Abstract
We created a multiplex, quantitative, real-time PCR assay that amplifies cytomegalovirus (CMV) and human DNA in the same reaction tube, allowing for a viral load determination that is normalized to measured human DNA. The assay targets a conserved region of the CMV DNA polymerase gene that is not affected by known drug resistance mutations. All 36 strains of CMV detected by culture or qualitative PCR in a population of lung transplant recipients were detected. The assay detected 1 to 10 copies of CMV plasmid DNA. The analytic sensitivity was not affected by the presence of DNA from 106 human cells but was reduced approximately 10-fold by alkaline lysates of leukocyte preparations. CMV quantitation was linear over a range of 101 to 106 copies. The intraassay and interassay coefficients of variation were 29 and 40%. Human DNA was regularly detected in patient plasma samples, and the amount was increased by storage of blood at room temperature before plasma separation and by plasma separation techniques that allowed leukocyte contamination. Applied to whole blood, the assay provides a measurement of CMV DNA in relation to cellular content without a need for cell counting procedures. Applied to plasma, the assay can reveal artifactual increases in plasma CMV levels resulting from leukocyte contamination. Further study of the utility of this assay to monitor patient populations at risk for CMV disease is warranted.
Cytomegalovirus (CMV) continues to be an important cause of morbidity and occasional mortality in immunocompromised patients. While the impact of CMV has been lessened by the use of ganciclovir, concerns including drug toxicity, cost, and drug resistance have stimulated efforts to find the most efficient strategies for using the drug, some of which involve using a diagnostic test to monitor for infection (15). The ideal diagnostic test should be sufficiently sensitive to detect infection at an early stage before clinically significant disease has occurred, but not so sensitive that only a small proportion of patients with a positive test would actually develop disease. Quantitative nucleic acid detection assays are the most likely to be useful in this way because they combine inherent analytic sensitivity with the ability to define threshold levels that could be used to initiate treatment. A number of studies have shown relationships between the level of CMV DNA in blood and the likelihood of disease (2, 5, 7, 10-13, 17, 20, 26). The stability of DNA is an additional advantage that is important because specimens may have to be transported before testing (18, 21).
The development of real-time PCR technology has simplified nucleic acid quantification and is coming into more widespread use. Real-time, quantitative PCR has several advantages over older forms of quantitative PCR assays. Experience to date has reported comparable sensitivity but superior reproducibility and precision compared to previous methods, with a wide dynamic range (14, 19). The fluorescence-based real-time assays also allow for multiple PCRs, which can be quantified independently (27), to occur in the same tube (multiplexing). This is accomplished by using probes that emit fluorescence at different wavelengths, allowing for independent detection.
We developed a multiplex, quantitative, real-time (MQR) PCR assay for CMV and human DNAs based on TaqMan technology. By performing simultaneous PCRs for CMV and human DNAs in the same reaction tube, we were able to independently quantify the amounts of CMV and human DNAs in a given specimen, making it possible to normalize the amount of CMV DNA to the amount of human DNA in the specimen. Because CMV is largely cell associated, this should decrease variability in CMV viral load measurements that is related to fluctuations in the white blood cell count. When we performed the assay on whole-blood or leukocyte specimens, we expressed the CMV viral load as the CMV copy number per microgram of human DNA. Applied to the analysis of CMV DNA in plasma, the multiplex assay can provide a measure of the contamination of plasma by leukocyte DNA that presumably occurs by cell breakage during processing. The CMV viral load in plasma could potentially be artifactually increased by such contamination.
MATERIALS AND METHODS
Real-time PCR primers and probes.
To establish the MQR PCR assay, we identified two sets of primers, one specific for CMV and the other specific for human DNA. The CMV primers were directed at a conserved region of the DNA polymerase gene of strain AD169 (GenBank accession number X17403) (6, 28). In selecting primers, we avoided regions to which ganciclovir resistance mutations have been mapped (6, 28). The forward primer (CPOL-F720) was 5′-GCT GAC GCG TTT GGT CAT C, and the reverse primer (CPOL-R780) was 5′-ACG ATT CAC GGA GCA CCA G. The internal probe (CPOL-741FAM), 5′-TCG GCG GAT CAC CAC GTT CG, was labeled at the 5′ end with the fluorescent dye 6-carboxyfluorescein and on the 3′ end with the quencher dye 6-carboxytetramethylrhodamine (TETRA). The real-time, quantitative CMV PCR assay did not amplify laboratory strains of any of the other seven human herpesviruses (data not shown).
