Abstract
In a multicenter comparison of PCR assays utilizing 120 quantitated samples of 16 Chlamydia pneumoniae isolates, an LCx research-use-only (RUO) PCR developed by Abbott Laboratories demonstrated 100% sensitivity on 48 samples with >1 copy of DNA per μl of specimen. The sensitivities of five in-house PCR assays ranged from 54 to 94% for the same samples. All six assays showed decreased sensitivities as the DNA copy numbers of the samples decreased. Overall, sensitivities ranged from 68% for the LCx PCR assay to 29% for one of the in-house tests. The LCx RUO PCR and three of the five in-house PCR tests reported no false positives with the 24 negative samples. Increasing the number of replicates tested increased the sensitivities of all of the assays, including the LCx PCR. The LCx RUO assay showed high reproducibility for a single technologist and between technologists, with a kappa agreement of 0.77. The within-center agreements of the five in-house PCR tests varied from 0.19 to 0.74 on two challenges of 60 specimens 1 month apart. The LCx C. pneumoniae RUO PCR shows excellent potential for use in clinical studies, which could enable standardization of results in the field.
The respiratory pathogen Chlamydia pneumoniae has been reported to be associated with respiratory disease, atherosclerosis, and other chronic diseases, originally by serological testing and more recently by the use of nucleic acid amplification (NAA) tests performed on several types of clinical specimens (3, 4, 11, 20). Reports from different laboratories have yielded conflicting results, such as a lack of interlaboratory consensus with regard to PCR results, poor correlation between PCR and serology, and a large range in laboratory results for the diseases mentioned above, which are of real concern to the scientific community (6). To date 19 in-house PCR assays have been published (6). A major outer membrane protein (MOMP)-based, single-step PCR that could identify Chlamydia trachomatis, Chlamydia psittaci, and C. pneumoniae by using three primer pairs and restriction enzyme digestion was originally developed (8). Subsequently, a single-step protocol for C. pneumoniae that amplified a conserved genus-specific target of the chlamydial MOMP gene, followed by restriction enzyme digestion and species identification, was developed (19). A MOMP-based C. pneumoniae PCR with species-specific differentiation, performed by hybridization of the amplified PCR products to internal probes, was also developed (22). The MOMP-based nested PCR described by Tong and Sillis (23) has been more widely employed than the others, since it has proved to be both more sensitive and more specific than other C. pneumoniae diagnostic methods. Another target for PCR amplification of C. pneumoniae has been the 16S rRNA gene (7). This is a single-step PCR with detection by enzyme immunoassay after hybridization to a biotinylated RNA probe, complementary to a fragment of the amplified 16S rRNA gene. The assay has been further developed into a nested format (2, 17, 26) and has also been multiplexed (13, 16). Another frequently used PCR for detection of C. pneumoniae DNA was derived from a cloned 474-bp C. pneumoniae-specific PstI fragment targeted by the HL and HR primers and the HM probe in a nonnested format (5) which has been modified and nested (9, 12). Other amplification targets for C. pneumoniae DNA detection are genes coding for 60-kDa (18, 25) and 53-kDa (10) proteins.
Current approaches to help interpret variable findings in clinical studies using PCR assays for C. pneumoniae have focused on standardizing the PCR protocols, including choice of target genes, primers, PCR conditions, detection systems, and nucleic acid extraction techniques (1, 14). It has been recognized that a commercially produced PCR assay might enable standardization of C. pneumoniae PCR results. To test the agreement or variability of different assays produced, five laboratories in North America, experienced in conducting PCR assays for C. pneumoniae, participated in this study. They donated positive and negative clinical specimens which were propagated, titrated, and used to create panels of coded specimens. The panel specimens were first DNA extracted and then sent to the participating laboratories to be tested according to their standard routine PCR protocols. An industry-developed LCx C. pneumoniae PCR assay kit (Diagnostics Division, Abbott Laboratories, North Chicago, Ill.) intended for research use only (RUO) was used in one center to test the same panels in a blinded fashion and was compared to the “in-house” PCR tests. We report comparisons of sensitivity, specificity, and reproducibility from these studies.
