Abstract
Over a 9-month period, 14 strains of Ralstonia pickettii were isolated from various biological samples inoculated in a blood culture medium. Molecular epidemiological investigation confirmed the relatedness of the strains. The source of the contamination proved to be the blood culture bottle caps.
Ralstonia pickettii (formerly Pseudomonas picketti and Burkholderia pickettii) is a nonfermenting gram-negative bacillus of relatively low virulence that is often associated with pseudobacteremia or asymptomatic colonization of patients. Contamination of water supplies, skin disinfectants, and saline solutions used either for patient care (1, 2, 3, 5, 7, 8) or for laboratory diagnosis (4, 9) have previously been incriminated. We report on a new source of contamination caused by R. pickettii in biological samples of patients hospitalized in different units of Antoine Béclère Hospital, Clamart, France.
Between March and November 2000, 14 strains of R. pickettii were isolated from various biological samples (five pleural fluid, three ascitic fluid, three pus, and two biopsy samples and one blood sample) inoculated in an enrichment broth (Vital system; BioMérieux, Marcy l'Etoile, France) generally used for blood culture. All samples except the blood sample were inoculated in the microbiology laboratory in both aerobic and anaerobic bottles by removing the caps to open the bottles, a procedure chosen to protect laboratory technicians from the risk of contamination through needle-stick injuries. Growth of R. pickettii was detected in 12 anaerobic bottles and 2 aerobic bottles after 48 to 72 h of incubation in the Vital system at 37°C. All strains were identified by use of the API NE identification system (BioMérieux). Antibiotic susceptibility patterns were determined by the disk diffusion method on Mueller-Hinton agar (Bio-Rad, Marnes-la-Coquette, France) and were interpreted according to the standards of the Comité de l'Antibiogramme de la Société Française de Microbiologie. The clonalities of the strains were established by random amplified polymorphic DNA (RAPD) analysis, as described previously (6), with primers P3 (5′-AGACGTCCAC-3′) and P15 (5′-AATGGCGCAG-3′) (Genset, Paris, France). Briefly, DNA extraction was performed with chloroform and isopropanol after cell lysis with lysozyme and proteinase K. The extracted DNA was amplified in a Trio-Thermoblock system (Biometra). The amplification products were then analyzed by electrophoresis in a 1.5% agarose gel, stained with ethidium bromide, and detected by UV transillumination. For pulsed-field gel electrophoresis (PFGE), DNA embedded in agarose plugs was digested with the SpeI restriction enzyme (4). Separation of the DNA fragments was carried out on a 1.2% agarose gel in a CHEF-DR III system (Bio-Rad). The separated fragments were stained with ethidium bromide and visualized by UV transillumination.
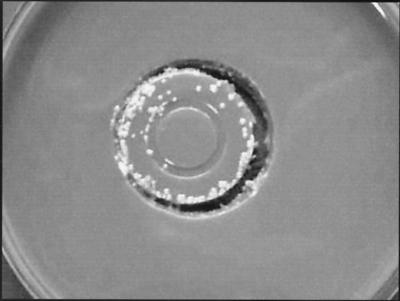
An epidemiological investigation was undertaken to elucidate the source of contamination, since isolation of these strains did not correlate with the medical status of any of the samples from patients. This investigation evaluated patients, disinfectants, and enrichment broth media. Antiseptic solutions could not be incriminated, as different disinfectants were used in the different clinical units. Cultures of the enrichment broth media from five unopened bottles were made by flooding 5 ml of broth on chocolate agar and incubating the rest of the broth, and the cultures remained negative after 5 days of incubation at 30°C. Since all except one of the positive samples were from anaerobic bottles inoculated in the microbiology laboratory, the contamination could have originated from our inoculation protocol. For this reason, cultures of 15 caps of unused bottles, chosen at random from bottles from five different lots, were made by imprinting the caps for 1 min on chocolate agar medium before incubation at 30°C. After 48 h of incubation, a pure growth of R. pickettii was obtained from seven bottle caps originating from three different lots (Fig. 1).
FIG. 1.
Imprint of a bottle cap on chocolate agar medium showing R. pickettii colonies.
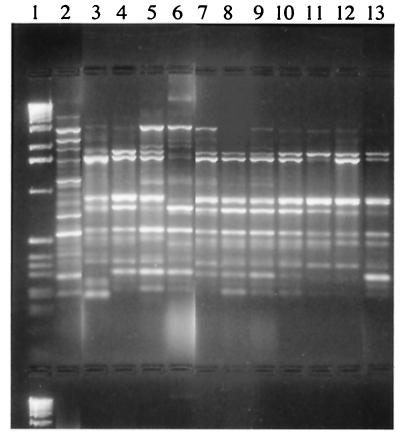
All of the R. pickettii isolates detected had the same antibiotic susceptibility patterns as the strains obtained from the biological fluids tested. They were susceptible to ureidopenicillins, cefotaxime, imipenem, ciprofloxacin, and trimethoprim-sulfamethoxazole. They were resistant or had decreased susceptibilities to aminopenicillins, aztreonam, cefotetan, aminoglycosides, colistin, and fosfomycin. The patterns obtained by RAPD analysis and PFGE for strains isolated from the biological samples and those recovered from the bottle caps were highly similar (the patterns differed by three bands or less). Our results showed that RAPD analysis, which is less time-consuming than PFGE (2 days versus 5 days), was an adequately discriminatory technique for detection of an outbreak. Representative products obtained by RAPD analysis with primer P3 for eight strains isolated from various biological samples and for three strains isolated from different lots of blood culture bottle caps are shown in Fig. 2. Additionally, we confirmed the previous observations of Maroye et al. (6) that antibiotyping is very helpful in the initial discrimination of isolates when there is a suspicion of an outbreak.