The human primers and internal probe were purchased from Perkin-Elmer Applied Biosystems (Foster City, Calif.) and targeted a region of the human apoprotein B (HAPB) gene. The forward primer (HAPB-F8087) was 5′-TGA AGG TGG AGG ACA TTC CTC TA, and the reverse primer (HAPB-R8185) was 5′-CTG GAA TTG CGA TTT CTG GTA A. The internal probe (HAPB-801VIC) was 5′-CGA GAA TCA CCC TGC CAG ACT TCC GT and was labeled on the 5′ end by the fluorescent dye VIC and on the 3′ end by the fluorescent dye TETRA.
Real-time PCR conditions.
All multiplex PCR mixtures contained 50 μl and were performed by using the ABI Prism 7700 Sequence Detection System. Each reaction mixture contained CMV (CPOL) primers and probe at 200 nM (1 μl of a 10 μM solution of each). The HAPB probe was also at 200 nM, while the primers were at 30 nM. This concentration of HAPB primers promoted consistent amplification of human DNA past the threshold cycle (CT), allowing for its quantification, while limiting the reaction so that it did not compete with simultaneous CMV amplification for resources. TaqMan Universal PCR master mix (2×), containing AmpliTaq Gold DNA polymerase, deoxynucleoside triphosphates with dUTP, AmpErase UNG, Passive Reference 1, and optimized buffers, was purchased from Perkin-Elmer Applied Biosystems. Each 50-μl mixture contained 25 μl of master mix and 4.3 μl of primers and probes. When leukocyte lysate samples were analyzed, the volume of sample added to the reaction mixture was 8 μl. This volume was chosen to facilitate comparison with results of a qualitative CMV PCR assay which had been performed previously on the same samples using an 8-μl sample volume (20). When whole blood was analyzed, the sample volume was 10 μl. PCR cycle parameters were 2 min of incubation at 50°C and 10 min at 95°C, followed by 45 cycles of 95°C for 15 s and 60°C for 1 min.
Quantitative standards.
To establish the quantitative assay, we created a plasmid (pCPOL) containing the CMV DNA polymerase amplicon by using the TOPO TA cloning procedures with plasmid pcDNA3.1/V5/His-TOPO (Invitrogen, Carlsbad, Calif.). Insertion of the CMV amplicon was confirmed by nucleotide sequencing, and the purified recombinant plasmid was quantified by spectrophotometry. Quantification of CMV DNA in clinical samples was achieved by using serial 10-fold dilutions of the previously quantified plasmid standards. Plasmid standards and test samples were run in duplicate, and the average values were used for calculation of the CMV viral load. The prequantitated plasmid standards were diluted in 10 mM Tris (pH 9) and stored at −20°C. CMV viral loads in whole-blood and leukocyte specimens were expressed as CMV copies per microgram of human DNA, based on the measured copy numbers of CMV and human DNAs and using the conversion factor of 150,000 human cells (300,000 genomes) per μg of human DNA. The CMV viral load in plasma specimens was expressed as CMV copies per milliliter of plasma.
Specimens.
The clinical specimens used in this evaluation included leukocyte lysate preparations that were collected as part of a study of CytoGam prophylaxis in adult lung transplant recipients at risk for CMV infection (20) plus whole-blood and plasma samples collected in EDTA tubes from patients enrolled in a study of CMV kinetics who were CMV positive by culture and/or qualitative PCR. Both studies were approved by the Human Studies Committee at the Washington University School of Medicine. Leukocyte lysate preparations were prepared by boiling 106 leukocytes in 80 μl of lysate buffer (40 mM NaOH-100 mM Tris [pH 7.6]) for 10 min. PCR testing was performed on 8-μl aliquots. DNA from 200 μl of whole blood was prepared by using QIAamp DNA Mini Kits (Qiagen, Valencia, Calif.) and was resuspended in 100 μl of 10 mM Tris (pH 9). Two hundred microliters of plasma was processed for DNA by using the QIAamp DNA Mini Kits, and the DNA was resuspended in 100 μl of 10 mM Tris (pH 9). Testing was performed on 10-μl aliquots. All specimens were stored at −20°C prior to quantitative PCR testing.
RESULTS
PCR conditions.