MATERIALS AND METHODS
Study design.
We conducted a multicenter, parallel comparison of six NAA PCR tests for detection of C. pneumoniae DNA in 60 preextracted samples. The six assays were performed independently by experienced research technologists in five laboratories who were blinded to the true sample status and to each other's results. Participants were instructed to test the samples as per their routine PCR protocol. Three participants performed a nested MOMP-based PCR protocol, one performed a single 16S rRNA PCR, and one performed a single cloned PstI-based PCR test. A technologist at the St. Joseph's Hospital, McMaster University, site performed the LCx C. pneumoniae PCR assay.
The first shipment of coded specimens consisted of two panels (A and B), each of which had 30 samples containing 75 μl each. After submission of the results for the 60 samples in panels A and B, each laboratory received two similar panels (C and D), which contained the same specimens as the previous panels A and B but with the order changed (for a total of 120 samples).
Specimen panels.
C. pneumoniae isolates from humans (collected under the guidelines of approved institutional ethics review boards) or animals were solicited from the participating Chlamydia research laboratories. The panels included respiratory isolates (n = 5), lung tissues (n = 2), bronchial washes (n = 3), nasopharyngeal swabs (n = 2), buffycoats (n = 2), carotid tissues (n = 2), and sputa (n = 2) collected from the laboratory sites participating in this study; ATCC C. pneumoniae strains VR-1310 and AR39; and strain PS32 from the University of Washington Research Foundation. The isolates were shipped to the McMaster University Chlamydiology Research Laboratory at St. Joseph's Hospital, Hamilton, Ontario, Canada, and inoculated into HEp-2 cell lines for propagation (14). C. pneumoniae was detected by staining of the monolayers with a genus-specific anti-lipopolysaccharide monoclonal antibody (Pathfinder; Bio-Rad Diagnostics, Montreal, Quebec, Canada). Contamination with Mycoplasma species or C. trachomatis was excluded by PCR (15, 24). Of the 48 specimens received, 16 were successfully propagated in HEp-2 cells and confirmed as uncontaminated C. pneumoniae. DNA was extracted from all samples with the QIAamp DNA Mini-kit (Qiagen, Mississauga, Ontario, Canada) according to the manufacturer's instructions and was eluted in a final volume of 50 μl. Tenfold serial dilutions (10−1 to 10−8) of the purified DNA from all the C. pneumoniae-positive samples were subsequently prepared and tested in triplicate by an established nested PCR (23) to determine an approximate end point. The last two positive dilutions and the following two negative dilutions from each end point and titration were integrated into the panels. The propagated specimens were used at four different dilutions, representing different DNA copy numbers, as specimens for the multicenter study.
In addition to the 48 C. pneumoniae specimens, 12 negative specimens were incorporated into the panel. These included specimens with water only, with related organisms such as C. trachomatis and Simkania negevensis (ATCC VR1471), and clinical specimens (sputum or blood) which had tested negative in multiple determinations (10 or more) by an established nested PCR (23). Aliquots (75 μl) for each panel specimen were sent, frozen on dry ice, to each participating laboratory. Data report sheets were included in all shipments, as were instructions for testing and reporting panels A and B (60 specimens), followed 1 month later by panels C and D (60 specimens).
The orders of the two sets of 60 specimens were randomized and coded, and all participating laboratory technologists were blinded to the specimen status. The study analysis was initially performed, and ambiguities in reporting were resolved, before the code was broken.
Quantitation of panel specimens.
