Abstract
Hepatitis B virus (HBV) surface gene variants have been associated with diagnostic escape and immune escape following vaccination. The most common mutation observed in these variants is a glycine-to-arginine substitution at amino acid 145 (G145R). In order to sensitively detect the presence of this mutant in serum, a new molecular detection system was developed; in this new system, a gap ligase chain reaction (gLCR) assay was coupled with electrochemiluminescence detection of reaction products. The gLCR assay could detect approximately 10 copies of mutant DNA and could discriminate low levels of mutant DNA in the presence of excess wild-type DNA. Detection of the G145R mutant in clinical specimens was evaluated by testing 56 suspect serum specimens. The G145R mutation was observed in 18 of 28 HBV-DNA-positive samples. The approximate percentage of mutant present in each specimen was calculated by comparison with a standard curve of an increasing ratio of mutant DNA to wild-type DNA. Most samples contained a very low percentage of mutant virus (approximately 5%), with an observed range of approximately 3 to 74%. The G145R mutation was most frequently observed in specimens producing a diagnostic anomaly or from transplant patients but was also observed in specimens from vaccinated individuals and specimens in which HBsAg diagnostic escape was suspected. Therefore, the gLCR assay is a sensitive and specific method for detection of G145R mutants, which could be modified to include the detection of other HBV mutants.
During infection with hepatitis B virus (HBV), exposure of the viral surface protein (HBsAg) to host immunity provides an important basis for protective responses against HBV. However, this exposure of HBsAg, coupled with the fact that HBV demonstrates a 10-fold-greater mutation rate than other DNA viruses (14), provides an opportunity for the appearance of point mutations, insertions, and deletions in the S gene. Related to the appearance of surface gene variations is the emergence of immune escape variants in individuals who become infected with HBV despite the presence of anti-HBs antibody derived either through vaccination or hepatitis B immunoglobulin (HBIg) prophylaxis (10, 17, 20). One of the most frequently observed mutations in these variants is a change from glycine to arginine at amino acid 145 (G145R) within the major hydrophilic domain of the S protein (6). Although other nucleotide substitutions have been associated with immune escape, the G145R mutation is the most common and appears to be the most stable, as it does not readily revert to the wild type (13). The G145R mutation has also been shown in chimpanzee studies to be readily transmissible and pathogenic (23).
This mutant and other HBsAg mutants may pose a significant risk to the blood supply, as certain HBsAg diagnostic test kits are either unable to detect mutant HBsAg or they have decreased sensitivity (15, 29). Also, vaccinated individuals may be susceptible to infection with the mutant HBV virus, as vaccine-elicited antibodies may not fully recognize the mutant S protein, although this is a controversial issue (25, 30). These points highlight the requirement for a sensitive detection system to allow close monitoring and surveillance of these mutants. A gap ligase chain reaction (gLCR) assay was developed; the gLCR assay allows detection of the G145R mutant from serum specimens following electrochemiluminescence (ECL) detection of reaction products. The gLCR assay was able to sensitively distinguish the G145R mutant in a mixed HBV population.
MATERIALS AND METHODS
Serum specimens.
Serum specimens were selected from routine diagnostic specimens sent from laboratories across Canada for detection of serological HBV markers and HBV DNA. The complete HBV serological status of each specimen was not known. Specimens from the years 1997 through 2000 were chosen on the basis of clinical or laboratory information provided with the specimen that would suggest the presence of a possible HBsAg mutant (n = 53) or were randomly chosen from chronic hepatitis patients (n = 3). Specimens suggesting the presence of a possible HBsAg mutant were selected from the following individuals or specimens: liver (n = 28), kidney (n = 2), and bone marrow (n = 1) transplant recipients; individuals in whom vaccine failure was suspected (n = 6), including two vaccinated individuals born to HBV carrier mothers; individuals undergoing HBIg therapy (n = 3); individuals undergoing interferon treatment (n = 1); individuals with transfusion-associated hepatitis (n = 1); individuals with nonspecific hepatitis (n = 2); and specimens producing a diagnostic anomaly (weak HBsAg with strong positive PCR; anti-HBe and HBeAg positive; anti-HBs and HBsAg positive; diagnostic failure) (n = 9).
DNA extraction and PCR.
