digested groEL DNA of the S. enterica serovar Typhimurium strains produced four HaeIII-restricted bands of between 150 and 850 bp. The five serovar Typhimurium strains exhibited PCR-RFLP profile 1 (Table 2 and Fig. 1). Denatured digested groEL DNA produced a similar PCR-SSCP profile (profile 1) for all of the serovar Typhimurium strains (Table 2 and Fig. 2). It should be noted that the double-stranded DNA which reannealed from complementary single-stranded DNA under the experimental conditions used migrated faster than the denatured single-stranded DNA. These double-stranded DNAs are not shown in Fig. 2 because they did not increase the discriminatory level of the SSCP profiles between the strains studied.
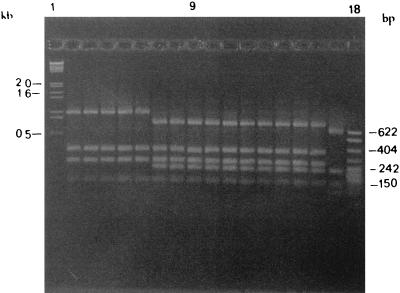
FIG. 1.
PCR-RFLP profiles of groEL DNAs after HaeIII digestion for the 41 strains from 10 serovars run on a 1.5% TBE gel. Lanes 1 and 18, molecular weight markers (New England Biolabs Marker II). The strains and RFLP profiles are as follows: lane 2, S. enterica serovar Typhimurium strain 1, profile 1; lanes 3 and 4, serovar Infantis strains 17 and 18, respectively, profile 1; lane 5, serovar Newport strain 25, profile 1; lane 6, serovar Virchow strain 34, profile 1; lanes 7, 8, and 9, serovar Enteritidis strains 6, 7, and 8, respectively, profile 2; lane 10, serovar Infantis strain 22, profile 2; lane 11, serovar Newport strain 24, profile 2; lane 12, serovar Hadar strain 29, profile 2; lane 13, serovar Paratyphi A strain 37, profile 2; lane 14, serovar Paratyphi B strain 38, profile 2; lanes 15 and 16, S. enterica serovar Typhi strains 39 and 40, respectively, profile 2; and lane 17, serovar Arizona strain 41, profile 3. Strain numbers are as in Table 2.
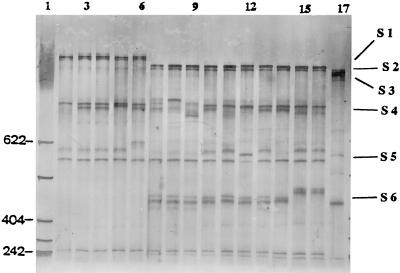
FIG. 2.
PCR-SSCP profiles of groEL DNAs after HaeIII digestion for the strains from 10 serovars run on a CleanGel (10% polyacrylamide gel). Lane 1, New England Biolabs Marker II (with sizes in base pairs). The strains and SSCP profiles are as follows: lane 2, S. enterica serovar Typhimurium strain 1, profile 1; lanes 3 and 4, serovar Infantis strains 17 and 18, respectively, profile 11; lane 5, serovar Newport strain 25, profile 12; lane 6, serovar Virchow strain 34, profile 13; lanes 7, 8, and 9, serovar Enteritidis strains 6, 7, and 8, respectively, profiles 2, 3, and 4; lane 10, serovar Infantis 22, profile 7; lane 11, serovar Newport strain 24, profile 5; lane 12, serovar Hadar strain 29, profile 6; lane 13, serovar Paratyphi A strain 37, profile 8; lane 14, serovar Paratyphi B strain 38, profile 9; lanes 15 and 16, serovar Typhi strains 39 and 40, respectively, profile 10; and lane 17, serovar Arizona strain 41, profile 14. Strain numbers are as in Table 2. S1, single-strand bands of the denatured 850-bp HaeIII groEL fragment; S2, denatured 630-bp HaeIII groEL fragment; S3, denatured 620-bp HaeIII groEL fragments; S4, denatured 350-bp HaeIII groEL fragment; S5, denatured 250-bp HaeIII groEL fragment; S6, denatured 238-bp HaeIII groEL fragment.
