Abstract
A PCR assay that uses primers whose sequences were obtained from the published sequence of the cdt-III gene was developed to determine the frequencies of the cdt-I, cdt-II, and cdt-III genes in Escherichia coli isolates from humans and animals. E. coli isolates producing cytolethal distending toxin (CDT) were infrequently detected. The cdt-I gene was preferentially detected in strains with the cnf1 gene, while the cdt-III gene was found in strains carrying the cnf2 gene. The cdt-III genotype was more prevalent in animal isolates, while the cdt-I and cdt-II genotypes were more evident in human isolates. The presence of further cdt gene variants was indicated by the presence of toxin activity in cell culture in the absence of PCR amplification of the cdt-I, cdt-II, or cdt-III gene.
Cytolethal distending toxin (CDT) is a potent bacterial exotoxin that has dramatic effects on target cells in culture (23). Intoxication of eukaryotic cells results in blockage of the cell cycle at the G2/M transition (2, 22) by a mechanism involving prevention of cdc2 protein kinase dephosphorylation and activation (6). Cellular effects include accumulation of F-actin assemblies resembling stress fibers (2), progressive cell distension, and eventual cell death (14, 15). Epidemiologic studies have not found a statistically significant difference in the incidence of CDT-producing Escherichia coli in children with diarrhea and healthy controls (23). However, animal studies with toxigenic E. coli strains (18) and Campylobacter jejuni CDT-knockout mutants (26) suggest that CDT may be a virulence factor in vivo.
Three genes, cdtA, cdtB, and cdtC, are required for the production of an active toxin in E. coli (24, 28), Shigella dysenteriae (19), Campylobacter spp. (25), Haemophilus ducreyi (7), Helicobacter hepaticus (32), and Actinobacillus actinomycetemcomitans (16). Although these genes appear to be homologous in all bacteria, the degree of relatedness at the genetic level varies widely (23). Three cdt genetic variants, designated cdt-I, cdt-II, and cdt-III, have been identified and cloned from E. coli (22, 24, 28). However, PCR methods have been developed only for the detection of the cdt-I and cdt-II variants (19). The distribution of the cdt-I and cdt-II genes within E. coli is not well characterized (23), and little is known about the incidence or prevalence of cdt-III. PCR primers were developed for the detection of cdt-III. A number of E. coli isolates from humans and animals were tested for the presence of all three cdt variants.
A group of 46 CDT-producing E. coli isolates was collected between 1986 and 1998 from Canadian and international sources. CDT-positive animal isolates collected during this time were also characterized. Enhanced surveillance for cdt genes was accomplished by testing 151 non-Shiga toxin-producing non-O157:H7 E. coli isolates collected between October 1998 and the end of 2000 together with a randomly chosen subset of 53 E. coli O157:H7 isolates. In addition, 72 non-O157 E. coli isolates from a series of consecutive blood cultures obtained in the St. Boniface Hospital, Winnipeg, Manitoba, Canada, were examined for CDT.
Culture media for E. coli, Aeromonas spp., Campylobacter spp., Arcobacter spp., and Helicobacter spp. were optimized for CDT production (14, 15). Bacterial supernatants were obtained by centrifugation and filtration through 0.2-μm-pore-size filters. After a maximum of eight twofold dilutions, the supernatants were added to nonconfluent Chinese hamster ovary (CHO) cells seeded at 2 × 104 cells/ml. Observations for the characteristic effects of CDT were made between 24 and 120 h of incubation. For neutralization studies, rabbit polyclonal antibodies were prepared from partially purified CDT-I. Filtrates were also added to Vero, Y-1 adrenal, and HeLa cells to check for the presence of other toxins (14).