DNA was extracted from 150 μl of serum by proteinase K-sodium dodecyl sulfate lysis and phenol-chloroform extraction methods (18), and resuspended in a final volume of 30 μl of sterile, nuclease-free water. Extracted DNA or control vector DNA was amplified by nested PCR using primers specific for the S gene of HBV for the first stage (sense, Spr1A [5′ GTTCAGGAACAGTAAGCCC 3′]; antisense, Spr2A [5′ ACTTTCCAATCAATAGGCC 3′]) and primers specific for the a determinant of the S protein for the second stage (sense, adetpr1 [5′ CCCGTTTGTCCTCTAMTTCCAGG 3′]; antisense, adetpr2 [5′ YGATGGGATGGGAATACARGTGC 3′]). All oligonucleotides used for amplification were custom synthesized by the DNA Core Facility at the National Microbiology Laboratory. Reaction tubes for both stages of PCR contained 5 μl of DNA extract or first-stage PCR product, AmpliTaq Gold reaction buffer (Applied Biosystems, Foster City, Calif.), 0.2 mM (each) deoxynucleoside triphosphates (Invitrogen Life Technologies, Burlington, Ontario, Canada), 2.5 mM MgCl2, 25 pmol of each primer, and 2.5 U of AmpliTaq Gold polymerase. Thermal cycling parameters for the first stage of amplification involved touchdown PCR (2 cycles at each annealing temperature, with 1 cycle consisting of 30 s at 94°C, 30 s at 59°C [with 1°C touchdown to 55°C], and 30 s at 72°C), followed by 35 cycles, with 1 cycle consisting of 30 s at 94°C, 30 s at 54°C, and 30 s at 72°C (8). Nested PCR cycling parameters involved 35 cycles, with 1 cycle consisting of 30 s at 94°C, 30 s at 58°C, and 30 s at 72°C. In order to prevent PCR carryover contamination during nested PCR, each step of the procedure was performed in a separate room with dedicated equipment, with a directional flow from the beginning of the procedure to the end. Negative controls containing serum or water were also included in each extraction run, and an extra negative control containing water was included at each stage of PCR. Amplified products were gel purified using a Wizard DNA Clean-Up System (Promega Corporation, Madison, Wis.).
gLCR.
To develop the gLCR assay, vector constructs containing S genes having either a guanine (wild type) or adenine (G145R mutant) at nucleotide 587 were prepared. The adenine-587 construct (pA587) was prepared by ligation of an S-gene PCR product derived from amplification of DNA extracted from serum known to contain a mixture of wild-type and G145R mutant virus (a kind gift from Robert Purcell, Hepatitis Viruses Section, National Institute of Allergy and Infectious Diseases, National Institutes of Health) (23). The guanine-587 construct (pG587) was prepared by ligation of an S-gene PCR product derived from amplification of DNA extracted from an HBV-DNA-positive, HBsAg-positive, HBeAg-positive, anti-HBe antibody-negative, clinical specimen. All stock preparations of cloned DNA used for gLCR experiments were sequenced to confirm a wild-type or mutant genotype at nucleotide 587.
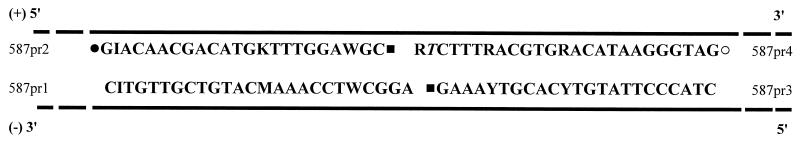
The gLCR assay was performed by addition of 5 μl of gel-purified amplicon to 45 μl of gLCR premix, followed by incubation at 94°C for 3 min, 25 thermocycles (1 cycle consisting of 30 s at 94°C and 60 s at 65°C), and a final incubation at 99°C for 10 min. Each gLCR tube contained the following components in a final volume of 50 μl: 1 μM concentrations (each) of dTTP, dCTP, and dATP; 150 nM concentrations (each) of probes 587pr1 to 587pr4 (Fig. 1); reaction buffer (50 mM Tris-HCl [pH 7.6], 25 mM potassium acetate, 10 mM magnesium acetate, 10 mM magnesium chloride, 10 mM dithiothreitol, 1 mM NAD, 0.1% Triton X-100; all obtained from Sigma-Aldrich Canada Ltd., Oakville, Ontario, Canada); 0.5 μl of salmon sperm DNA (Invitrogen Life Technologies); 1 U of Vent (exo−) DNA polymerase (New England BioLabs, Mississauga, Ontario, Canada); and 20 U of Taq DNA ligase (New England BioLabs). Certain chemical modifications were included on probes 587pr2, 587pr3, and 587pr4 to permit ECL detection of gLCR products. Probe 587pr2 had both a 5′ phosphate addition and 3′ biotin label, while probe 587pr3 had a 5′ phosphate addition (both prepared by the DNA Core Facility of the National Microbiology Laboratory), and probe 587pr4 had a 5′ ruthenium label (prepared by IGEN International, Inc., Gaithersburg, Md.). Probes 587pr2 and 587pr4 hybridize to the sense strand of genomic HBV DNA, while probes 587pr1 and 587pr3 hybridize to the antisense strand of genomic HBV DNA (Fig. 1). A mismatch in 587pr4 specific for the G145R substitution was included at the penultimate 3′ end instead of the ultimate 3′ end of the probe. This placement was based on previous evidence for gLCR detection of Chlamydia trachomatis and human immunodeficiency virus which suggested that a mismatch position at the penultimate 3′ end is discriminated better than a mismatch at the ultimate 3′ end (1).