Undenatured, digested groEL DNAs of the 11 S. enterica serovar Enteritidis strains produced five HaeIII-restricted bands of between 150 and 630 bp. The 11 serovar Enteritidis strains all exhibited PCR-RFLP profile 2 (Table 2 and Fig. 1). Denatured, digested groEL DNAs from the same 11 strains produced 3 different PCR-SSCP profiles (Table 2 and Fig. 2); nine strains exhibited profile 2, and one strain each exhibited profiles 3 and 4.
Undenatured, digested groEL DNAs of the seven S. enterica serovar Infantis and five S. enterica serovar Newport strains produced two PCR-RFLP profiles. Profile 1 was represented by six serovar Infantis and two serovar Newport strains which had four HaeIII-restricted bands of between 150 and 850 bp, while profile 2 was represented by a single serovar Infantis strain and three serovar Newport strains which had five restricted bands ranging from 150 to 630 bp (Table 2 and Fig. 1). Denatured, digested groEL DNAs also produced two different PCR-SSCP profiles for each of these serovars. Six serovar Infantis strains exhibited PCR-SSCP profile 11 and a single strain had profile 7 (Table 2 and Fig. 2), while two serovar Newport strains were represented by PCR-SSCP profile 12 and the other three strains were represented by profile 5 (Table 2 and Fig. 2).
Undenatured, digested groEL DNAs of the five S. enterica serovar Hadar, two S. enterica serovar Typhi, 1 S. enterica serovar Paratyphi A, and 1 S. enterica serovar Paratyphi B strains produced five HaeIII-restricted bands of between 150 and 630 bp; thus, all the nine strains from these four serovars exhibited PCR-RFLP profile 2 (Table 2 and Fig. 1). Denatured, digested groEL DNAs produced the same PCR-SSCP profile (profile 6) for the five serovar Hadar strains, profile 10 for both of the S. enterica serovar Typhi strains, and profiles 8 and 9, respectively, for serovar Paratyphi A and serovar Paratyphi B (Table 2 and Fig. 2).
Undenatured, digested groEL DNA of the single S. enterica serovar Arizona strain produced four HaeIII-restricted bands of 100 to 620 bp. This strain had PCR-RFLP profile 3, which was not observed in any of the other Salmonella serovars (Table 2 and Fig. 1). The denatured, digested groEL gene of this serovar Arizona strain also produced PCR-SSCP profile 14, which was not exhibited by any of the other Salmonella serovars studied (Table 2 and Fig. 2).
It was noted that some of the denatured fragments, designated S1 to S6 in Fig. 2, showed more than two single strands of DNA. This reflects the fact that single-stranded DNA may have multiple conformers. These multiple conformers could be divided into one or two stronger and one or two weaker SSCP bands. Multiple conformers of single-stranded DNA have been observed by other researchers as well (10).
The reproducibility of PCR-RFLP and PCR-SSCP profiles was also confirmed in a separate experiment (data not shown).
DISCUSSION
Major factors that induce genome variation in Enterobacteriaceae are chromosomal rearrangements (inversions, translocations, deletions, and duplications), horizontal gene transfers, mobile genetic elements, and base pair mutations (16, 29). Since base pair mutations play an important role in genetic variation, it is necessary to use techniques capable of detecting them when searching for mutations or sequence polymorphisms in genes, gene systems, or whole genomes. DNA sequencing is a reliable way of identifying such mutations, but sequencing to detect base pair changes is somewhat cumbersome and costly when large numbers of samples need to be rapidly analyzed. This technique is not presently feasible for epidemiological studies.
Due to this, two PCR-based techniques (PCR-SSCP and PCR-RFLP) were evaluated to try to detect polymorphisms within the groEL gene to discriminate between strains of different Salmonella serovars. SSCP is different from typing methods that involve RFLP analysis, which tests for heterogeneity or polymorphisms at restriction endonuclease cut sites, for it analyzes heterogeneity or polymorphisms at the base pair level of a DNA sequence. PCR-SSCP analysis is one of the simplest and most sensitive methods based on PCR technology for detection of mutations (22).