Possible inhibitory effects of leukocyte lysate and whole-blood DNA preparations on detection of CMV were evaluated by testing dilutions of CMV plasmid (pCPOL) in reaction mixtures containing either 8 μl of leukocyte lysate (containing DNA equivalent to 105 human cells), 10 μl of a whole-blood DNA preparation (containing 8 × 104 to 2 × 105 leukocytes for normal white blood cell counts), or water. Both the cell lysates and human DNA preparations were derived from healthy CMV-negative volunteers. Compared to analysis of pCPOL in water with no added background DNA, CT values for pCPOL were significantly increased at each plasmid copy number tested when whole-blood DNA or leukocyte lysate was added to the reaction mixture (Table 1). Because of this effect of the background matrix on CMV detection, quantitative standards for all subsequent reactions were prepared to include 105 cell equivalents of leukocyte lysate or purified human DNA, selected to match the samples being analyzed.
TABLE 1.
Effect of human DNA prepared from whole blood or leukocyte lysate on detection of CMV DNA
| No. of CMV plasmid copies | Mean CT in water | Mean CT (ΔCT)a in:
|
|
|---|---|---|---|
| Whole-blood DNA | Leukocyte lysate | ||
| 106 | 26.12 | 27.27 (1.15)b | 30.59 (4.47)b |
| 103 | 33.79 | 36.08 (2.29)b | 40.08 (6.29)b |
| 102 | 36.97 | 39.75 (2.78)b | 42.90 (5.93)b |
Mean CT values are from samples run in triplicate. The CT is the PCR cycle at which the CMV detection threshold was exceeded. ΔCT is the difference between the mean CT of samples in the indicated backgrounds and the mean CT of samples in water.
The difference from the mean CT in water is significant (P < 0.01 by an unpaired t test). The mean differences between the CT in whole-blood DNA and the CT in leukocyte lysate are also statistically significant (P < 0.01 by an unpaired t test).
Sensitivity.
The analytic sensitivity of the MQR PCR assay for detection of CMV DNA was evaluated by testing serial dilutions of the CMV plasmid pCPOL, added to reaction mixtures containing either 8 μl of leukocyte lysate, 10 μl of a whole-blood preparation, or water. The MQR assay regularly detected 10 copies of plasmid pCPOL per reaction in a background of water or whole-blood DNA, and it intermittently detected 1 copy. In the background of leukocyte lysate, 100 copies of pCPOL per reaction were regularly detected and 10 copies were intermittently detected. Therefore, the sensitivity of the MQR PCR assay was considered to be 1 to 10 CMV plasmid copies when the reaction matrix was water or whole-blood DNA and 10 to 100 CMV plasmid copies when the matrix was leukocyte lysate.
The ability of the MQR PCR assay to detect a broad range of CMV isolates was evaluated by using a collection of leukocyte lysate specimens from 43 adult lung transplant recipients obtained at weekly intervals after transplantation. Each of the specimens had previously been cultured for CMV in the virology laboratory at St. Louis Children's Hospital. In addition, each specimen had also been tested for CMV DNA by using a qualitative PCR assay that detects a segment of the CMV pp65 gene (24). The MQR PCR assay detected CMV DNA in one or more specimens from all 29 patients who were positive by culture and in specimens from all 7 additional patients who were positive only by qualitative CMV PCR.
Linearity.
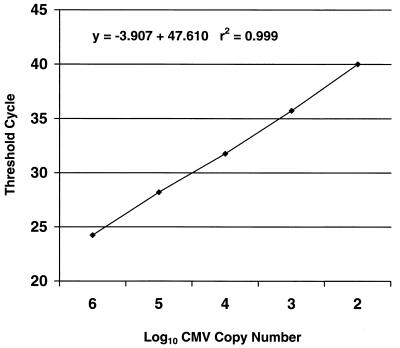
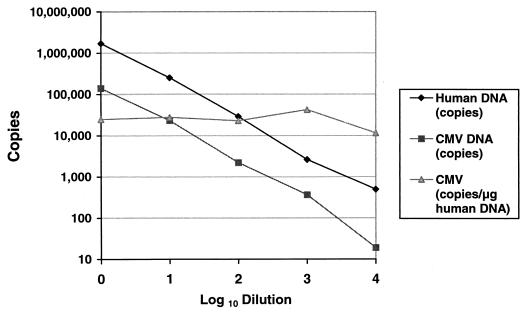
The dynamic range of the quantitative assay for CMV was evaluated with serial dilutions of the CMV plasmid and with dilutions of CMV-positive clinical specimens. As shown in Fig. 1, the relationship between the CT and the log10 plasmid pCPOL copy number was linear over the range of 102 to 106 copies. Figure 2 shows serial dilutions of a CMV-positive patient specimen, demonstrating the linearity of both the CMV and human DNA PCR assays. The calculated CMV viral load (CMV copies per microgram of human DNA) remained stable despite dilution of the original specimen.