Template DNA was prepared either by alkaline lysis with phenol-chloroform extraction (27) or by boiling two to three single colonies of the target isolate in 100 μl of distilled water for 5 min. PCR primers were designed to amplify the sequence between nucleotides 1066 and 3296 of the cdt-IIIA sequence (22). The forward primer (primer cdt-III-f) was 5′-AAA CAG GAC GGT AAT AAT GAC TAA TA-3′, and the reverse primer (primer cdt-III-r) was 5′-GTG ATC TCC TTC CAT GAA AAT ATA GT-3′. PCR was performed in a mixture that consisted of 1× PCR buffer, 2 mM deoxynucleoside triphosphates, 1.5 mM MgCl2, 20 μM primer oligonucleotides, and 1.25 U of Taq polymerase with a Gene Amp PCR System 2400 (Perkin-Elmer Applied Biosystems Ltd., Mississauga, Ontario, Canada). Initial denaturation at 94°C for 2 min was followed by 30 cycles of 94°C for 2 min, 54°C for 2 min, and 72°C for 1 min and 2 cycles of extension at 72°C for 7 min. The amplicon was 2,230 bp long and contained the entire cdt-III-coding region. The cdt-III PCR product was digested with SmaI to give three fragments of 187, 260 and 1,783 bp. PCR for cdt-I and cdt-II was done by the protocol of Okuda et al. (19).
The cells were evaluated by PCR for the presence of the following other virulence factors: Shiga toxin genes stx1 and stx2 (17, 21); the eae gene (11, 21); the elt gene, which encodes the E. coli heat-labile toxin (10, 20); the est gene, which encodes the E. coli heat-stable toxin (20, 30); the invasion-associated locus (ial) of the enteroinvasive E. coli virulence plasmid or invasion genes (20, 29); cytotoxic necrotizing factor genes cnf1 and cnf2 (4, 20); and a gene associated with the enteroaggregative phenotype (20).
The cdt-III PCR product was subjected to automated fluorescent cycle sequencing with a Wizard Miniprep kit (Promega; Fisher Scientific Ltd., Ottawa, Ontario, Canada) and an ABI Prism Big Dye Terminator Cycle Sequencing Ready Reaction kit (version 2.0; Applied Biosystems) on an ABI PRISM 3100 Genetic Analyzer (Applied Biosystems). Primers cdt-III-f and cdt-III-r were used for the initial sequencing reactions. The internal primer sets used to complete the sequencing were cdt-III seq (5′-GGC TCC ACA TTG TTA GAA AGC-3′), cdtIII seq-int3 (5′-GTC TAC AGG TTT ATG CAT CAG CAG AGC-3′), cdtIII seq-int4′ (5′-CCA TTG CTT GGC ATA CGG ATT GGC-3′), cdtIII seq-3′ (5′-CAT TCC GCA ATC TCC AAA GTG GGC-3′), cdt-III seq-f (nucleotide 1724; 5′-GAA ACT TCG AAT TTA TGT GG-3′), and cdtIII seq-r (nucleotide 2629; 5′-AAG ACA TCT GAG TCC TTC TG-3′). LaserGene-DNAStar software was used for sequence editing and comparisons.
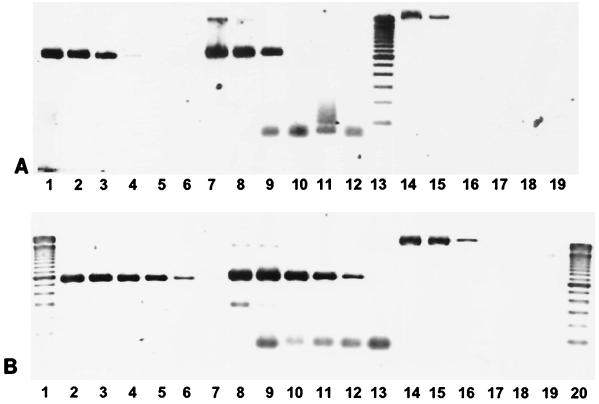
Evaluation of the PCR method for detection of the cdt-III gene indicated that the primer sets were specific. Of 315 E. coli isolates examined, 24 possessed the cdt-I gene alone, 5 carried the cdt-II gene alone, and 21 carried the cdt-III gene alone. No isolates carried combinations of cdt genes. Isolates of Campylobacter (11 isolates), Helicobacter (8 isolates), Aeromonas (31 isolates), Salmonella (4 isolates), and Arcobacter (4 isolates) did not amplify a product by use of primers specific for cdt-III even when they were positive for CDT by the CHO cell assay. Detection of the cdt-III gene required 1.25 × 104 bacterial cells, while detection of the cdt-I and cdt-II genes required 1.25 × 102 and 1.25 × 103 bacterial cells, respectively (Fig. 1A). Both the cdt-I-specific and cdt-II-specific primers had high degrees of sensitivity when pure DNA was used, detecting 0.05 ng of DNA, while with the cdt-III-specific primers a minimum of 5 ng of DNA was required for detection (Fig. 1B). The sequence of the cdt-III PCR product obtained from three representative isolates had 100% homology with the published sequence (data not shown). Digestion of the amplified products with SmaI gave the 187-, 260-, and 1,783-bp DNA fragments expected, confirming the high degree of sequence homology.