Selected specimens were quantitated by real-time PCR. An aliquot of 5 μl of extracted DNA from the panel specimens was added to 15 μl of a reaction mixture containing 4 mM MgCl2, 0.5 μM primers CPNA and CPNB (7), and SYBR Green dye (LC DNA FastSTART Master SYBR Green 1 kit). Specimens were amplified on a Lightcycler (Roche Molecular Biochemicals) under the following cycling conditions: an initial 10 min at 95°C for FastStart Taq DNA polymerase activation, followed by 40 cycles of 2 s of denaturation at 95°C, 5 s of annealing at 55°C, and 19 s of extension at 72°C. Data were obtained after the extension period in the “single” mode. Serially diluted cloned plasmid controls containing the CPNA-CPNB PCR product (10 to 106 copies) were used to generate a standard curve for quantitation of C. pneumoniae DNA.
PCR methods.
Table 1 summarizes the characteristics of the five in-house PCR methods used by the study participants. Each laboratory director was instructed to detail the laboratory's method in advance. Interpretation of results and photographs of the agarose gels were submitted with the results reported for each panel.
TABLE 1.
Summary of in-house PCR methods for detection of C. pneumoniae
| Variable | Laboratory site
|
||||
|---|---|---|---|---|---|
| A | B | C | D | E | |
| PCR format | Single | Nested | Single | Nested | Nested |
| Target gene | 16S rRNA | ompA | Cloned PstI | ompA | ompA |
| Primers | CPNA-CPNB | CP1-CP2, CPC-CPD | HL1-HR1 | CP1-CP2, CPC-CPD | CP1-CP2, CPC-CPD |
| Amplicon size (bp) | 463 | 207 | 474 | 207 | 207 |
| No. of replicates tested | 1 | 3 | 4 | 2 | 1 or 2 |
| Criterion for positivity | 1/1 | 1/3 | 1/4 | 1/2 | 1/1 or 1/2 |
| Size of PCR mixture (μl) | 50 | 50 | 50 | 50 | 100 |
| Vol of original specimen used in PCR (μl) | 15 | 10 | 0.1, 0.5, 1, 5 | 10 | 10 |
| Vol of amplicon transferred in nested PCR (μl) | N/Aa | 2.5b | N/A | 5c | 10d |
| Detection method | AGEe | AGE | AGE and hybridization | AGE | AGE |
| Reference | 7 | 23 | 5 | 23 | 23 |
N/A, not applicable.
The amplicon was diluted 1:10, and 2.5 μl of the dilution was added to 25 μl of the round 2 reaction mixture.
The amplicon was diluted 1:2, and 5 μl of the dilution was added to 50 μl of the round 2 reaction mixture.
Ten microliters of undiluted amplicon was added to 100 μl of the round 2 reaction mixture.
AGE, agarose gel electrophoresis.
LCx C. pneumoniae PCR (RUO) kit.
Panel samples were tested with the LCx C. pneumoniae PCR (RUO) assay as outlined by the manufacturer's protocol. Briefly, the activation mixture was prepared by mixing equal volumes of LCx Activation Reagent II and LCx C. pneumoniae Oligo Mix. A 40-μl volume of the freshly prepared activation mixture and 20 μl of the purified DNA sample were subsequently added to the appropriate LCx amplification vial. Amplification was carried out with a 480 thermocycler (Perkin-Elmer, Norwalk, Conn.) under the following conditions: 97°C for 2 min; 40 cycles of 97°C for 30 s, 59°C for 30 s, and 72°C for 30 s; and finally, 1 cycle of 97°C for 5 min and 12°C for 5 min. PCR products were detected with the LCx Analyzer. Samples yielding a rate over 100 cps per second (c/s/s) were considered C. pneumoniae positive. This cutoff was determined by testing titrated C. pneumoniae isolates and uninfected HEp-2 cell DNA multiple times. Separation between high and low signals was arbitrarily assigned at a cutoff of 100 c/s/s and was validated as follows: the mean rate of 41 known positive specimens was 1,304.6 ± 256.7 c/s/s; the mean rate of 22 known negative specimens was 12.9 ± 1.8 c/s/s.
Statistical methods.