FIG. 1.
gLCR probes for the specific detection of the G145R substitution. Labels and probe additions are indicated as follows: ▪, 5′ phosphate addition; •, biotin label; ○, ruthenium label. HBV genomic DNA is denoted by the black lines. Probes 587pr2 and 587pr4 hybridize to the sense strand (+) of HBV genomic DNA, while probes 587pr1 and 587pr3 hybridize to the antisense strand (−) of HBV genomic DNA. The penultimate 3′ nucleotide of probe 587pr4 (T) is the mismatch specific for the G145R substitution.
Negative controls for gLCR included a template-free reaction tube as well as 5 ng of gel-purified, nested PCR amplicon amplified from the pG587 construct, run in triplicate or more. A positive control of 5 ng of gel-purified, nested PCR amplicon amplified from the pA587 construct was also included in each gLCR assay.
ECL detection.
Products resulting from gLCR were analyzed by an ORIGEN analyzer (IGEN International, Inc.). ORIGEN technology utilizes streptavidin-coated, paramagnetic beads as a support phase to capture biotin-labeled products. Captured products are then detected through electrical excitation of an additional ruthenium label to produce a light signal. Therefore, gLCR products should be detected only if probes 587pr2 and 587pr4 had ligated following hybridization and extension of probe 587pr4 during gLCR cycling. To detect gLCR products, 11 μl of the completed gLCR mixture was added to 50 μl (0.125 μg/ml) of streptavidin-coated paramagnetic beads (Dynabeads M-280 Streptavidin Coated Beads; IGEN International, Inc.), and the mixture was vortexed for 20 min. Following vortexing, 240 μl of ORIGEN Assay Buffer (IGEN International, Inc.) was added per tube, and the mixture was analyzed.
gLCR standard curve.
A standard curve of ECL values derived from gLCR of samples having an increasing percentage of pA587 mixed with pG587 was prepared. Gel-purified, nested PCR products from pA587 and pG587 were quantified, and the two were mixed in an increasing ratio of pA587/pG587. Mixtures were then tested in triplicate for the G145R mutation by the gLCR assay (50 ng of total DNA added per gLCR mixture) with ECL detection of gLCR products. Clinical specimens processed for gLCR were included in the same standard curve assay run, which permitted calculation of an approximate percentage of mutant DNA by linear regression analysis.
Cloning and sequencing of PCR products.
PCR products specific for the a determinant of the S protein were sequenced. All DNA was sequenced with a LI-COR DNA Sequencer Long ReadIR 4200 (LI-COR, Inc., Lincoln, Nebr.) using infrared dye-labeled primers in a cycle sequencing reaction. Certain PCR products were cloned into the pCRII-TOPO cloning vector (Invitrogen Life Technologies) by direct ligation of Taq polymerase-amplified products with vector 3′ T overhangs. Multiple clones of each PCR product were sequenced in order to approximate the ratio of G145R-containing clones to wild-type clones. G145R mutants were distinguished by an adenine at nucleotide 587 (A587), while wild-type sequence was distinguished by a guanine at nucleotide 587 (G587).
RESULTS
gLCR assay.
gLCR assay conditions were optimized incrementally by altering buffer and reaction conditions. Increased concentrations of Tris and magnesium salts were observed to increase assay specificity, as did omission of the deoxynucleoside triphosphate complementary to the wild-type nucleotide for the gap extension reaction on the reverse strand (data not shown).
The cutoff value of the gLCR assay was calculated for each assay run in order to determine the positive or negative status of each sample. Cutoff values were calculated from the mean plus 3 standard deviations of replicate (at least three) pG587 ECL values (Table 1). ECL values of pG587 gLCR products were consistently between 15 to 30 times lower than the ECL values of pA587 gLCR products (Table 1). Specimens were considered positive for the G145R mutation if their ECL value was above the cutoff value. An arbitrary ECL value of between one and two times the assay cutoff was chosen as a possible false-positive value; therefore, any samples falling within this range were assayed again. Samples repeatedly having an ECL value between one and two times the assay cutoff were considered true-positive values.
TABLE 1.
Example of typical ECL values for the gLCR assay and calculation of the cutoff levela
| Sample | ECL value | G145R |
|---|---|---|
| IGEN assay buffer | 170 | |
| PCR negative control | 756 | |
| Sample 21 | 27,083 | + |
| Sample 25 | 12,013 | − |
| Sample 32 | 16,720 | Repeatb |
| Sample 33 | 18,090 | Repeatb |
| pG587 | 12,137 | |
| pG587 | 11,792 | |
| pG587 | 11,746 | |
| pA587 | 234,429 |
Cutoff level equal to the mean of negative-control pG587 ECL values (11,891.7) plus 3 standard deviations (174.5) above the mean equals 12,415.
