Abstract
We report on a case of human gastroenteritis caused by the pathogen Campylobacter hyointestinalis. Recurrent watery diarrhea and intermittent vomiting were the most significant symptoms of the previously healthy patient. Whole-cell protein electrophoresis and 16S rRNA gene sequencing were used to identify this Campylobacter species. Investigation of the patient's surroundings led to the recovery of a second C. hyointestinalis strain originating from porcine feces. Subsequent typing of the human and the porcine isolates by pulsed-field gel electrophoresis revealed similar macrorestriction profiles, indicating transmission of this pathogen.
Campylobacter species are important agents of gastroenteritis and invasive disease in humans. These proteobacteria are microaerobic, spiral rods displaying a characteristic corkscrew-like motion. The genus Campylobacter comprises 16 validated species, most of which are potent human and/or animal pathogens. Their natural hosts are typical food animals (e.g., chicken, cattle, and pigs), and these play a central role in their transmission to humans. Campylobacter jejuni is the leading cause of bacterial food-borne diarrhea throughout the world (1). Together with C. coli it is the best-characterized Campylobacter, and for this reason routine laboratories mainly focus on these two taxa. While isolation procedures, including procedures that use selective media and growth at 42°C, reduce the high proportion of the natural fecal flora during examination of stool specimens, their use also results in the counterselection of campylobacters other than C. jejuni and C. coli. Moreover, recovery of these fastidious species requires incubation under specialized atmospheric conditions (e.g., in a hydrogen-enriched atmosphere) or at lower temperatures. As a consequence, it has been suggested that infections caused by other taxa of campylobacters may be underdiagnosed (4), resulting in a significant underestimation of the number of non-C. jejuni and non-C. coli Campylobacter species that are the causes of human disease (8).
C. hyointestinalis was first described in 1983 by Gebhart et al. (6) as a possible cause of porcine proliferative enteritis. The organism has subsequently been isolated from the feces of humans with gastroenteritis and, in a few cases, from the blood of patients with bacteremia (7). Here we report on the isolation of C. hyointestinalis and the obvious transmission of this pathogen from a pig to a woman.
Case report.
An 88-year-old white female suffered from persistent diarrhea, abdominal pain, and intermittent vomiting for more than 1 month. A stool specimen appeared watery but not bloody. Examination did not show any other enteric pathogen except a Campylobacter strain (strain H00/108), which displayed unusual phenotypic behavior that did not correlate with that of C. jejuni, C. coli, or C. lari. Subsequent 16S ribosomal DNA (rDNA) sequence analysis identified the strain as C. hyointestinalis. Twenty-two days after the initial stool examination the patient was visited at home. She lived and worked on a farm with animals, including several pigs, chickens, cats, and a dog. The patient was confined to bed with persistent diarrhea. Two fresh stool specimens were collected. In addition, 16 specimens were taken from pigs in the pigsty. The patient was hospitalized on the same day. Vomiting, abdominal cramps, and watery diarrhea were persistent, but she had no fever. Physical examination revealed signs of dehydration and a herpetic corneal lesion. Laboratory examinations provided a leukocyte count of 11.2 × 109/liter (11% lymphocytes, 12% monocytes, 77% neutrophils) and decreased serum electrolyte levels (130 mmol of sodium per liter, 2 mmol of potassium per liter, 95 mmol of chloride per liter). Other irregular values for serum included 7.8 mg of C-reactive protein per dl, 197 mg of urea per dl, 2.2 mg of creatinine per dl, 43 U of γ-glutamyl transferase per liter, 271 U of alkaline phosphatase per liter, and 432 U of lipase per liter. The patient was parenterally rehydrated, and electrolytes were substituted. The initial ciprofloxacin treatment was switched to clarithromycin because of the persisting diarrhea. On hospital day 18 the patient was discharged with an improved condition, although the diarrhea did not completely resolve. A final stool examination was negative for any enteric pathogen.
The specimens taken at the patient's residence were examined. C. hyointestinalis (strains CS00/14 and CS00/15) and C. jejuni (strains CS00/11 and CS00/12) were simultaneously identified in both of the patient's samples. Examination of the porcine specimens revealed two that were positive for C. hyointestinalis (strains CS00/10 and CS00/13) and seven that were positive for C. coli (strains CS00/2 to CS00/8).