The groEL gene encodes a heat shock protein (GroEL) which is a member of the stress response protein (HSP60) family (36). Heat shock proteins are produced to protect prokaryotic cells from various stressful conditions, both intracellular and extracellular (36). Heat shock proteins function to stabilize essential and virulence-related proteins in bacterial pathogenesis during exposure to environmental stress (25). Stress proteins affect virulence regulation in many pathogens, e.g., Vibrio cholerae and Listeria monocytogenes (5).
Previous work in our lab documented that heat shock proteins are induced and expressed in S. enterica serovar Typhi following thermal stress (31). This led us to investigate whether there were any polymorphisms or heterogeneity within the 1.6-kb groEL gene between serovar Typhi strains. PCR-SSCP analysis of 44 serovar Typhi isolates from various geographical regions, using the groEL gene, proved to be insignificant, as no polymorphisms were detected in this gene between all of the serovar Typhi strains studied (data not shown).
Due to the base pair conservation of the groEL gene in S. enterica serovar Typhi, the techniques of PCR-RFLP and PCR-SSCP were then applied to analyze different Salmonella serovars. Interesting results were generated when PCR-RFLP and PCR-SSCP analyses were carried out on the 1.6-kb groEL gene from strains of 10 different Salmonella serovars. In general, the 41 strains exhibited three HaeIII RFLP profiles on a 1.5% TBE gel. Strains belonging to S. enterica serovar Typhimurium and serovar Virchow were grouped in PCR-RFLP profile 1. Serovar Hadar, serovar Paratyphi A, serovar Paratyphi B, serovar Typhi exhibited PCR-RFLP profile 2, while strains of serovar Newport and serovar Infantis had a mixture of RFLP profiles 1 and 2. The serovar Arizona strain had a completely different RFLP profile (profile 3). This is not surprising, as this group of strains is reported to be a distant relative of S. enterica subspecies in terms of biochemical and genetic classification (13). The data generated by PCR-RFLP analysis indicate that there are differences in the HaeIII restriction sites within the groEL genes of strains belonging to different serovars and strains within a given serovar. However, no clear discrimination between the serovars and serogroups could be made by this technique.
PCR-SSCP analysis of the 41 strains from 10 different serovars (of which serovars Typhimurium, Newport, Infantis, Hadar, and Virchow were the most common serovars in Switzerland at the time of the study) produced 14 PCR-SSCP profiles, with the differences clearly being seen in the denatured 350-, 250-, and 238-bp HaeIII-restricted fragments. The interesting observation here was that each Salmonella serovar and serogroup had its own unique SSCP profile(s) (Table 2), indicating the possibility of this technique in being used for Salmonella interserovar differentiation. The other interesting aspect was that strains within some serovars (intraserovar) could be differentiated as well (e.g., three SSCP profiles were produced for the 11 serovar Enteritidis strains, and two SSCP profiles each were generated for the 7 serovar Infantis and 5 serovar Newport strains). The epidemiological data in Table 1 show that the strains from these three serovars in most cases were isolated from different sources and therefore are considered to be sporadic cases. The strains were deposited to the Swiss Reference Laboratory, University of Berne. Thus, based on the results of this study, there is a possibility of using PCR-SSCP as a subtyping tool to differentiate sporadic strains within these serovars. It should be noted that it would have been ideal to study outbreak-related strains that are more clonal by nature to assess the discriminatory power of PCR-SSCP as a typing tool. Further assessment of this method has to be carried out using more strains from various serovars and also strains from different epidemiological settings. Like all efficient typing methods, the strong point of SSCP is its reproducibility. PCR-SSCP was done in a separate experiment using the same 41 strains from the 10 serovars, which produced 14 identical SSCP profiles as shown in Table 2 and Fig. 2.
Our goal was not to replace the classical typing methods for Salmonella serovars, such as serotyping and phage typing, but rather to complement these methods with a rapid, simple, and sensitive molecular technique such as PCR-SSCP. There is no doubt that established typing methods such as PFGE and ribotyping could be used in the differentiation of Salmonella serovars, although it may come as a surprise that PFGE has primarily been used only for intraserovar molecular characterization of salmonellae (18, 20, 32). Ribotyping, on the other hand, has been carried out to characterize Salmonella serotypes Reading, Senftenberg, and Typhimurium (3). PCR-SSCP has an advantage compared to both of these methods because of its rapidity and simplicity. The total time needed for PCR-SSCP from the extraction of genomic DNA to the visualization of the gel is less than 24 h. The slight drawback of PCR-SSCP is that it detects the occurrence of base pair mutations in segments of DNA but does not give any information on the type of base changes, which has to be confirmed by sequencing. Recently, multiple-fluorescence-based PCR-SSCP has provided the capacity for simultaneous labeling of several fragments as well as the potential for an automated data collection system (11).