Samples with ECL values falling between one and two times the assay cutoff level were assayed again.
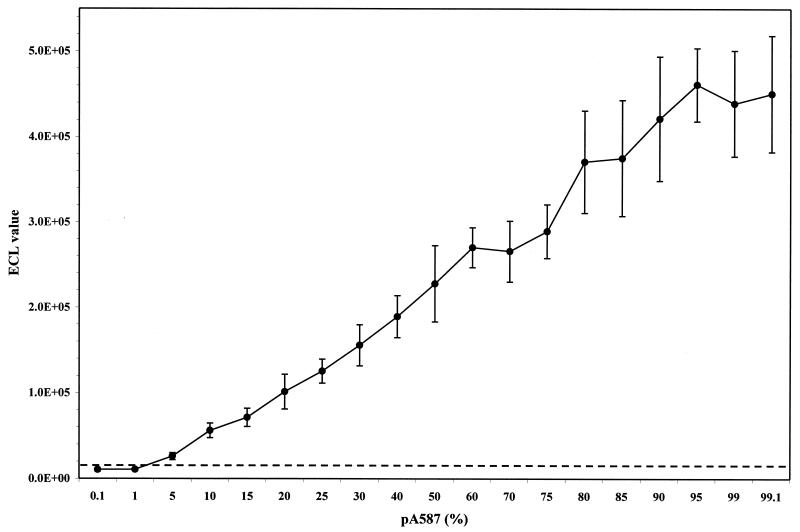
The limit of detection of pA587 copies was determined in order to measure the sensitivity of the gLCR assay. Tenfold serial dilutions (106 to 0.1 copies) of pA587 were prepared and processed as described above. A limit of detection of approximately 10 copies of the mutant clone was reproducibly observed following ECL detection. The ability of the assay to detect and discriminate the G145R mutation in the presence of excess wild-type DNA was also tested. Gel-purified, nested PCR product from each of the cloned DNAs was quantified and mixed in increasing ratios, which were then tested in duplicate or triplicate by gLCR and ECL detection. The limit of detection was observed at a ratio of 1 to 5% mutant DNA to 99 to 95% wild-type DNA (Fig. 2).
FIG. 2.
gLCR standard curve of an increasing percentage of pA587 mixed with pG587. Gel-purified, nested PCR product from pA587 was mixed in an increasing ratio with gel-purified, nested PCR product from pG587. Mixtures were then tested in duplicate or triplicate for the G145R mutation by the gLCR assay with ECL detection of gLCR products. The mean ECL value plus standard deviation is plotted at each percentage of pA587 in a background of pG587 to a total of 100%. The broken line represents the cutoff value for the assay. 1.0E+05, 1 × 105; 2.0E+05, 2 × 105; 3.0E + 05, 3 × 105; 4.0E + 05, 4 × 105; 5.0E + 05, 5 × 105.
Clinical specimen analysis.
Fifty-six serum samples were analyzed for the presence of the G145R mutant by gLCR assay (Table 2). Twenty-eight specimens were determined to be positive for HBV DNA, and of these, there were 18 specimens having reproducible ECL values above the assay cutoff. Eleven samples were considered indeterminate, and after repeated analysis, ten were confirmed to have ECL values above the cutoff level, while one sample fell below the cutoff level (sample 49). To further investigate the sensitivity of detection of the G145R variant within the mixed population of a specimen, three samples (samples 23, 32, and 33) were investigated by cloning and sequencing of the nested PCR product. For sample 23, only one mutant sequence was identified from 50 sequenced, while no mutant sequences were identified from the 27 and 39 clones chosen for samples 32 and 33, respectively. In keeping with this result, only the wild-type sequence was observed following sequencing of the original PCR product of samples 23, 32, and 33.
TABLE 2.