Bacterial strains and antimicrobial susceptibility.
All specimens were directly plated onto solid modified charcoal-cefoperazone deoxycholate agar (Oxoid, Basingstoke, United Kingdom), and the plates were incubated at 37 and 42°C in a microaerophilic atmosphere (Genbox microaer; bioMerieux, Marcy l'Etoile, France) for 48 h. The growth of bacterial colonies and their spiral morphology by Gram staining initiated subcultivation on Columbia-blood agar plates (bioMerieux). Biochemical characterization included assays for oxidase, catalase, indoxyl acetate, and hippurate hydrolysis by previously described protocols (11, 15). In addition, every isolate was subjected to analysis with the API Campy system (bioMerieux). Antimicrobial susceptibility testing was done by disk diffusion on Mueller-Hinton agar with 5% sheep blood (bioMerieux) in a microaerophilic atmosphere (Genbox microaer; bioMerieux) at 37°C, and the results were interpreted after 24 and 48 h. The following zone diameters were used: for nalidixic acid (30 μg), ≥19 mm for susceptible (S) and ≤13 mm for resistant (R); for ciprofloxacin (5 μg), ≥21 mm for S and ≤15 mm for R; for erythromycin (15 μg), ≥23 mm for S and ≤13 mm for R; and for tetracycline (30 μg), ≥19 mm for S and ≤14 mm for R. Cephalothin MICs were determined by the E-test (AB Biodisk, Solna, Sweden).
The 14 Campylobacter strains identified in this study are listed in Table 1. All isolates were microaerophilic, gram-negative, curved, motile rods. Five isolates (isolates H00/108, CS00/10, CS00/13, CS00/14, and CS00/15) displayed unusual biochemical characteristics, as shown in Table 1. They grew at 25, 37, and 42°C; and the cephalothin MICs ranged between 16 and 32 μg/ml. Analysis with the API Campy system did not give reliable results. It specified strains CS00/11 and CS00/12 as C. jejuni and strains CS00/2 to CS00/8 as C. coli.
TABLE 1.
Bacterial strains identified in this studya
| Strain | Species | Source | Catalase activity | Nitrate reduction | Hippurate hydrolysis | Indoxyl acetate activity | Susceptibility to:
|
|||
|---|---|---|---|---|---|---|---|---|---|---|
| NAL | CIP | ERY | TET | |||||||
| H00/108b | C. hyointestinalis | Human stool | + | + | − | − | R | S | S | S |
| CS00/14c | C. hyointestinalis | Human stool | + | + | − | − | R | S | S | S |
| CS00/15c | C. hyointestinalis | Human stool swab | + | + | − | − | R | S | S | S |
| CS00/10c | C. hyointestinalis | Porcine stool | + | + | − | − | R | S | S | S |
| CS00/13c | C. hyointestinalis | Porcine stool | + | + | − | − | R | S | S | S |
| CS00/11c | C. jejuni | Human stool | + | + | + | + | S | S | S | S |
| CS00/12c | C. jejuni | Human stool swab | + | + | + | + | S | S | S | S |
| CS00/2c | C. coli | Porcine stool | + | + | − | + | S | S | R | S |
| CS00/3c | C. coli | Porcine stool | + | + | − | + | S | S | R | S |
| CS00/4c | C. coli | Porcine stool | + | + | − | + | S | S | R | S |
| CS00/5c | C. coli | Porcine stool | + | + | − | + | S | S | R | S |
| CS00/6c | C. coli | Porcine stool | + | + | − | + | S | S | R | S |
| CS00/7c | C. coli | Porcine rectal swab | + | + | − | + | S | S | R | S |
| CS00/8c | C. coli | Porcine rectal swab | + | + | − | + | S | S | R | S |
Abbreviations and symbols: NAL, nalidixic acid; CIP, ciprofloxacin; ERY, erythromycin; TET, tetracycline; +, positive reaction; −, negative reaction.
Initial isolate.
Isolated 22 days after isolation of the initial isolate.
16S rDNA analysis.