FIG. 1.
Sensitivities of cdt-specific primers. (A) Different concentrations of boiled cell preparations. The isolates in lanes 1 to 6 were amplified with cdt-I-specific primers, the isolates in lanes 7 to 12 were amplified with cdt-II-specific primers, and the isolates in lanes 14 to 19 were amplified with cdt-III-specific primers. The numbers of boiled bacterial cells serving as templates for each lane were as follows: lanes 1, 7, and 14, 1.25 × 105; lanes 2, 8, and 15, 1.25 × 104; lanes 3, 9, and 16, 1.25 × 103; lanes 4, 10, and 17, 1.25 × 102; lanes 5, 11, and 18, 12.5; lanes 6, 12, and 19, 1.25. Lane 13, 100-bp ladder. (B) Different concentrations of extracted DNA preparations. The isolates in lanes 2 to 7 were amplified with cdt-I-specific primers, the isolates in lanes 8 to 13 were amplified with cdt-II-specific primers, and the isolates in lanes 14 to 19 were amplified with cdt-III-specific primers. The amounts of DNA in each lane were as follows: lanes 2, 8, and 14, 500 ng; lanes 3, 9, and 15, 50 ng; lanes 4, 10, and 16, 5 ng; lanes 5, 11, and 17, 0.5 ng; lanes 6, 12, and 18, 0.05 ng. Lanes 7, 13, and 19, negative controls containing water instead of the DNA template used for each PCR; lanes 1 and 20, 100-bp ladders.
The distribution of the cdt genes among the isolates and their association with other virulence factors are shown in Table 1. cdt-III was strongly associated with cnf2, and cdt-I was strongly associated with cnfI. All cdt-III-positive, cnf2-positive strains were isolated from cattle. Several of these strains also carried the stx1 gene. All cdt-I-positive and cnf1-positive strains were derived from humans, as were all except one of the other strains carrying cdt-I; the strain that was the exception was isolated from a pig. Similarly, all cdt-II-positive E. coli were from human patients. A minority (2 of 50) of the CDT-producing E. coli isolates were found to carry the eae gene. While some cdt-positive E. coli that also express the heat-stable toxin have been found (5), none were found in this study.
TABLE 1.
Association of other E. coli virulence genes with cdt genes
| cdt variant (no. of isolates) | No. of isolates with each combination of genesa
|
|||||||
|---|---|---|---|---|---|---|---|---|
| A cdt gene alone | eae | stx1 + stx2 | stx2 | cnf1 | cnf2 | cnf1 + eae | cnf2 + stx1 | |
| cdt-I (24) | 10 | 1 | 1 | 0 | 12 | 0 | 0 | 0 |
| cdt-II (5) | 4 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
| cdt-III (21) | 1 | 0 | 0 | 1 | 0 | 10 | 0 | 9 |
Fifty of 322 E. coli strains were positive for one or more variants of cdt.
Evaluation of 151 non-O157:H7 stx-negative E. coli strains from humans, animals, and the environment for the presence of the cdt-I, cdt-II, and cdt-III genes showed that 5 (3.3%) of the strains carried a cdt gene variant. Carriage of cdt genes was more frequent among the non-O157 blood isolates (5 of 72; 6.9%). None of 53 randomly selected E. coli O157:H7 isolates were found to carry any of the cdt variant genes. The results of CHO cell assays for CDT correlated well with those of PCR for the detection of cdt gene variants. However, among 46 CDT-producing human isolates collected between 1986 and 1998 and used to estimate the frequency of carriage of cdt gene variants, 3 isolates (7%) carried the cdt-I gene but did not produce detectable toxin. In addition, seven strains (15%) were CDT positive only by the cell culture assay. Toxin identity was confirmed in these seven strains by neutralization with specific anti-CDT antiserum.