Analysis of clinical specimens by gLCR assay
| Sample no.a | Marker or identifier | DNA | G145R | Factor > cutoff | % G145Rb |
|---|---|---|---|---|---|
| 1 | Liver transplant | − | |||
| 2 | Vaccine failure | − | |||
| 3 | Kidney transplant | − | |||
| 4 | Liver transplant | − | |||
| 5 | Vaccine failure | − | |||
| 6 | HBIg treatment | − | |||
| 7 | Liver transplant | − | |||
| 8 | Liver transplant | + | − | ||
| 9 | Interferon therapy | + | + | 2.2 | 5.6 |
| 10 | Liver transplant | − | |||
| 11 | Vaccine failure | − | |||
| 12 | Diagnostic anomaly | + | − | ||
| 13 | Bone marrow transplant | + | + | 1.1 | |
| Repeat | + | + | 1.3 | 2.7 | |
| 14 | Diagnostic anomaly | + | + | 1.6 | |
| Repeat | + | + | 2.3 | 5.1 | |
| 15 | Liver transplant | + | + | 24.7 | 73.6 |
| 16 | Diagnostic anomaly | + | + | 1.3 | |
| Repeat | + | + | 1.4 | 3.2 | |
| 17 | Blood transfusion | + | + | 1.3 | |
| Repeat | + | + | 1.3 | 2.9 | |
| 18 | Liver transplant | + | − | ||
| 19 | Diagnostic anomaly | − | |||
| 20 | Liver transplant | + | + | 2.2 | 5.6 |
| 21 | Pre-liver transplant | + | + | 2.2 | 5.5 |
| 22 | Kidney transplant | + | + | 2.6 | 5.8 |
| 23 | Liver transplant | + | + | 2.9 | 7.2 |
| 24 | HBIg treatment | + | − | ||
| 25 | HBIg treatment | + | − | ||
| 26 | Diagnostic anomaly | − | |||
| 27 | Diagnostic anomaly | + | + | 1.5 | |
| Repeat | + | + | 2.0 | 4.9 | |
| 28 | Vaccine failure | + | − | ||
| 29 | Nonspecific hepatitis | − | |||
| 30 | Nonspecific hepatitis | − | |||
| 31 | Liver transplant | − | |||
| 32 | Vaccine failure | + | + | 1.4 | 3.2 |
| Repeat | + | + | 1.3 | ||
| 33 | Vaccine failure | + | + | 1.5 | |
| Repeat | + | + | 1.5 | 3.4 | |
| 34 | Liver transplant | − | |||
| 35 | Liver transplant | − | |||
| 36 | Liver transplant | − | |||
| 37 | Liver transplant | + | + | 6.3 | 18.1 |
| 38 | Liver transplant | − | |||
| 39 | Liver transplant | − | |||
| 40 | Liver transplant | − | |||
| 41 | Liver transplant | − | |||
| 42 | Liver transplant | − | |||
| 43 | Liver transplant | − | |||
| 44 | Liver transplant | − | |||
| 45 | Liver transplant | + | + | 1.4 | 3.2 |
| Repeat | + | + | 1.1 | ||
| 46 | Liver transplant | − | |||
| 47 | Liver transplant | − | |||
| 48 | Liver transplant | − | |||
| 49 | Liver transplant | + | + | 1.1 | |
| Repeat | + | − | 0.9 | ||
| 50 | Liver transplant | − | |||
| 51 | Chronic hepatitis | + | − | ||
| 52 | Diagnostic anomaly | + | + | 2.4 | 7.6 |
| 53 | Diagnostic anomaly | + | + | 1.4 | |
| Repeat | + | + | 2.4 | 7.6 | |
| 54 | Diagnostic anomaly | + | + | 1.7 | |
| Repeat | + | + | 3.0 | 7.7 | |
| 55 | Chronic hepatitis | + | − | ||
| 56 | Chronic hepatitis | + | − |
Samples 32 and 33 were from HBV chronically infected individuals born to HBV carrier mothers and vaccinated at birth. Samples 34 to 50 were submitted from a single individual over a 4-year period in chronological order.
Approximate percentage of mutant DNA within the total DNA population as calculated by linear regression from a standard curve (Fig. 2).
Sequencing results were obtained for original PCR products from 20 of 28 HBV-DNA-positive specimens (8 samples could not be sequenced due to insufficient DNA). Twelve of these were from gLCR-identified G145R-positive specimens, while eight were determined to be wild type by gLCR. All eight specimens found to be wild-type by gLCR were also found to be wild type by sequencing. Interestingly, many specimens testing positive for G145R by gLCR were also determined to have a wild-type sequence by sequencing of the original PCR product (six G587; five mixed G/A587; one A587). As the approximate percentage of mutant DNA increased, the likelihood of observing an adenine at position 587 during sequencing also increased (for example, sample 15 was A587 and samples 37, 53, and 54 were mixed G/A587), although this correlation was not observed for all samples (for example, sample 21 was mixed G/A587 [approximately 5.5% mutant]; samples 9 and 20 were both G587 [approximately 5.6% mutant]).
The G145R mutation was observed in various specimens including specimens from seven transplant recipients (kidney, liver, and bone marrow), specimens producing a diagnostic anomaly (weak HBsAg with strong positive PCR; anti-HBe and HBeAg positive; anti-HBs and HBsAg positive; diagnostic failure), and in two vaccinated individuals born to HBV carrier mothers (Table 2). The study also included 17 consecutive specimens from one liver transplant recipient submitted over a 4-year period (samples 34 to 50). All samples submitted from this individual were consistently negative for HBV markers other than antibody to HBsAg. Upon nested PCR, three of the samples were found to be HBV DNA positive (samples 37, 45, and 49 [Table 2]) and positive for the G145R mutation, although the apparent G145R level declined over time, until the last HBV-DNA-positive sample, which was not reproducibly positive for the G145R mutant.