SPSS for Windows, version 10.0 (SPSS Inc., Chicago, Ill.), was used for all statistical analyses. For the 24 negative panel specimens, specificity was determined for each of the tests. For the 96 potentially positive specimens, analyses were performed to examine the sensitivity by a measure for correlated dichotomous outcomes (Cochrane Q), followed by pairwise comparisons between the LCx C. pneumoniae PCR RUO assay and all other tests by use of the McNemar χ2 test. For the Cochrane Q test, which measures overall differences between the six tests, an alpha of 0.05 was established for determining statistical significance, whereas for the five pairwise comparisons, an alpha of 0.01 was established to account for multiple testing (Bonferroni's correction).
Reproducibility was examined by contingency table methods, overall agreement, and Cohen's kappa (agreement beyond chance). Multiple linear regression models were constructed to examine the relationship between C. pneumoniae concentration, test positivity, test volume, and number of replicates.
The study was designed, and all statistical analyses were performed, independently of the manufacturer of the LCx RUO PCR.
RESULTS
Test sensitivity.
Based on testing of 96 samples in four panels on two separate occasions at different dilutions of C. pneumoniae, the sensitivity of the LCx C. pneumoniae PCR assay was 100% (48 of 48) when the samples contained >1 copy of C. pneumoniae DNA per μl (Table 2), whereas the sensitivities of the five in-house PCR assays ranged from 54 to 94%. The sensitivity of the LCx RUO assay decreased to 63% (15 of 24) when the DNA copy number ranged from 0.1 to 0.9/μl and decreased to 8% at <0.1/μl, yielding an overall sensitivity of 68%. The overall sensitivities of the five in-house tests ranged from 29 to 63%. Laboratory C, testing four different volumes of the sample to achieve at least one positive, detected a larger number of positives than the other laboratories at the most dilute level (17% [4 of 24]) but performed no better than the others when the samples contained higher DNA copy numbers.
TABLE 2.
Comparison of sensitivities of C. pneumoniae PCRs by copy number for 96 coded samples
| DNA copy no./μl of sample | % of specimens testing positive (no. testing positive/no. of true positives) in the indicated laboratory or testa
|
|||||
|---|---|---|---|---|---|---|
| A (1 of 1) | B (1 of 3) | C (1 of 4) | D (1 of 2) | E (1 of 1) | RUO (1 of 3) | |
| ≥1.00 | 79 (38/48) | 94 (45/48) | 75 (36/48) | 94 (45/48) | 54 (26/48) | 100 (48/48) |
| 0.10-0.99 | 13 (3/24) | 54 (13/24) | 13 (3/24) | 33 (8/24) | 8 (2/24) | 63 (15/24) |
| <0.10 | 0 (0/24) | 8 (2/24) | 17 (4/24) | 0 (0/24) | 0 (0/24) | 8 (2/24) |
| Total | 43 (41/96) | 63 (60/96) | 45 (43/96) | 55 (53/96) | 29 (28/96) | 68 (65/96)b |
Numbers in parentheses after each laboratory or test designation are the minimum number of replicates testing positive and the total number of replicates tested per specimen.
The sensitivity of the LCx RUO PCR was similar to that of the laboratory B PCR and greater than that of the PCR of laboratory A, C, D, or E (P < 0.01).
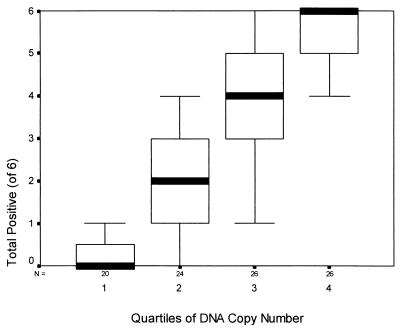
Serial 10-fold dilutions of each panel sample and quantitation by real time PCR in the Lightcycler provided relative numbers of DNA copies for detection. Figure 1 is a box plot of the DNA copy number (in quartiles) versus the number of test centers reporting positives (six centers). By linear regression, quartile DNA copy numbers explained most of the variability in detection (SPSS; R2 = 0.75, P < 0.001).
FIG. 1.