CDT production has been associated with enteropathogenic E. coli (EPEC) serogroups (4), and an association between the cdt gene and the eae gene has been demonstrated (1, 5, 12). The frequency of cdt gene carriage by EPEC strains was found to be 2.8% in Brazil (12) and 6.4% in India (5). Similar frequencies of carriage were found among the non-O157 E. coli isolates in the present study. However, carriage of cdt genes is not strongly correlated with the presence of the eae gene alone. Both the low prevalence of cdt genes and their association with other virulence genes suggest that the cdt genes are acquired independently in a number of E. coli lineages, possibly as the result of horizontal gene transfer. Among blood culture isolates, cdt was detected in only a minority of strains within the E. coli B2 virulence group (13). Furthermore, analysis of the sequence surrounding the A. actinomycetemcomitans cdt genes and of naturally occurring cdt deletions suggested that the cdt genes are contained within a virulence-associated region resembling a pathogenicity island (16). Whether this is also true for cdt-positive E. coli strains is not known.
In some strains, the genes encoding cdt-III and cnf2 are carried on the pVir plasmid (22), suggesting they may be readily transferred among E. coli strains. These observations support the hypothesis that cdt genes may have been acquired recently by a small proportion of E. coli strains. The strains were not assessed for the presence of the pVir plasmid in the present study; however, there was a strong association (19 of 21 isolates) between the presence of cdt-III and the presence of cnf2. In contrast to a previous study (13), there was a strong association between carriage of the cdt-I variant and carriage of cnf1. The origins of the E. coli strains in this study and the previous study (13) differ, in that many strains in the present study were obtained from stools of patients with diarrhea. Further investigations into the association of these genes in different E. coli subpopulations are required to resolve this issue.
Previous studies indicated a strong association of cnf1-positive strains with humans and cnf2-positive strains with animals (3). The association of cdt-III with cattle was therefore not surprising given that cdt-III is carried on the same plasmid as cnf2. The presence of the cdt-III gene was associated with the stx1 genotype. E. coli strains carrying only stx1 are also prevalent in cattle (8, 9, 31). In contrast, carriage of the cdt-I gene was not strongly correlated with the presence of stx genes. Although the underlying mechanisms responsible for the associations seen in the present study are not known, the results imply that cattle and humans provide ecological niches for E. coli strains that impose different selective pressures on the organism, resulting in the carriage of different virulence gene sets. In this case selection may be for genes other than cdt-III on the pVir virulence plasmid.
The cdt-III-specific PCR described here was effective in detecting the cdt-III gene from isolated DNA and should prove useful for studies aimed at characterizing the distribution of cdt genes in E. coli populations. However, CDT was detected by cell culture assays in strains that were negative by PCR with primers specific for the cdt-I, cdt-II, and cdt-III gene variants. This supports the hypothesis of Pérès and colleagues (22) that there are additional cdt gene variants that cannot be found by the available PCR methods and suggests that both PCR and cell culture assays are necessary for enumeration of CDT-positive E. coli strains. Characterization of these additional variant genes will aid in the development of improved PCR-based assays for the detection of cdt.
Acknowledgments
We thank Claude Ouellette for the sequencing data, David Woodward for providing strains from the LCDC collection, Michelle Alfa from the St. Boniface Hospital for contributing the panel of human blood isolates, and the provincial public health laboratories for submission of isolates to National Laboratory for Enteric Pathogens. Thanks also go to Dave Spreitzer for technical assistance and to Louis Bryden for DNA sequencing.