G145R mutant levels were approximated in positive specimens by use of a standard curve of an increasing percentage of pA587 mixed with pG587 (Fig. 2). Clinical specimens processed for gLCR were included in the same standard curve assay run which permitted calculation of an approximate percentage of mutant DNA by linear regression analysis (Table 2). The percentage range of G145R for the mutant positive samples was from approximately 3 to 74% of the total mixed population, with the majority of specimens having mutant DNA levels of approximately 3 to 7%.
DISCUSSION
The present study describes a new method for the sensitive detection of HBsAg mutants. By gLCR-coupled ECL detection, the G145R mutation was detectable to a limit of 10 copies and in the presence of a majority of wild-type sequence (limit of 1 to 5% of the total DNA population). These limits are comparable to or more sensitive than other highly sensitive HBsAg mutant detection assays (21, 23). The sensitivity was also demonstrated by the detection of a single mutant clone out of 50 clones sequenced of a specimen PCR product. Additional mutant sequences were probably not detected due to the low level of mutant present within the entire HBV population. Although the percentage of mutant necessary for detection by sequencing was not directly addressed in this study, mutant sequence was inconsistently observed when present at approximately 3 to 20%. Therefore, sequencing may fail to detect low-level mutants in a dependable manner. The gLCR assay has the added advantage that nucleotide mismatches close to the 3′ end of each probe are well tolerated and will not interfere with gLCR detection. Such mismatches may interfere with PCR-based detection methods during hybridization or subsequent restriction digestion steps. Additionally, the gLCR assay can be automated (1) similar to methods used for the commercial detection of Chlamydia trachomatis (5). Other identified immune or diagnostic escape mutations, such as T/I126A/S, Q129H, K/L141E, and D144A (24, 27), could also be identified by including specific probes in a gLCR mixture with the a determinant amplified product.
The gLCR is a modification of the LCR requiring an initial template-dependent extension of one probe to fill in a gap of 2 to 3 nucleotides prior to ligation occurring between the adjacent probes. The first-stage extension has been shown to decrease the number of false-positive results, thus leading to greater specificity (1, 28). LCR has been used previously to detect HBV precore mutants (19) and HBsAg mutants (K141E) (4), whereas the present study is the first to detect the G145R mutation by this method.
The present study is also the first to investigate a wide variety of diagnostic serum specimens, including specimens from transfusion or transplant patients having recurrent hepatitis; individuals on interferon therapy or passive immunoprophylaxis (HBIg); individuals characterized as having HBV vaccine failure (including two individuals born to HBV carrier mothers and vaccinated at birth), nonspecific hepatitis, or chronic hepatitis; and specimens giving anomalous results with various diagnostic marker assays. Sixty-four percent (18 of 28) of HBV-DNA-positive samples were determined to be positive for the G145R mutation by the gLCR assay. Approximately one-half (7 of 15) of the transplant recipients having recurrent hepatitis were positive for the mutation, including 5 of 12 liver transplant recipients. This study also included 17 consecutive samples from one liver transplant recipient; the 17 samples were submitted to our laboratory over a 4-year period. Although HBV DNA levels periodically fell below the limit of detection, those samples positive for HBV DNA were also positive for the G145R mutation. Total G145R mutant levels within these specimens appeared to decrease over time until the mutant could not be reproducibly detected (sample 49). Although treatment information for this patient was not available, it has been shown that HBIg treatment-associated HBsAg mutants disappear following cessation of treatment in post-orthotopic liver transplant patients (2, 7).
The gLCR assay highlighted the presence of the G145R mutation among specimens demonstrating a diagnostic anomaly. The G145R mutation has been associated with loss of detection by monoclonal antibody-based HBsAg detection (3, 26). Loss of detection due to mutation is also suspected with several serum specimens (samples 53 and 54) in this study. Upon gLCR assay, both were found to contain mutant DNA which may explain the observed loss of HBsAg detection, as has been found with some HBsAg diagnostic test kits (15, 29). Of nine samples demonstrating a serological diagnostic anomaly, six (67%) had the G145R mutation. Therefore, assaying for the G145R mutation by gLCR could be important in providing further diagnostic information on particular specimens, especially in those cases of weak HBsAg (samples 14 and 16) or cocirculating HBsAg and anti-HBs antibody (sample 52).
The value of a system able to sensitively detect HBsAg mutants is contingent upon the true clinical significance of these mutants. S-gene mutants have been associated with an aggressive or worsening clinical course (16, 20). The matter of whether the current vaccine is effective in preventing infection with these mutants, particularly in infants born to HBV carrier mothers, is under debate (9, 11, 25, 30). Of particular concern is the observed emergence of a determinant mutants occurring in geographic regions where universal vaccination has been instituted (12, 24). Although the absolute number of new infections has been reduced in these regions, infections are most often observed in vaccinated infants born to HBV carrier mothers, suggesting that infants exposed to maternal mutant virus may select for this strain during development of vaccine-induced antibodies. In addition, these infants become chronically infected and continue to harbor the mutant HBV despite reaching protective levels of anti-HBs antibody (10), as do liver transplant patients on long-term posttransplant HBIg treatment (7). Therefore, although research has shown that chimpanzees are protected against infection with mutant HBV by vaccine-induced antibody (22), infection with HBsAg mutants may depend largely upon the level of protective antibody at the time of exposure to the mutant virus. In this respect, an assay able to sensitively detect HBV immune escape mutants could assist physicians with pregnant HBV carrier patients or patients awaiting liver transplants in determining the possibility of an infant breakthrough infection or posttransplantation relapse, respectively.