The 16S rDNA of strain H00/108 was sequenced since biochemical characterization was not sufficient for strain identification. DNA was rapidly isolated with Chelex 100 resin (Bio-Rad, Hercules, Calif.) (19). Bacterial colonies from agar plates were suspended in 50 μl of phosphate-buffered saline. A total of 150 μl (wt/vol) of 20% Chelex 100 suspended in a buffer containing 10 mM Tris-HCl (pH 8), 0.1 mM EDTA, and 0.1% NaN3 was then added. The mixture was vortexed, boiled for 10 min, vortexed for 30 s, and placed on ice for 1 min. Samples were centrifuged at 13,000 rpm (centrifuge model 5417R [Eppendorf, Hamburg, Germany]) for 2 min, and 10 μl of the supernatant was directly subjected to PCR with three pairs of oligonucleotide primers whose sequences correspond to the Campylobacter 16S rRNA gene. The following primer combinations were used: Ps5/1 (5′-TATGGAGAGTTTGATCCTGG-3′) and Ps3/1 (5′-GTTAAGCTGTTAGATTTCAC-3′), Ps5/2 (5′-AGCGTTACTCGGAATCACTG-3′) and Ps3/2 (5′-ACAGCCGTGCAGCACCTGTC-3′), and Ps5/3 (5′-AACCTTACCTGGGCTTGATA-3′) and Ps3/3 (5′-AAGGAGGTGATCCAGCCGCA-3′). Fifty microliters of the PCR mixture contained 10 mM Tris-HCl (pH 8.8) at 25°C, 2 mM MgCl2, 50 mM KCl, 0.1% Triton X-100, 100 μM deoxynucleoside triphosphates, 25 pmol of each 5′ primer and 3′ primer, and 2 U of Dynazyme DNA polymerase (Finnzymes Oy, Espoo, Finland). Conditions for PCR, performed in a Mastercycler gradient thermocycler (Eppendorf, Hamburg, Germany), were as follows: hot start at 95°C for 2 min, followed by 35 cycles of a program of 95°C for 15 s, 54°C for 30 s, and 72°C for 1.5 min, plus a final extension at 72°C for 2 min. The amplification products were purified with a QIAquick Spin PCR purification kit (Qiagen, Hilden, Germany) and were quantified photometrically. Ninety nanograms of the PCR product was submitted to the sequencing reaction with the BigDye Terminator Cycle Sequencing Ready Reaction kit (Applied Biosystems, Foster City, Calif.), according to the specifications of the supplier. The extension products were purified by ethanol precipitation and were directly sequenced on an ABI Prism 310 automated sequencer (Applied Biosystems). Sequence analysis was performed with the BLAST server available on the National Center for Biotechnology Information website (http://www.ncbi.nlm.nih.gov/BLAST/).
The 16S rDNA sequence data for strain H00/108 were available over a length of 1,437 bp. A BLAST search revealed 99.7% identity (1,419 of 1,423 bp) to the 16S rDNA of C. hyointestinalis subsp. hyointestinalis SVS3035 (GenBank accession number AF097690). Hence, strain H00/108 was identified as a member of the species C. hyointestinalis.
Whole-cell protein analysis.
Protein fingerprinting with strains CS00/13 and CS00/14 was performed for species identification by a laboratory of the Belgian Coordinated Collection of Microorganisms (BCCM/LMG). Preparation of cell extracts and protein gel electrophoresis (sodium dodecyl sulfate [SDS]-polyacrylamide gel electrophoresis) were carried out as described previously (17). The normalized and digitized protein patterns were analyzed numerically and were found to cluster with a subset of fingerprints for reference strains and recent isolates of relevant taxa of campylobacters. Cluster analysis identified strains CS00/13 and CS00/14 as C. hyointestinalis (data not shown).
Genotyping by PFGE.