Box-and-whisker plot of the number of test centers (n = 6) reporting a specimen positive versus the quartile of the C. pneumoniae DNA copy number for 96 specimens. N, number of specimens tested in each quartile. Thick lines, median values; rectangular boxes, 25th to 75th percentiles; “whiskers,” 10th and 90th percentiles (SPSS for Windows 10.0). Quartile 1, <0.05 copies/μl; quartile 2, 0.05 to 0.49 copies/μl; quartile 3, 0.5 to 4.99 copies/μl; quartile 4, ≥5.0 copies/μl.
Pairwise comparison of the overall sensitivities of the six PCR tests (Cochrane Q = 71.3; P < 0.001) demonstrated that the LCx RUO PCR was superior to the PCR assays of centers A (P < 0.001), C (P < 0.001), D (P = 0.007), and E (P < 0.001) and equivalent to that of center B (P = 0.36).
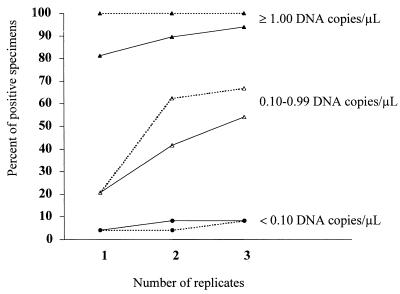
Impact of replicate testing on the sensitivity of the RUO assay.
The data in Table 2 suggested that laboratories doing replicate testing had a better chance of identifying positives with a low level of C. pneumoniae nucleic acid. We performed an analysis of the impact of replicate testing on the LCx C. pneumoniae PCR assay and the in-house PCR from laboratory B. Both assays tested three aliquots of each sample. Figure 2 illustrates the impact of the number of replicates on the percentages of samples positive in the LCx C. pneumoniae RUO assay and the in-house PCR from laboratory B according to the DNA copy number in each sample. When the 24 samples contained higher numbers of C. pneumoniae DNA copies (≥1.0 DNA copy/μl), testing more replicates resulted in a minimal increase in the percentage of positive results with the in-house PCR of laboratory B and failed to increase that percentage in the LCx PCR (Fig. 2). Our observations were similar when the samples were very dilute (<0.1 copy/μl). The advantage of replicate testing was demonstrated by an increased percentage of positive samples in the 0.1-to-0.99 copy/μl interval. For these samples, the sensitivity of the in-house PCR test increased from 21% (5 of 24) for single testing to 42% (10 of 24) for a criterion of at least one positive out of two replicates and 54% (13 of 24) for at least one positive out of three replicates. For the LCx C. pneumoniae PCR assay, the results were: 21% (5 of 24), 63% (15 of 24), and 67% (16 of 24), respectively, indicating that a single LCx RUO test could have missed more than half of the positive specimens containing these levels of C. pneumoniae DNA.
FIG. 2.
Impact of number of replicates tested on the percentage of samples testing positive in the Abbott LCx C. pneumoniae RUO assay (broken lines) and the in-house PCR from laboratory B (solid lines), by DNA copy number per microliter. A sample was considered to test positive if one or more positive results were obtained for one, two, or three replicates tested.
Between the first round of testing panels (A and B) and the second (C and D), laboratory E increased testing from single to duplicate replicates. Examination of this maneuver in relation to the number of positives identified showed some improvement in detection. For panels A and B, laboratory E detected 25% (12 of 48) of the positive samples; in panels C and D, 16 of the 48 positive samples were found positive (33.3%).
Test specificity.
The LCx C. pneumoniae PCR assay demonstrated a sample specificity of 100% (24 of 24) for the negative specimens. In total, this assay was performed six times on the 12 negative specimens during the multicenter panel challenges (72 determinations) and an additional three times in a reproducibility panel (36 tests), and all 108 tests were scored negative. The specificities of the five in-house assays on the negative specimens were as follows: 100% (24 of 24) for centers B, D, and F and 95.8% (23 of 24) for centers A and E. Approaching test specificity from a test point of view, during the first challenge these negative samples were tested one to four times per center (total, 11 × 12 = 132) and a single run was positive; thus, the overall specificity for all the tests was 99.2% (131 of 132). In the second challenge, laboratory D had two false-positive replicates of one sample, laboratory A had one false-positive result, and laboratory E switched to double testing. Therefore, the overall test specificity in the second challenge was 97.9% (141 of 144).