REFERENCES
- 1.Ansaruzzaman, M., M. J. Albert, S. Nahar, R. Byun, M. Katouli, I. Kühn, and R. Möllby. 2000. Clonal groups of enteropathogenic Escherichia coli isolated in case-control studies of diarrhoea in Bangladesh. J. Med. Microbiol. 49:177-185. [DOI] [PubMed] [Google Scholar]
- 2.Aragon, V., K. Chao, and L. A. Dreyfus. 1997. Effect of cytolethal distending toxin on F-actin assembly and cell division in Chinese hamster ovary cells. Infect. Immun. 65:3774-3780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Blanco, J., M. Blanco, M. Alonso, J. E. Blanco, J. I. Garabal, and E. A. González. 1992. Serogroups of Escherichia coli strains producing cytotoxic necrotizing factors CNF1 and CNF2. FEMS Microbiol. Lett. 96:155-160. [DOI] [PubMed] [Google Scholar]
- 4.Blanco, M., J. E. Blanco, J. Blanco, M. P. Alonso, C. Balsalobre, M. Mourino, C. Madrid, and A. Juarez. 1996. Polymerase chain reaction for detection of Escherichia coli strains producing cytotoxic necrotizing factor type 1 and type 2 (CNF1 and CNF2). J. Microbiol. Methods 26:95-101. [Google Scholar]
- 5.Bouzari, S., and A. Varghese. 1990. Cytolethal distending toxin (CLDT) production by enteropathogenic Escherichia coli (EPEC). FEMS Microbiol. Lett. 71:193-198. [DOI] [PubMed] [Google Scholar]
- 6.Comayras, C., C. Tasca, S. Y. Pérès, B. Ducommun, E. Oswald, and J. De Rycke. 1997. Escherichia coli cytolethal distending toxin blocks the HeLa cell cycle at the G2/M transition by preventing cdc2 protein kinase dephosphorylation and activation. Infect. Immun. 65:5088-5095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cope, L. D., S. Lumbley, J. L. Latimer, J. Klesney-Tait, M. K. Stevens, L. S. Johnson, M. Purven, R. S. Munson, Jr., T. Lagergard, J. D. Radolf, and E. J. Hansen. 1997. A diffusible cytotoxin of Haemophilus ducreyi. Proc. Natl. Acad. Sci. USA 94:4056-4061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cray, W. C., Jr., L. A. Thomas, R. A. Schneider, and H. W. Moon. 1996. Virulence attributes of Escherichia coli isolated from dairy heifer feces. Vet. Microbiol. 53:369-374. [DOI] [PubMed] [Google Scholar]
- 9.Dorn, C. R., D. H. Francis, E. J. Angrick, J. A. Willgohs, R. A. Wilson, J. E. Collins, B. H. Jenke, and S. J. Shawd. 1993. Characteristics of Vero cytotoxin producing Escherichia coli associated with intestinal colonization and diarrhea in calves. Vet. Microbiol. 36:149-159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Frankel, G., J. A. Giron, J. Valmassoi, and G. K. Schoolnik. 1989. Multi-gene amplification: simultaneous detection of three virulence genes in diarrhoeal stool. Mol. Microbiol. 3:1729-1734. [DOI] [PubMed] [Google Scholar]
- 11.Gannon, V. P. J., M. Rashed, R. K. King, and E. Golsteyn-Thomas. 1993. Detection and characterization of the eae gene of Shiga-like toxin-producing Escherichia coli using polymerase chain reaction. J. Clin. Microbiol. 31:1268-1274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Guth, B. E. C., R. Giraldi, T. A. T. Gomes, and L. R. M. Marques. 1994. Survey of cytotoxin production among Escherichia coli strains characterized as enteropathogenic (EPEC) by serotyping and the presence of EPEC adherence factor (EAF) sequences. Can. J. Microbiol. 40:341-344. [DOI] [PubMed] [Google Scholar]
- 13.Johnson, J. R., and A. L. Stell. 2000. Extended virulence genotypes of Escherichia coli strains from patients with urosepsis in relation to phylogeny and host compromise. J. Infect. Dis. 181:261-272. [DOI] [PubMed] [Google Scholar]
- 14.Johnson, W. M., and H. Lior. 1988. A new heat-labile cytolethal distending toxin (CLDT) produced by Escherichia coli isolates from clinical material. Microb. Pathog. 4:103-113. [DOI] [PubMed] [Google Scholar]
- 15.Johnson, W. M., and H. Lior. 1988. A new heat-labile cytolethal distending toxin (CLDT) produced by Campylobacter spp. Microb. Pathog. 4:115-126. [DOI] [PubMed] [Google Scholar]