Macrorestriction profiling was used to determine the epidemiological relationships of the isolates. Standard methods for macrorestriction and pulsed-field gel electrophoresis (PFGE) were used, with minor modifications (3). The bacterial cells were embedded in 1% low-melting-point agarose (SeaPlaque GTG; FMC BioProducts, Rockland, Maine), and the mixture was incubated in 1.5 ml of lysis buffer (1% SDS, 0.25 mM EDTA [pH 8], 0.75 mg of proteinase K per ml) at 56°C for 16 h. The agar blocks were washed three times for 30 min each time with 0.5 ml of Tris-EDTA buffer (pH 7) containing 2 mM Pefabloc (Roche, Mannheim, Germany) at 37°C, followed by three washings for 30 min each time with 1.5 ml of Tris-EDTA buffer (pH 7) at 37°C. Digestions with restriction endonucleases SalI and SmaI were carried out according to the specifications of the supplier (New England Biolabs, Beverly, Mass.). DNA fragments were separated in 1% UltraPure agarose (Life Technologies, Paisley, Scotland) with 0.5× Tris-borate-EDTA buffer for 24 h at 175 V and 8°C, with pulse times ranging from 15 to 35 s, by using a contour-clamped homogeneous electric field electrophoresis unit (GeneNavigator; Amersham Pharmacia Biotech, Freiburg, Germany). The sizes of the DNA fragments were determined by using the bacteriophage lambda ladder PFG marker (New England Biolabs).
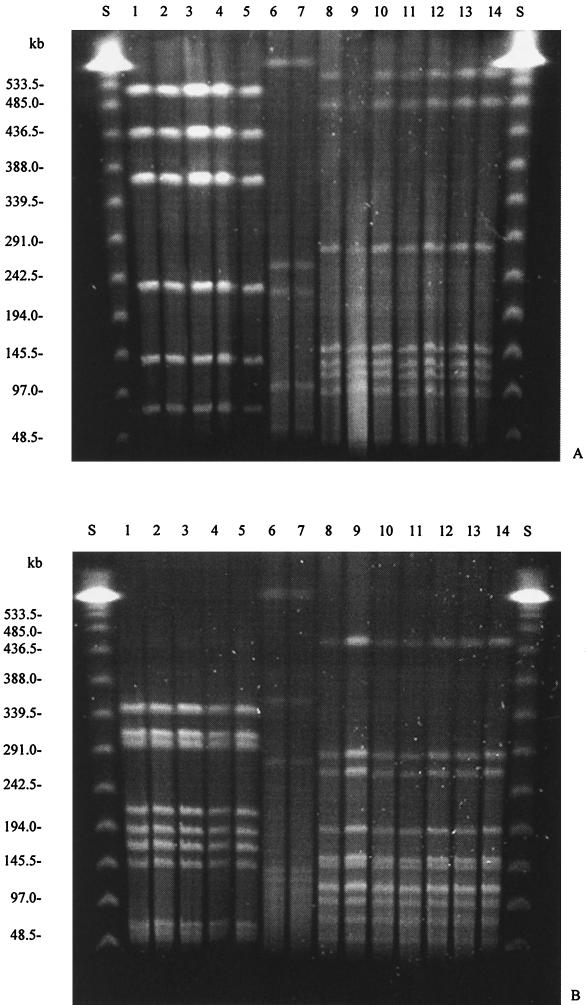
The C. hyointestinalis isolates obtained from the patient's stool (isolates H00/108, CS00/14, and CS00/15) and from the porcine samples (isolates CS00/10 and CS00/13) revealed the same banding patterns by SmaI- and SalI-derived macrorestriction (Fig. 1). Therefore, these isolates can be considered genotypically identical and to belong to the same clone of C. hyointestinalis. Both strains of C. jejuni (strains CS00/11 and CS00/12) isolated from the patient's stool were genotypically identical. In addition, all seven strains of C. coli (strains CS00/2 to CS00/8) obtained from different porcine fecal samples were also genotypically identical (Fig. 1).
FIG. 1.
Genotyping of the strains isolated in the present study by PFGE with restriction endonucleases SalI (A) and SmaI (B). Lanes: S, bacteriophage λ PFGE standard; 1, H00/108; 2, CS00/14; 3, CS00/15; 4, CS00/10; 5, CS00/13; 6, CS00/11; 7, CS00/12; 8, CS00/2; 9, CS00/3; 10, CS00/4; 11, CS00/5; 12, CS00/6; 13, CS00/7; 14, CS00/8.
Discussion.