FIG. 1.
Relationship between the CT and log10 CMV copy number, showing linearity. The CMV copy number represents copies of plasmid pJSCPOL.
FIG. 2.
Serial dilutions of a CMV-positive patient specimen, demonstrating the linearity of both the CMV and human DNA PCR assays. The calculated CMV viral load (CMV copies per microgram of human DNA) remained stable despite dilution of the original specimen.
Reproducibility.
To evaluate interassay (between-runs) and intraassay (within-run) variability, six leukocyte lysate specimens were tested in duplicate on four successive days. We made separate evaluations for CMV DNA, human DNA, and the calculated CMV viral load (CMV copy number per microgram of human DNA). The mean CMV level in the six samples ranged from 2.0 × 102 to 1.7 × 105 CMV copies. Intraassay variability was calculated for each duplicate evaluation of the six samples on all four days and was expressed as the coefficient of variation (CV, calculated as the standard deviation divided by the mean). The mean intraassay CVs were 26% for CMV DNA measurements, 18% for human DNA measurements, and 29% for the CMV viral load. The mean interassay CVs for the six samples were 34% for CMV DNA, 18% for human DNA, and 40% for the CMV viral load.
Analysis of plasma.
Initial application of the MQR PCR assay to plasma specimens indicated that human DNA was regularly detected in these specimens. We performed the following experiments to investigate the effects of specimen preparation on the amount of human DNA present in plasma. The reason for these experiments was the concern that some or all of the plasma DNA might be derived from lysis of leukocytes present in the specimen. If this was true and if the leukocytes contained CMV DNA, cell lysis would artifactually increase the amount of CMV DNA detected in plasma.
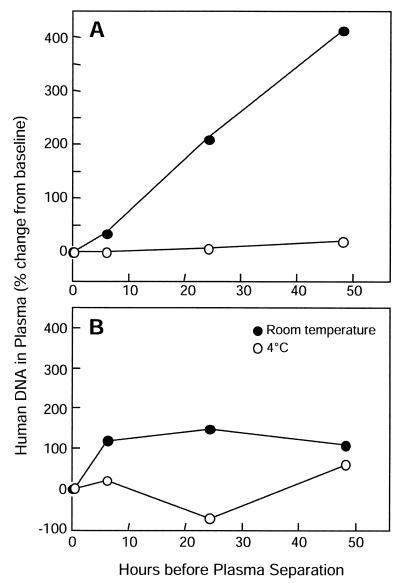
To evaluate the effects of time and temperature of storage of whole blood before plasma separation, we obtained blood specimens in EDTA tubes from two healthy human volunteers. A baseline aliquot was removed, and then each blood specimen was divided, with half of the specimen stored at room temperature and the other half stored at 4°C. Processing of aliquots consisted of removing 1.5 ml at baseline and at 6, 24, and 48 h, separating plasma by centrifugation for 10 min, and carefully collecting 200 μl of plasma from the top layer.
Corrected for concentration during processing, baseline plasma specimens contained 1.3 × 104 and 1.0 × 104 human DNA copies per ml. As shown in Fig. 3, storage at room temperature resulted in a substantial increase in the amount of human DNA detectable in the plasma of both volunteers. Storage at 4°C was not associated with significant increases in the human DNA content in plasma from either volunteer.
FIG. 3.
Effects of interval from blood draw to plasma separation and storage temperature on levels of human DNA in plasma. Panels A and B show data for two different volunteers.