- 16.Mayer, M. P. A., L. C. Bueno, E. J. Hansen, and J. M. DiRienzo. 1999. Identification of a cytolethal distending toxin gene locus and features of a virulence-associated region in Actinobacillus actinomycetemcomitans. Infect. Immun. 67:1227-1237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Meng, J., S. Zhao, M. P. Doyle, S. E. Mitchell, and S. Kresovich. 1996. A multiplex PCR for identifying Shiga-like toxin-producing Escherichia coli O157:H7. Lett. Appl. Microbiol. 24:172-176. [DOI] [PubMed] [Google Scholar]
- 18.Okuda, J., M. Fukumoto, Y. Takeda, and M. Nishibuchi. 1997. Examination of diarrheagenicity of cytolethal distending toxin: suckling mouse response to the products of the cdtABC genes of Shigella dysenteriae. Infect. Immun. 65:428-433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Okuda, J., H. Kurazono, and Y. Takeda. 1995. Distribution of the cytolethal distending toxin A gene (cdtA) among species of Shigella and Vibrio, and cloning and sequencing of the cdt gene from Shigella dysenteriae. Microb. Pathog. 18:167-172. [DOI] [PubMed] [Google Scholar]
- 20.Pass, M. A., R. Odedra, and R. M. Batt. 2000. Multiplex PCRs for identification of Escherichia coli virulence genes. J. Clin. Microbiol. 38:2001-2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Paton, A. W., and J. C. Paton. 1997. Detection and characterization of Shiga toxigenic Escherichia coli by using multiplex PCR assays for stx1, stx2, eaeA, enterohemorrhagic E. coli hlyA, rfb0111, and rfb0157. J. Clin. Microbiol. 36:598-602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pérès, S. Y., O. Marchès, F. Daigle, J. Nougayrède, F. Hérault, C. Tasca, J. De Rycke, and E. Oswald. 1997. A new cytolethal distending toxin (CDT) from Escherichia coli producing CNF2 blocks HeLa cell division in G2/M phase. Mol. Microbiol. 24:1095-1107. [DOI] [PubMed] [Google Scholar]
- 23.Pickett, C. L., and C. A. Whitehouse. 1999. The cytolethal distending toxin family. Trends Microbiol. 7:292-297. [DOI] [PubMed] [Google Scholar]
- 24.Pickett, C. L., D. L. Cottle, E. C. Pesci, and G. Bikah. 1994. Cloning, sequencing and expression of the Escherichia coli cytolethal distending toxin genes. Infect. Immun. 62:1046-1051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pickett, C. L., E. C. Pesci, D. L. Cottle, G. Russell, A. N. Erdem, and H. Zeytin. 1996. Prevalence of cytolethal distending toxin production in Campylobacter jejuni and relatedness of Campylobacter sp. cdtB genes. Infect. Immun. 64:2070-2078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Purdy, D., C. M. Buswell, A. E. Hodgson, K. McAlpine, I. Henderson, and S. A. Leach. 2000. Characterisation of cytolethal distending toxin (CDT) mutants of Campylobacter jejuni. J. Med. Microbiol. 49:473-479. [DOI] [PubMed] [Google Scholar]
- 27.Sambrook, J., E. F. Fritsch, and T. Maniatis, ed. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 28.Scott, D. A., and J. B. Kaper. 1994. Cloning and sequencing of the genes encoding Escherichia coli cytolethal distending toxin. Infect. Immun. 62:244-251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sethabutr, O., M. Venkatesan, G. S. Murphy, B. Eampokalap, C. W. Hoge, and P. Echeverria. 1993. Detection of shigellae and enteroinvasive Escherichia coli by amplification of the invasion plasmid antigen H DNA sequence in patients with dysentery. J. Infect. Dis. 167:458-461. [DOI] [PubMed] [Google Scholar]
- 30.Stacy-Phipps, S., J. J. Mecca, and J. B. Weiss. 1995. Multiplex PCR assay and simple preparation method for stool specimens detect enterotoxigenic Escherichia coli DNA during course of infection. J. Clin. Microbiol. 33:1054-1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tokhi, A. M., J. S. M. Peiris, S. M. Scotland, G. A. Willshaw, H. R. Smith, and T. Cheasty. 1993. A longitudinal study of Vero cytotoxin producing Escherichia coli in cattle calves in Sri Lanka. Epidemiol. Infect. 110:197-208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Young, V. B., K. A. Knox, and D. B. Schauer. 2000. Cytolethal distending toxin sequence and activity in the enterohepatic pathogen Helicobacter hepaticus. Infect. Immun. 68:184-191. [DOI] [PMC free article] [PubMed] [Google Scholar]