We report on a case of campylobacteriosis affecting an 88-year-old female. The disease presented as persistent diarrhea with intermittent vomiting and was caused by the pathogen C. hyointestinalis. Two more C. hyointestinalis strains originating from pigs were isolated from the patient's surroundings. Genotyping by PFGE, which has proved to be a highly discriminatory approach for the typing of Campylobacter (13, 16, 18), enabled the determination of the epidemiological relationship among the patient's and the porcine isolates. All C. hyointestinalis isolates of human and porcine origin were genotypically identical. We conclude that infection was acquired through direct or indirect contact of the patient with the pigs on the farm, most likely via the fecal-oral route, since the patient had frequent contact with these animals.
In 1986, the first human infection caused by C. hyointestinalis was described, wherein this pathogen was associated with proctitis (5). Since then, the association of C. hyointestinalis with human gastrointestinal disease has been verified in several cases (8). In a study of 3,877 Campylobacter isolates from stool samples from South African pediatric patients, C. hyointestinalis strains were detected in 51 (1.3%) of the samples (9). The main symptom that results from infection with C. hyointestinalis is nonbloody, watery diarrhea, although some investigators have reported evidence of blood in the stools of C. hyointestinalis-infected patients (8). Since most cases of infection were evident in hosts with impaired immunity (2, 8), C. hyointestinalis is proposed to be an opportunistic pathogen (10). Other reports suggest that this organism can also infect immunocompetent people (2). Consistent with the latter findings, some C. hyointestinalis strains produce a cytotoxin, which may contribute to their virulence (14). For the patient described here, we conclude that C. hyointestinalis H00/108 was the cause of gastroenteritis on the basis of its exclusive detection in the first stool examination. Although we did not determine any immune defect in this patient, the patient's advanced age (88 years), the extended course of her clinical symptoms, and the herpetic corneal lesion might indicate a weakened immunity. The simultaneous recovery of C. hyointestinalis and C. jejuni in the second examination might have been due to superinfection caused by poor hygiene at the patient's farm. In that case, C. jejuni probably worsened the patient's condition, which subsequently led to her hospitalization.
The clinical significance of non-C. jejuni, non-C. coli Campylobacter species is thought to be widely underestimated as a consequence of inappropriate isolation procedures (8). Cephalothin is a constituent of many Campylobacter selective media (12); thus, cephalothin-sensitive campylobacters, such as C. hyointestinalis, C. fetus, and C. upsaliensis are underdetected. To improve the range of detection, alternative preparative methods (e.g., filtration techniques) or PCR-based strategies can be used for the diagnosis of Campylobacter infections (15). Nonetheless, most routine laboratories do not apply these techniques for the initial steps in stool examination because of their elevated costs and time requirements.
In summary, we advocate the following considerations. (i) Campylobacters are the most common cause of human bacterial diarrhea, and medical microbiologists should become aware of campylobacters other than C. jejuni and C. coli as potential agents of human disease. (ii) It is necessary to improve techniques for the recovery of these pathogens in routine stool examinations to assess their levels of involvement in human illness.
Nucleotide sequence accession number.
The 16S rDNA sequence of strain H00/108 has been deposited in GenBank (accession number AF499005).
Acknowledgments
We thank S. Häusler for excellent technical assistance, D. Janssens from the Belgian Coordinated Collections of Microorganisms (BCCM/LMG) for performing the whole-cell protein analysis, and C. Schober for critically reviewing the manuscript.
REFERENCES
- 1.Blaser, M. J. 1997. Epidemiologic and clinical features of Campylobacter jejuni infections. J. Infect. Dis. 176:103-105. [DOI] [PubMed] [Google Scholar]
- 2.Breynaert, J. P., P. Vandamme, and S. Lauwers. 1998. Review of 9 cases of human infection with Campylobacter hyointestinalis, p. 428-431. In A. J. Lastovica, D. G. Newell, and E. E. Lastovica (ed.), Campylobacter, Helicobacter and related organisms. Institute of Child Health, Cape Town, South Africa.