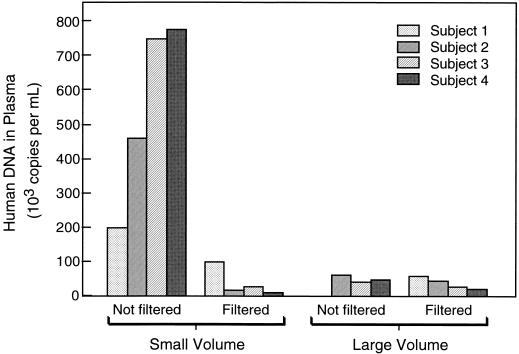
We performed an additional experiment to determine whether the technique used for plasma separation affected the amount of human DNA in plasma. Whole-blood specimens were obtained from four healthy volunteers, and plasma was separated by two different methods, one using a microcentrifuge for small volumes and one using a standard centrifuge for larger volumes. Small-volume plasma separation consisted of spinning 1.2 ml of blood in the microcentrifuge at 1,600 rpm (170 × g) for 10 min with 400 μl of plasma collected from the top layer. Large-volume plasma separation consisted of spinning 3 ml of blood in 10-ml plastic tubes at 1,600 rpm (315 × g) for 10 min and removing 1 ml of plasma from the top layer. To evaluate the possibility that intact leukocytes were contaminating the separated plasma, each plasma specimen was equally divided into two aliquots, one of which was passed through a 0.22-μm-pore-size filter (Millex-GV; Millipore Corporation, Bedford, Mass.) prior to processing. For technical reasons, no result was available for the specimen from volunteer 1 processed by the large-volume method without filtration. As shown in Fig. 4, the amounts of plasma DNA in samples from all four volunteers separated by the small-volume protocol without filtration were increased over those in samples separated by the large-volume protocol without filtration (P = 0.05 by a paired t test). The excess amount of human DNA in plasma separated by the small-volume protocol was reduced by filtration, implying that the source was intact cells.
FIG. 4.
Effect of plasma separation method on the amount of human DNA in plasma. The human DNA copy number was measured in specimens from four different volunteers after each specimen was processed according to each of four different procedures.
DISCUSSION
The MQR PCR assay is a novel test with several attributes that distinguish it from other assays for CMV. It employs TaqMan real-time PCR technology that is semiautomated and has high throughput, precision, and reproducibility. It provides sensitive detection of CMV, with primers and probe that bind to a conserved region of the CMV DNA polymerase gene that is not affected by known ganciclovir resistance mutations (6, 28). The most notable aspect of this assay, however, is its ability to determine the CMV viral load based on the content of human DNA in the specimen, reflecting the number of cells in the specimen. This ability has important implications for its use on a variety of specimens, including leukocyte preparations, whole blood, and plasma.
The most appropriate component of blood for measuring the CMV level is still undefined. However, the concept that CMV is a highly cell associated virus leads to the idea that quantitation of CMV levels within leukocytes is likely to be the measurement that most directly reflects the pathophysiology of systemic CMV infection. This has led some laboratories, including our own, to isolate, count, and adjust the number of leukocytes before performing analyses for CMV in order to take into account and correct for the wide variability in the number of leukocytes per volume unit of blood. The MQR PCR assay is able to independently quantify CMV and human DNAs in the same specimen, allowing for normalization of the CMV viral load to the measured amount of human DNA present. An important practical advantage is that the assay can be carried out on whole blood without any need for time-consuming procedures for isolation of leukocytes. As illustrated in Fig. 2, it also provides a viral load measurement that is not affected by inadvertent alteration of the specimen volume, such as dilution or desiccation, which may occur with handling or storage. Finally, since human DNA was detected in all of our specimens (including plasma), the human DNA component of the multiplex assay serves as an internal control for inhibition of PCR.
The MQR PCR assay is sensitive for the detection of CMV, with the ability to amplify 1 to 10 CMV copies per reaction tube. The addition of crude leukocyte lysate, but not purified DNA extracted from whole blood, resulted in an ∼10-fold decrease in analytic sensitivity. We accounted for this effect in the quantitative assay by ensuring that the quantitative plasmid standards contained comparable amounts of leukocyte lysate or whole-blood DNA (from CMV-negative individuals) to match the specimens being tested in the assay run. The MQR PCR assay proved to be reliable for CMV detection in a variety of patients known to have CMV infection by viral culture or qualitative PCR. Although no formal comparison of sensitivity for CMV detection was made to the qualitative CMV PCR assay in clinical use at St. Louis Children's Hospital, in the course of analyzing a series of samples from lung transplant recipients, we encountered five blood specimens from patients with active, biopsy-proven CMV pneumonitis (two of whom also had positive CMV blood cultures) that were positive by the MQR assay but negative by the qualitative assay.