Another observation from the SSCP analysis of the groEL gene involving different serovars is as follows: unlike the conserved nature of the groEL gene among S. enterica serovar Typhi strains (data not shown), it was noted that base pair mutations do occur in this gene for strains belonging to serovars Enteritidis, Infantis, and Newport, where more than one groEL SSCP profile was noted within each serovar (intraserovar discrimination). It was also evident that polymorphisms in the groEL gene sequence varied from serovar to serovar (interserovar discrimination) based on the SSCP results. Sequence data obtained from GenBank have complemented these SSCP results to a certain extent. For instance, it is interesting that the groEL gene of S. enterica serovar Typhi strain Ty21, isolated around 1912, had only one base pair difference from the groEL gene of S. enterica serovar Typhi strain CT18, isolated in Vietnam in 1994 (Sanger Center Blast Server results [http://www.Sanger.ac.uk/cgi-bin/nph-Blast_server.html]). The recently sequenced genomes of S. enterica serovar Typhimurium LT2 (17) and S. enterica serovar Typhi CT18 (24) have revealed 98% DNA homology between the groEL genes (groEL is known as mopA in S. enterica serovar Typhimurium) of both these serovars (Entrez-Pubmed Blast Search [http://www3.ncbi.nlm-nih.gov/Entrez/index.html]). The groEL sequences of the other Salmonella serovars are not known. Why does this conservation occur in some serovars and is more relaxed in others? One reason could be that the mutations detected do not necessarily result in amino acid substitution and do not disrupt the essential heat shock protein product. Another possible reason is that the host specificity of strains belonging to particular serovars (as they encounter different levels of intracellular and extracellular environmental stress such as varying temperature, nutrient availability, pH, levels of O2, inorganic ion concentrations, presence of toxic chemicals, levels of antibiotics or vaccines, and host immune and genetic factors) plays a role in determining the base pair sequence of the groEL gene and the outcome of the stress protein produced, which facilitates survival under adverse conditions. This will be an interesting subject to assess in the future.
Acknowledgments
The study was funded by IRPA (Intensification of Research in Priority Areas) grant no. 06-02-03-0750.
We thank Kenneth E. Sanderson for his useful comments and help in generating the manuscript.
REFERENCES
- 1.Aarts, H. J. M., L. A. J. T. Vanlith, and J. Keljer. 1998. High-resolution genotyping of Salmonella strains by AFLP fingerprinting. Lett. Appl. Microbiol. 26:131-135. [DOI] [PubMed] [Google Scholar]
- 2.Cowan, S. T., and J. Steel. 1993. Cowan and Steel's manual for the identification of medical bacteria, 3rd ed. Cambridge University Press, Cambridge, United Kingdom.