- 3.Chang, N., and D. E. Taylor. 1990. Use of pulsed-field agarose gel electrophoresis to size genomes of Campylobacter species and to construct a SalI map of Campylobacter jejuni UA580. J. Bacteriol. 172:5211-5217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Engberg, J., S. L. W. On, C. S. Harrington, and P. Gerner-Schmidt. 2000. Prevalence of Campylobacter, Arcobacter, Helicobacter, and Suturella spp. in human fecal samples as estimated by reevaluation of isolation methods for campylobacters. J. Clin. Microbiol. 38:286-291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fenell, C. L., A. M. Rompalo, P. A. Totten, K. L. Bruch, B. M. Flores, and W. E. Stamm. 1986. Isolation of “Campylobacter hyointestinalis” from a human. J. Clin. Microbiol. 24:146-148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gebhart, C. J., G. E. Ward, K. Chang, and H. J. Kurtz. 1983. Campylobacter hyointestinalis (new species) isolated from swine with lesions of proliferative enteritis. Am. J. Vet. Res. 44:361-367. [PubMed] [Google Scholar]
- 7.Lastovica, A. J. 1996. Campylobacter/Helicobacter bacteremia in Cape Town, South Africa, 1977-1995, p. 475-479. In D. G. Newell, J. M. Ketley, and R. A. Feldman (ed.), Campylobacters, Helicobacters and related organisms. Plenum Press, New York, N.Y.
- 8.Lastovica, A. J., and M. B. Skirrow. 2000. Clinical significance of Campylobacter and related species other than Campylobacter jejuni and C. coli, p. 89-120. In I. Nachamkin and M. J. Blaser (ed.), Campylobacter, 2nd ed. American Society for Microbiology, Washington, D.C.
- 9.Le Roux, E., and A. J. Lastovica. 1998. The Cape Town protocol: how to isolate the most campylobacters for your dollar, pound, franc, yen, etc., p. 30-33. In A. J. Lastovica, D. G. Newell, and E. E. Lastovica (ed.), Campylobacter, Helicobacter and related organisms. Institute of Child Health, Cape Town, South Africa.
- 10.Minet, J., B. Grosbois, and F. Megraud. 1988. Campylobacter hyointestinalis: an opportunistic enteropathogen? J. Clin. Microbiol. 26:2659-2660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nachamkin, I. 1995. Campylobacter and Arcobacter, p. 483-491. In P. R. Murray, E. J. Baron, M. A. Pfaller, F. C. Tenover, and R. H. Yolken (ed.), Manual of clinical microbiology, 6th ed. American Society for Microbiology, Washington, D.C.
- 12.Nachamkin, I., J. Engberg, and F. M. Aarestrup. 2000. Diagnosis and antimicrobial susceptibility of Campylobacter species, p. 45-66. In I. Nachamkin and M. J. Blaser (ed.), Campylobacter, 2nd ed. American Society for Microbiology, Washington, D.C.
- 13.Newell, D. G., J. A. Frost, B. Duim, J. A. Wagenaar, R. H. Madden, J. van der Plas, and S. L. W. On. 2000. New developments in the subtyping of Campylobacter species, p. 27-44. In I. Nachamkin and M. J. Blaser (ed.), Campylobacter, 2nd ed. American Society for Microbiology, Washington, D.C.
- 14.Ohya, T., and M. Nakazawa. 1992. Production and some properties of cytotoxins produced by Campylobacter species isolated from proliferative enteropathy in swine. J. Vet. Med. Sci. 54:1031-1033. [DOI] [PubMed] [Google Scholar]
- 15.On, S. L. W. 1996. Identification methods for campylobacters, helicobacters, and related organisms. Clin. Microbiol. Rev. 9:405-422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.On, S. L. W., and P. Vandamme. 1997. Identification and epidemiological typing of Campylobacter hyointestinalis subspecies by phenotypic and genotypic methods and description of novel subgroups. Syst. Appl. Microbiol. 20:238-247. [Google Scholar]
- 17.Pot, B., P. Vandamme, and K. Kersters. 1994. Analysis of electrophoretic whole-organism protein fingerprints, p. 493-521. In M. Godfellow and A. G. O'Donnell (ed.), Modern microbial methods. Chemical methods in prokaryotic systematics. John Wiley & Sons, Chichester, United Kingdom.
- 18.Salama, S. M., H. Tabor, M. Richter, and D. E. Taylor. 1992. Pulsed-field gel electrophoresis for epidemiologic studies of Campylobacter hyointestinalis isolates. J. Clin. Microbiol. 30:1982-1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Walsh, P. S., D. A. Metzger, and R. Higuchi. 1991. Chelex®100 as a medium for simple extraction of DNA for PCR-based typing from forensic material. BioTechniques 10:506-513. [PubMed] [Google Scholar]