The intraassay and interassay variabilities of the MQR PCR assay were comparable to the corresponding figures for a quantitative competitive PCR assay for CMV that we had developed previously (17). This was surprising, because high reproducibility is considered to be characteristic of real-time PCR assays. The reproducibility studies were performed using leukocyte lysate samples, and it is possible that the inhomogeneity of these samples may have contributed to the observed variability. It is likely that variability will be decreased when this assay is performed on DNA purified from whole-blood samples.
Because CMV is highly cell associated, samples of whole blood or leukocyte preparations provide for more sensitive detection of virus than assays that exclude leukocytes (4, 9, 22, 30). However, the presence of CMV DNA in plasma may reflect active viral replication and thus could be more associated with clinically significant disease (25, 29). Applied to plasma samples, the MQR assay reveals the amount of human DNA present, which can be an indicator of contamination of plasma by DNA derived from leukocytes. The detection of human DNA in plasma is not necessarily problematic, since cell-free DNA is regularly present in human plasma and is increased in a variety of disease states, especially cancer (1). However, as shown in Fig. 3 and 4, we found that aspects of plasma separation and storage could artifactually increase the amount of human DNA in plasma samples. Likewise, Schafer et al. described the potential for false-positive detection of CMV DNA in plasma when sample preparation is delayed (23). The potential magnitude of an artifactual increase in plasma CMV DNA levels resulting from leukocyte contamination is shown by the following example. If a patient with a white blood cell count of 5,000 cells/μl and a hematocrit of 40% had a CMV viral load of 105 copies per μg of human DNA (150,000 cells) and had lysis of 1% of the leukocytes in whole blood prior to separation of plasma, the resulting increase in plasma viral load would be 55,000 copies per ml. Further studies are required to refine the interpretation of the level of CMV DNA in plasma, but it is now evident that attention to the details of blood handling, storage, and plasma separation is required for standardization of plasma CMV DNA assays.
Important principles to protect the integrity of plasma samples include minimizing lysis of leukocytes after blood collection and minimizing contamination by leukocytes during plasma separation. Based on our findings, practical suggestions to improve the accuracy of measurement of CMV DNA levels in plasma include (i) prompt separation of plasma following phlebotomy, (ii) standardization of plasma separation procedures, at least within an individual laboratory, and (iii) filtration of plasma through a non-DNA-binding filter when leukocyte contamination is a concern. In our study, storage of blood at 4°C prior to plasma separation resulted in less contamination than storage at room temperature. However, another study found no effect of storage temperature (23). Further study is required to resolve this discrepancy.
Evidence is accumulating from a variety of patient populations that CMV viral load is an important predictor of the symptomatic disease (2, 5, 7, 10-13, 17, 20, 26). Measurement of CMV DNA levels has advantages of analyte stability, sensitivity, and a large range of values compared to the pp65 antigenemia assay and assays for CMV mRNA (3, 8, 16, 18, 21). The question of which blood fraction is best for measuring CMV DNA is not resolved. The MQR PCR assay described here has advantages of real-time PCR and simultaneous measurement of CMV and human DNAs that are applicable to either whole-blood or plasma testing.
Acknowledgments
We are grateful to Charles Rice and Alexander A. Kolykhalov for providing access to the TaqMan instrument and helping us establish the assay, to Monique Gaudreault-Keener and Richard Buller for advice on laboratory techniques and assistance with specimen storage and retrieval, to the staff of the Virology Laboratory at St. Louis Children's Hospital for assistance with specimen processing, and to Barbara Hartman for assistance with preparation of the manuscript and figures.