- 3.Esteban, E., K. Snipes, and D. Hird, D. 1993. Use of ribotyping for characterization of Salmonella serotypes. J. Clin. Microbiol. 31:233-237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ezquerra, E., A. Burnens, C. Jones, and J. Stanley. 1993. Genotypic typing and phylogenetic analysis of Salmonella paratyphi B and S. java with IS200. J. Gen. Microbiol. 139:2409-2414. [DOI] [PubMed] [Google Scholar]
- 5.Gross, R. 1993., Signal transduction and virulence regulation in human and animal pathogens. FEMS Microbiol. Rev. 104:301-326. [DOI] [PubMed] [Google Scholar]
- 6.Hayashi, K. 1992. PCR-SSCP: a method for detection of mutations. Genet. Anal. Tech. Appl. 9:73-79. [DOI] [PubMed] [Google Scholar]
- 7.Hoorfar, J., P. Ahrens, and P. Radstrom. 2000. Automated 5′ nuclease PCR assay for identification of Salmonella enterica. J. Clin. Microbiol. 38:3429-3435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ivanoff, B. 1995. Typhoid fever: global situation and W. H. O. recommendations. Southeast Asian J. Trop. Med. Public Health 26(Suppl. 2):1-6. [Google Scholar]
- 9.Jensen, M. A., and R. J. Hubner. 1996. Use of homoduplex ribosomal DNA spacer amplification products and heteroduplex cross-hybridization products in the identification of Salmonella serovars. Appl. Environ. Microbiol. 62:2741-2746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jiang, C., C. Li, D. S. Chi, D. A. Ferguson, T. Ha, J. J. Laffan, and E. Thomas. 1998. Combination of single- and double-stranded conformational polymorphism for direct discrimination of gastric Helicobacter pylori. J. Microbiol. Methods 34:1-8. [Google Scholar]
- 11.Jordon, S. J., C. E. R. Dodd, and G. S. A. B. Stewart. 1999. Use of single-strand conformation polymorphism analysis to examine the variability of the rpoS sequence in environmental isolates of salmonellae. Appl. Environ. Microbiol. 65:3582-3587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lagatolla, C., L. Dolzani, E. Tonin, A. Lavenia, M. Dimichele, T. Tommasin, and C. Monti-Bragadin. 1996. PCR-ribotyping and characterizing Salmonella isolates of different serotypes. J. Clin. Microbiol. 34:2440-2443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.LeMinor, L. 1984. Salmonella lignieres, p. 427-458. In N. R. Kreig and J. G. Holt (ed.), Bergey's manual of systematic bacteriology, vol. I. Williams and Wilkins, Baltimore, Md. [Google Scholar]
- 14.Lindberg, A. A., and L. LeMinor. 1984. Serology of Salmonella. Methods Microbiol. 15:1-141.
- 15.Lindler, L. E., and J. M. Hayes. 1994. Nucleotide sequence of the Salmonella typhi groEL heat shock gene. Microb. Pathog. 17:271-275. [DOI] [PubMed] [Google Scholar]
- 16.Liu, S. L., and K. E. Sanderson. 1995. Genomic cleavage map of Salmonella typhi Ty2. J. Bacteriol. 177:5099-5107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.McClelland, M., K. E. Sanderson, J. Spieth, S. W. Clifton, P. Latrieille, L. Courtney, S. Porwollik, J. Ali, M. Dante, F. Du, S. Hou, D. Layman, S. Leonard, C. Nguyen, K. Scott, A. Holmes, N. Grewal, E. Mulvaney, E. Ryan, H. Sun, L. Florea, W. Miller, T. Stoneking, M. Nhan, R. Waterston, and R. K. Wilson. 2001. Complete genome sequence of Salmonella enterica serovar Typhimurium LT2. Nature 413:852-856. [DOI] [PubMed] [Google Scholar]
- 18.Mhand, R. A., N. Brahimi, N. Moustaoui, N. E. Mdaghri, H. Amarouch, F. Grimont, E. Bingen, and M. Benbachir. 1999. Characterization of extended spectrum β-lactamase producing Salmonella typhimurium by phenotypic and genotypic typing methods. J. Clin. Microbiol. 37:3769-3773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mohabeer, A. J., A. L. Hiti, and W. J. Martin. 1991. Non-radioactive single-strand conformation polymorphism (SSCP) using the Pharmacia “Phastsystem.” Nucleic Acids Res. 19:3154.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nair, S., C. L. Poh, Y. S. Lim, L. Tay, and K. T. Goh. 1994. Genome fingerprinting of Salmonella typhi by pulsed-field gel electrophoresis for subtyping common phage types. Epidemiol. Infect. 113:391-402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nair, S., E. Schreiber, T. Kwai-Lin, T. Pang, and M. Altwegg. 2000. Genotypic characterization of Salmonella typhi by amplified fragment length polymorphism fingerprinting provides increased discrimination as compared to pulsed-field gel electrophoresis and ribotyping. J. Microbiol. Methods 41:35-43. [DOI] [PubMed] [Google Scholar]