Acknowledgments
I am grateful to Manna Zhang and Tim Booth for valuable comments and suggestions.
REFERENCES
- 1.Abravaya, K., J. J. Carrino, S. Muldoon, and H. H. Lee. 1995. Detection of point mutations with a modified ligase chain reaction (gap-LCR). Nucleic Acids Res. 23:675-682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Carman, W. F., C. Trautwein, F. J. van Deursen, K. Colman, E. Dornan, G. McIntyre, J. Waters, V. Kliem, R. Muller, H. C. Thomas, and M. P. Manns. 1996. Hepatitis B virus envelope variation after transplantation with and without hepatitis B immune globulin prophylaxis. Hepatology 24:489-493. [DOI] [PubMed] [Google Scholar]
- 3.Cooreman, M. P., M. H. van Roosmalen, R. te Morsche, C. M. G. Sunnen, E. M. E. Schoondermark-van de Ven, J. B. M. J. Jansen, G. N. J. Tytgat, P. L. M. de Wit, and W. P. Paulij. 1999. Characterization of the reactivity pattern of murine monoclonal antibodies against wild-type hepatitis B surface antigen to G145R and other naturally occurring “a” loop escape mutations. Hepatology 30:1287-1292. [DOI] [PubMed] [Google Scholar]
- 4.Devi Karthigesu, V., M. Mendy, M. Fortuin, H. C. Whittle, C. R. Howard, and L. M. C. Allison. 1995. The ligase chain reaction distinguishes hepatitis B virus S-gene variants. FEMS Microbiol. Lett. 131:127-132. [DOI] [PubMed] [Google Scholar]
- 5.Dille, B. J., C. C. Butzen, and L. G. Birkenmeyer. 1993. Amplification of Chlamydia trachomatis DNA by ligase chain reaction. J. Clin. Microbiol. 31:729-731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Francois, G., M. Kew, P. Van Damme, M. J. Mphahlele, and A. Meheus. 2001. Mutant hepatitis B viruses: a matter of academic interest only or a problem with far-reaching implications? Vaccine 19:3799-3815. [DOI] [PubMed] [Google Scholar]
- 7.Ghany, M. G., B. Ayola, F. G. Villamil, R. G. Gish, S. Rojter, J. M. Vierling, and A. S. F. Lok. 1998. Hepatitis B virus S mutants in liver transplant recipients who were reinfected despite hepatitis B immune globulin prophylaxis. Hepatology 27:213-222. [DOI] [PubMed] [Google Scholar]
- 8.Hecker, K. H., and K. H. Roux. 1996. High and low annealing temperatures increase both specificity and yield in touchdown and stepdown PCR. BioTechniques 20:478-485. [DOI] [PubMed] [Google Scholar]
- 9.Hino, K., Y. Katoh, E. Vardas, J. Sim, K. Okita, and W. F. Carman. 2001. The effect of introduction of universal childhood hepatitis B immunization in South Africa on the prevalence of serologically negative hepatitis B virus infection and the selection of immune escape variants. Vaccine 19:3912-3918. [DOI] [PubMed] [Google Scholar]
- 10.Ho, M., C. Lu, J. Kuo, Y. Mau, and W. Chao. 1995. A family cluster of an immune escape variant of hepatitis B virus infecting a mother and her two fully immunized children. Clin. Diagn. Lab. Immunol. 2:760-762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ho, M., Y. Mau, C. Lu, S. Huang, L. Hsu, S. Lin, and H. Hsu. 1998. Patterns of circulating hepatitis B surface antigen variants among vaccinated children born to hepatitis B surface antigen carrier and non-carrier mothers-a population-based comparative study. J. Biomed. Sci. 5:355-362. [DOI] [PubMed] [Google Scholar]
- 12.Hsu, H., M. Chang, S. Liaw, Y. Ni, and H. Chen. 1999. Changes of hepatitis B surface antigen variants in carrier children before and after universal vaccination in Taiwan. Hepatology 30:1312-1317. [DOI] [PubMed] [Google Scholar]
- 13.Hsu, H., M. Chang, Y. Ni, H. Lin, S. Wang, and D. Chen. 1997. Surface gene mutants of hepatitis B virus in infants who develop acute or chronic infections despite immunoprophylaxis. Hepatology 26:786-791. [DOI] [PubMed] [Google Scholar]
- 14.Hunt, C. M., J. M. McGill, M. I. Allen, and L. D. Condreay. 2000. Clinical relevance of hepatitis B viral mutations. Hepatology 31:1037-1044. [DOI] [PubMed] [Google Scholar]
- 15.Ireland, J. H., B. O'Donnell, A. A. Basuni, J. D. Kean, L. A. Wallace, G. K. K. Lau, and W. F. Carman. 2000. Reactivity of 13 in vitro expressed hepatitis B surface antigen variants in 7 commercial diagnostic assays. Hepatology 31:1176-1182. [DOI] [PubMed] [Google Scholar]
- 16.Kalinina, T., A. Riu, L. Fischer, H. Will, and M. Sterneck. 2001. A dominant hepatitis B virus population defective in virus secretion because of several S-gene mutations from a patient with fulminant hepatitis. Hepatology 34:385-394. [DOI] [PubMed] [Google Scholar]
- 17.Lee, P., L. Chang, C. Lee, L. Huang, and M. Chang. 1997. Detection of hepatitis B surface gene mutation in carrier children with or without immunoprophylaxis at birth. J. Infect. Dis. 176:427-430. [DOI] [PubMed] [Google Scholar]
- 18.McCance, D. J. 1996. DNA viruses: DNA extraction, purification and characterization, p. 191-230. In B. W. Mahy and H. O. Kangro (ed.), Virology methods manual. Academic Press Limited, Toronto, Ontario, Canada.