REFERENCES
- 1.Anker, P. 2000. Quantitative aspects of plasma/serum DNA in cancer patients. Ann. N. Y. Acad. Sci. 906:5-7. [DOI] [PubMed] [Google Scholar]
- 2.Boeckh, M., and G. Boivin. 1998. Quantitation of cytomegalovirus: methodologic aspects and clinical applications. Clin. Microbiol. Rev. 11:533-554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Boivin, G., R. Belanger, R. Delage, C. Beliveau, C. Demers, N. Goyette, and J. Roy. 2000. Quantitative analysis of cytomegalovirus (CMV) viremia using the pp65 antigenemia assay and the COBAS AMPLICOR CMV MONITOR PCR test after blood and marrow allogeneic transplantation. J. Clin. Microbiol. 38:4356-4360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Boivin, G., J. Handfield, E. Toma, G. Murray, R. Lalonde, and M. G. Bergeron. 1998. Comparative evaluation of the cytomegalovirus DNA load in polymorphonuclear leukocytes and plasma of human immunodeficiency virus-infected subjects. J. Infect. Dis. 177:355-360. [DOI] [PubMed] [Google Scholar]
- 5.Bowen, E. F., C. A. Sabin, P. Wilson, P. D. Griffiths, C. C. Davey, M. A. Johnson, and V. C. Emery. 1997. Cytomegalovirus (CMV) viraemia detected by polymerase chain reaction identifies a group of HIV-positive patients at high risk of CMV disease. AIDS 11:889-893. [DOI] [PubMed] [Google Scholar]
- 6.Chou, S., N. S. Lurain, A. Weinberg, G. Y. Cai, P. L. Sharma, C. S. Crumpacker, and Adult AIDS Clinical Trials Group CMV Laboratories. 1999. Interstrain variation in the human cytomegalovirus DNA polymerase sequence and its effect on genotypic diagnosis of antiviral drug resistance. Antimicrob. Agents Chemother. 43:1500-1502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cope, A. V., C. Sabin, A. Burroughs, K. Rolles, P. D. Griffiths, and V. C. Emery. 1997. Interrelationships among quantity of human cytomegalovirus (HCMV) DNA in blood, donor-recipient serostatus, and administration of methylprednisolone as risk factors for HCMV disease following liver transplantation. J. Infect. Dis. 176:1484-1490. [DOI] [PubMed] [Google Scholar]
- 8.Gerna, G., F. Baldanti, D. Lilleri, M. Parea, E. Alessandrino, A. Pagani, F. Locatelli, J. Middeldorp, and M. G. Revello. 2000. Human cytomegalovirus immediate-early mRNA detection by nucleic acid sequence-based amplification as a new parameter for preemptive therapy in bone marrow transplant recipients. J. Clin. Microbiol. 38:1845-1853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gerna, G., M. Furione, F. Baldanti, and A. Sarasini. 1994. Comparative quantitation of human cytomegalovirus DNA in blood leukocytes and plasma of transplant and AIDS patients. J. Clin. Microbiol. 32:2709-2717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gor, D., C. Sabin, H. G. Prentice, N. Vyas, S. Man, P. D. Griffiths, and V. C. Emery. 1998. Longitudinal fluctuations in cytomegalovirus load in bone marrow transplant patients: relationship between peak virus load, donor/recipient serostatus, acute GVHD and CMV disease. Bone Marrow Transplant. 21:597-605. [DOI] [PubMed] [Google Scholar]
- 11.Griffiths, P. D., D. A. Clark, and V. C. Emery. 2000. Betaherpesviruses in transplant recipients. J. Antimicrob. Chemother. 45(Suppl. T3):29-34. [DOI] [PubMed] [Google Scholar]
- 12.Hassan-Walker, A. F., I. M. Kidd, C. Sabin, P. Sweny, P. D. Griffiths, and V. C. Emery. 1999. Quantity of human cytomegalovirus (CMV) DNAemia as a risk factor for CMV disease in renal allograft recipients: relationship with donor/recipient CMV serostatus, receipt of augmented methylprednisolone and antithymocyte globulin (ATG). J. Med. Virol. 58:182-187. [PubMed] [Google Scholar]
- 13.Humar, A., D. Gregson, A. M. Caliendo, A. McGeer, G. Malkan, M. Krajden, P. Corey, P. Greig, S. Walmsley, G. Levy, and T. Mazzulli. 1999. Clinical utility of quantitative cytomegalovirus viral load determination for predicting cytomegalovirus disease in liver transplant recipients. Transplantation 68:1305-1311. [DOI] [PubMed] [Google Scholar]
- 14.Nitsche, A., N. Steuer, C. Schmidt, O. Landt, and W. Siegert. 1999. Different real-time PCR formats compared for the quantitative detection of human cytomegalovirus DNA. Clin. Chem. 45:1932-1937. [PubMed] [Google Scholar]
- 15.Paya, C. 2001. Prevention of cytomegalovirus disease in recipients of solid-organ transplants. Clin. Infect. Dis. 32:596-603. [DOI] [PubMed] [Google Scholar]