FIG. 2.
Representative products of R. pickettii strains obtained by RAPD analysis with primer P3. Lane 1, molecular size marker; lane 2, unrelated strain; lanes 3 to 10, strains isolated from biological samples; lanes 11 to 13, strains isolated from blood culture bottle caps.
The manufacturer was rapidly alerted following the discovery of this contamination. Its investigation incriminated the water used to cool the bottles after sterilization as the source of contamination. Residual drops of water remaining between the plastic clip and the rubber cap were contaminated with R. pickettii. Therefore, contamination of the anaerobic blood culture medium with this aerobic microorganism had probably been promoted by the increased negative pressure created when the bottles were opened to inoculate the biological samples. In addition, R. pickettii is resistant to some antiseptics and is able to survive in an oligotrophic environment (9). The manufacturers were quick to disseminate to users strict recommendations concerning this issue, cautioning them not to open bottles before inoculation. This was quite important because in France, the bottles are used not only for cultivation of blood but also as enrichment broth for cultivation of samples other than blood. Since we have changed our inoculation protocols, we have observed no new cases of R. pickettii contamination.
In all cases cultures of specimens yielding R. pickettii should always be interpreted with caution and evaluated with the clinical data for each patient. As Labarca et al. (2) noted previously, when an outbreak of R. pickettii occurs, contamination of solutions should be suspected. An epidemiological investigation should be initiated promptly, and the first step should be to collect unopened bottles to culture the solutions in those bottles. All previous reports associated R. pickettii epidemics with extrinsic contamination of disinfectants, saline solution, sterile water, narcotic solutions, and other solutions used for patient care (1, 2, 3, 5, 7, 8) or to laboratory contamination (4, 9). Plastic vials contaminated with the water used to cool the vials after the moulding cycle had been suspected previously (3), but to our knowledge, this is the first time that the caps of the bottles were identified as the source of contamination with R. pickettii. In conclusion, the data presented here show that epidemiological investigations concerning contamination with R. pickettii should not be limited to water supplies, skin disinfectants, and saline solutions used for patient care or for laboratory diagnosis but should also include all manufactured products.
Acknowledgments
We thank Bénédict Savary for producing the photograph in Fig. 1 and the manufacturer for helping us to elucidate this problem.
REFERENCES
- 1.Gardner, S., and S. T. Shulman. 1984. A nosocomial common source outbreak caused by Pseudomonas pickettii. Pediatr. Infect. Dis. 3:420-422. [DOI] [PubMed] [Google Scholar]
- 2.Labarca, J. A., W. E. Trick, C. L. Peterson, L. A. Carson, S. C. Holt, M. J. Arduino, M. Meylan, L. Mascola, and W. R. Jarvis. 1999. A multistate nosocomial outbreak of Ralstonia pickettii colonization associated with an intrinsically contaminated respiratory care solution. Clin. Infect. Dis. 29:1281-1286. [DOI] [PubMed] [Google Scholar]
- 3.Lacey, S., and S. V. Want. 1991. Pseudomonas pickettii infections in a paediatric oncology unit. J. Hosp. Infect. 17:45-51. [DOI] [PubMed] [Google Scholar]
- 4.Luk, W. K. 1996. An outbreak of pseudobacteraemia caused by Burkholderia pickettii: the critical role of an epidemiological link. J. Hosp. Infect. 34:59-69. [DOI] [PubMed] [Google Scholar]
- 5.Maki, D. G., B. S. Klein, R. D. McCormick, C. J. Alvarado, M. A. Zilz, S. M. Stolz, C. A. Hassemer, J. Gould, and A. R. Liegel. 1991. Nosocomial Pseudomonas pickettii bacteremias traced to narcotic tampering. JAMA 265:981-986. [PubMed] [Google Scholar]
- 6.Maroye, P., H. P. Doermann, A. M. Rogues, J. P. Gachie, and F. Mégraud. 2000. Investigation of an outbreak of Ralstonia pickettii in a paediatric hospital by RAPD. J. Hosp. Infect. 44:267-272. [DOI] [PubMed] [Google Scholar]
- 7.McNeil, M. M., S. L. Solomon, R. L. Anderson, B. J. Davis, R. F. Spengler, B. E. Reisberg, C. Thornsberry, and W. J. Martone. 1985. Nosocomial Pseudomonas pickettii colonization associated with a contaminated respiratory therapy solution in a special care nursery. J. Clin. Microbiol. 22:903-907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Roberts, L. A., P. J. Collignon, V. B. Cramp, S. Alexander, A. E. McFarlane, E. Graham, A. Fuller, V. Sinickas, and A. Hellyar. 1990. An Australia-wide epidemic of Pseudomonas pickettii bacteraemia due to contamined “sterile” water for injection. Med. J. Aust. 152:652-655. [DOI] [PubMed] [Google Scholar]
- 9.Verschraegen, G., G. Claeys, G. Meeus, and M. Delanghe. 1985. Pseudomonas pickettii as a cause of pseudobacteremia. J. Clin. Microbiol. 31:278-279. [DOI] [PMC free article] [PubMed] [Google Scholar]