- 19.Minamitani, S., S. Nishiguchi, T. Kuroki, S. Otani, and T. Monna. 1997. Detection by ligase chain reaction of precore mutant of hepatitis B virus. Hepatology 25:216-222. [DOI] [PubMed] [Google Scholar]
- 20.Ngui, S. L., R. Hallet, and C. G. Teo. 1999. Natural and iatrogenic variation in hepatitis B virus. Rev. Med. Virol. 9:183-209. [DOI] [PubMed] [Google Scholar]
- 21.Ngui, S. L., S. O'Connell, R. P. Eglin, J. Heptonstall, and C. G. Teo. 1997. Low detection rate and maternal provenance of hepatitis B virus S gene mutants in cases of failed postnatal immunoprophylaxis in England and Wales. J. Infect. Dis. 176:1360-1365. [DOI] [PubMed] [Google Scholar]
- 22.Ogata, N., P. J. Cote, A. R. Zanetti, R. H. Miller, M. Shapiro, J. Gerin, and R. H. Purcell. 1999. Licensed recombinant hepatitis B vaccines protect chimpanzees against infection with the prototype surface gene mutant of hepatitis B virus. Hepatology 30:779-786. [DOI] [PubMed] [Google Scholar]
- 23.Ogata, N., A. R. Zanetti, M. Yu, R. H. Miller, and R. H. Purcell. 1997. Infectivity and pathogenicity in chimpanzees of a surface gene mutant of hepatitis B virus that emerged in a vaccinated infant. J. Infect. Dis. 175:511-523. [DOI] [PubMed] [Google Scholar]
- 24.Oon, C., G. Lim, Z. Ye, K. Goh, K. Tan, S. Yo, E. Hopes, T. J. Harrison, and A. J. Zuckerman. 1995. Molecular epidemiology of hepatitis B virus vaccine variants in Singapore. Vaccine 13:699-702. [DOI] [PubMed] [Google Scholar]
- 25.Purcell, R. H. 2000. Hepatitis B virus mutants and efficacy of vaccination. Lancet 356:769-774. [DOI] [PubMed] [Google Scholar]
- 26.Seddigh-Tonekaboni, S., J. Waters, S. Jeffers, R. Gehrke, B. Ofenloch, A. Horsch, G. Hess, H. C. Thomas, and P. Karayiannis. 2000. Effect of variation in the common “a” determinant on the antigenicity of hepatitis B surface antigen. J. Med. Virol. 60:113-121. [DOI] [PubMed] [Google Scholar]
- 27.Torre, F., and N. V. Naoumov. 1998. Clinical implications of mutations in the hepatitis B virus genome. Eur. J. Clin. Investig. 28:604-614. [DOI] [PubMed] [Google Scholar]
- 28.Wiedmann, M., W. J. Wilson, J. Czajka, J. Luo, F. Barany, and C. A. Batt. 1994.. Ligase chain reaction (LCR)-overview and applications. PCR Methods Appl. 3:S51-S64. [DOI] [PubMed]
- 29.Zaaijer, H. L., H. Vrielink, and M. Koot. 2001. Early detection of hepatitis B surface antigen and detection of HBsAg mutants: a comparison of five assays. Vox Sang. 81:219-221. [DOI] [PubMed] [Google Scholar]
- 30.Zuckerman, A. J. 2000. Effect of hepatitis B virus mutants on efficacy of vaccination. Lancet 355:1382-1384. [DOI] [PubMed] [Google Scholar]