- 22.Orita, M., H. Iwahana, H. Kanazawa, K. Hayashi, and T. Sekiya. 1989. Detection of polymorphisms of human DNA by gel electrophoresis as single-strand conformation polymorphism. Proc. Natl. Acad. Sci. USA 86:2766-2770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pang, T., Z. A. Bhutta, B. B. Finlay, and M. Altwegg. 1995. Typhoid fever and other salmonellosis: a continuing challenge. Trends Microbiol. 3:253-255. [DOI] [PubMed] [Google Scholar]
- 24.Parkhill, J., G. Dougan, K. D. James, N. R. Thompson, D. Pickard, J. Wain, C. Churcher, K. L. Mungall, S. D. Bentley, M. T. G. Holden, M. Sebaihia, S. Baker, D. Basham, K. Brooks, T. Chillingworth, P. Connerton, A. Cronin, P. Davis, R. M. Davies, L. Dowd, N. White, J. Farrar, T. Feltwell, N. Hamlin, A. Haque, T. T. Hien, S. Holroyd, K. Jagels, K. Krogh, T. S. Larsen, S. Leather, S. Moule, P. O'Gaora, C. Parry, S. Whitehead, and B. G. Barrell. 2001. Complete genome sequence of a multiple drug resistant Salmonella enterica serovar Typhi CT18. Nature 413:848-852. [DOI] [PubMed] [Google Scholar]
- 25.Parsons, C. M., R. J. Limberger, and M. Shayegani. 1997. Alterations in levels of DnaK and GroEL result in diminished survival and adherence of stressed Haemophilus ducreyi. Infect. Immun. 65:2413-2419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Philips, C. A., and J. T. George, J. T. 1994. Guess what's lurking in the lunch. Biologist 41:76-80. [Google Scholar]
- 27.Popoff, M. Y., J. Bockemuhl, and F. W. Brenner. 2000. Supplement 1998 (no. 42) to the Kauffmann-White scheme. Res. Microbiol. 151:63-65. [DOI] [PubMed] [Google Scholar]
- 28.Saito, H., and K. Miura. 1963. Preparation of transforming deoxyribonucleic acid by phenol treatment. Biochim. Biophys. Acta 72:619-629. [PubMed] [Google Scholar]
- 29.Sanderson, K. E., and S. L. Liu. 1998. Chromosomal rearrangements in Salmonella spp. Med. J. Indonesia Suppl. 1:30-37. [Google Scholar]
- 30.Shangkuan, Y. H., and H. C. Lin. 1998. Application of random amplified polymorphic DNA analysis to differentiate strains of Salmonella typhi and other Salmonella species. J. Appl. Microbiol. 85:693-702. [DOI] [PubMed] [Google Scholar]
- 31.Tang, S. W., S. Abubakar, S. Devi, S. Puthucheary, and T. Pang. 1997. Induction and characterization of heat shock proteins of Salmonella typhi in their reactivity with sera from patients with typhoid fever. Infect. Immun. 65:2983-2986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Thong, K. L., S. Nair, R. Chaudry, P. Seth, A. Kapil, D. Kumar, H. Kapoor, S. Puthucheary, and T. Pang. 1998. Molecular analysis of Salmonella paratyphi A from an outbreak in New Delhi, India. Emerg. Infect. Dis. 4:507-508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Threlfall, E. J. 1998. Development and application of molecular typing methods for Salmonella spp. The U. K. experience. Med. J. Indonesia Suppl. 1:151-154. [Google Scholar]
- 34.Wain, J., T. S. Diep, N. T. Chinh, H. Vinh, S. Fortune, N. T. T. Hoa, N. J. White, L. J. V. Piddock, C. M. Parry, and J. F. Farrar. 1998. Resistance to antimicrobial agents in Salmonella typhi in Vietnam. Clinical response to therapy and molecular mechanisms. Med. J. Indonesia Suppl. 1:5-8. [Google Scholar]
- 35.Widjojoatmodjo, M. N., A. C. Fluit, and J. Verhoet. 1994. Rapid identification of bacteria by PCR-single-strand conformation polymorphism. J. Clin. Microbiol. 32:3002-3007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zügel, U., and S. H. E. Kaufmann. 1999. Role of heat shock proteins in protection from and pathogenesis of infectious diseases. Clin. Microbiol. Rev. 12:19-39. [DOI] [PMC free article] [PubMed] [Google Scholar]