- 16.Preiser, W., S. Brauninger, R. Schwerdtfeger, U. Ayliffe, J. A. Garson, N. S. Brink, S. Franck, H. W. Doerr, and H. F. Rabenau. 2001. Evaluation of diagnostic methods for the detection of cytomegalovirus in recipients of allogeneic stem cell transplants. J. Clin. Virol. 20:59-70. [DOI] [PubMed] [Google Scholar]
- 17.Roberts, T. C., D. C. Brennan, R. S. Buller, M. Gaudreault-Keener, M. A. Schnitzler, K. E. Sternhell, K. A. Garlock, G. G. Singer, and G. A. Storch. 1998. Quantitative polymerase chain reaction to predict occurrence of symptomatic cytomegalovirus infection and assess response to ganciclovir therapy in renal transplant recipients. J. Infect. Dis. 178:626-635. [DOI] [PubMed] [Google Scholar]
- 18.Roberts, T. C., R. S. Buller, M. Gaudreault-Keener, K. A. Sternhell, K. Garlock, G. G. Singer, D. C. Brennan, and G. A. Storch. 1997. Effects of storage temperature and time on qualitative and quantitative detection of cytomegalovirus in blood specimens by shell vial culture and PCR. J. Clin. Microbiol. 35:2224-2228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ryncarz, A. J., J. Goddard, A. Wald, M. L. Huang, B. Roizman, and L. Corey. 1999. Development of a high-throughput quantitative assay for detecting herpes simplex virus DNA in clinical samples. J. Clin. Microbiol. 37:1941-1947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sanchez, J. L., R. M. Kruger, S. Paranjothi, E. P. Trulock, J. P. Lynch, C. Hicks, W. D. Shannon, and G. A. Storch. 2001. Relationship of cytomegalovirus viral load in blood to pneumonitis in lung transplant recipients. Transplantation 72:733-735. [DOI] [PubMed] [Google Scholar]
- 21.Schafer, P., W. Tenschert, K. Gutensohn, and R. Laufs. 1997. Minimal effect of delayed sample processing on results of quantitative PCR for cytomegalovirus DNA in leukocytes compared to results of an antigenemia assay. J. Clin. Microbiol. 35:741-744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schafer, P., W. Tenschert, L. Cremaschi, K. Gutensohn, and R. Laufs. 1998. Utility of major leukocyte subpopulations for monitoring secondary cytomegalovirus infections in renal-allograft recipients by PCR. J. Clin. Microbiol. 36:1008-1014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schafer, P., W. Tenschert, M. Schroter, K. Gutensohn, and R. Laufs. 2000. False-positive results of plasma PCR for cytomegalovirus DNA due to delayed sample preparation. J. Clin. Microbiol. 38:3249-3253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shibata, D., W. Martin, M. Appleman, D. Causey, J. Leedom, and N. Arnheim. 1988. Detection of cytomegalovirus DNA in peripheral blood of patients infected with human immunodeficiency virus. J. Infect. Dis. 158:1185-1192. [DOI] [PubMed] [Google Scholar]
- 25.Spector, S. A., R. Merrill, D. Wolf, and W. M. Dankner. 1992. Detection of human cytomegalovirus in plasma of AIDS patients during acute visceral disease by DNA amplification. J. Clin. Microbiol. 30:2359-2365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Spector, S. A., R. Wong, K. Hsia, M. Pilcher, and M. J. Stempien. 1998. Plasma cytomegalovirus (CMV) DNA load predicts CMV disease and survival in AIDS patients. J. Clin. Investig. 101:497-502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.von Ahsen, N., M. Oellerich, and E. Schutz. 2000. Use of two reporter dyes without interference in a single-tube rapid-cycle PCR: α(1)-antitrypsin genotyping by multiplex real-time fluorescence PCR with the LightCycler. Clin. Chem. 46:156-161. [PubMed] [Google Scholar]
- 28.Wirgart, B., M. Brytting, A. Linde, B. Wahren, and L. Grillner. 1998. Sequence variation within three important cytomegalovirus gene regions in isolates from four different patient populations. J. Clin. Microbiol. 36:3662-3669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wolf, D. G., and S. A. Spector. 1993. Early diagnosis of human cytomegalovirus disease in transplant recipients by DNA amplification in plasma. Transplantation 56:330-334. [DOI] [PubMed] [Google Scholar]
- 30.Zipeto, D., S. Morris, C. Hong, A. Dowling, R. Wolitz, T. C. Merigan, and L. Rasmussen. 1995. Human cytomegalovirus (CMV) DNA in plasma reflects quantity of CMV DNA present in leukocytes. J. Clin. Microbiol. 33:2607-2611. [DOI] [PMC free article] [PubMed] [Google Scholar]