Reproducibility of testing results between panels.
Since the samples in panels C and D were the same as those in A and B, but were numbered differently, we were able to determine the reproducibility of testing results for each in-house PCR and the RUO assay. Table 3 shows the reproducibility or agreement within each laboratory between the two panels of 60 specimens, tested 1 month apart. Calculation of the kappa values showed the LCx C. pneumoniae PCR assay to have the best agreement (0.77) and the in-house PCR in laboratory E to have the least agreement (0.19). The majority of laboratories showed agreement above 0.48. Examination of agreement according to amount of DNA in the specimen (data not shown for the in-house PCR tests) indicated the best agreement in all laboratories for specimens having the greatest or least amount of DNA in the samples, except for laboratory E. Analysis of intralaboratory reproducibility of the LCx C. pneumoniae PCR assay for the same day showed no statistically significant differences between three runs of 60 specimens each time (run AB yielded 29, 31, and 28 positives and a Cochrane Q of 1.6 on 2 df [P = 0.46]; run CD yielded 28, 28, and 26 positives and a Cochrane Q of 0.9 on 2 df [P = 0.64]). Analysis of intralaboratory reproducibility for the LCx RUO PCR assay 1 month apart was possible because three runs of 60 specimens were done at two different times (6 × 60). Testing reproducibility indicated no statistical difference between runs (Cochrane Q = 4.4 with 5 df; P = 0.49).
TABLE 3.
Within-center agreement between two C. pneumoniae panels of 60 specimens performed 1 month aparta
| Center or test | Copy no. | No. of specimens with the following resultsb:
|
Agreement | Kappac | |||
|---|---|---|---|---|---|---|---|
| +/+ | +/− | −/+ | −/− | ||||
| LCx | 5.06-121.08 | 12 | 0 | 0 | 0 | 12/12 | |
| 0.51-12.11 | 11 | 0 | 0 | 1 | 12/12 | ||
| 0.05-1.21 | 6 | 4 | 1 | 1 | 7/12 | ||
| 0.01-0.12 | 0 | 1 | 1 | 10 | 10/12 | ||
| 0.00 | 0 | 0 | 0 | 12 | 12/12 | ||
| LCx | Total | 29 | 5 | 2 | 24 | 53/60 | 0.77 |
| A | Total | 15 | 8 | 4 | 33 | 48/60 | 0.56 |
| B | Total | 17 | 9 | 2 | 32 | 49/60 | 0.61 |
| C | Total | 14 | 13 | 2 | 31 | 45/60 | 0.48 |
| D | Total | 23 | 0 | 8 | 29 | 52/60 | 0.74 |
| E | Total | 4 | 8 | 7 | 41 | 45/60 | 0.19 |
Technologists were blinded to the identity of samples, and the order was randomized separately for each panel.
Symbols: +, positive; −, negative. The two symbols listed are the result of the first test/result of the second test.
Kappa measures the agreement beyond change, with 0.0 indicating no agreement and 1.0 indicating perfect agreement. For all kappa values, P < 0.001.
Impact of an operator change on reproducibility of RUO assay results.
To examine the reproducibility of testing in the RUO assay with a change in the operator, the technologists performing the LCx PCR and the laboratory B in-house PCR switched assays for a third challenge of the 60 samples. Comparison of their results in the LCx C. pneumoniae PCR assay indicated excellent agreement by using the 1-positive-of-3-replicates criterion for positivity. Both technologists agreed on 30 positives and 23 negatives (kappa =0.76). Of the seven discordant results, the technologist with more experience in using the RUO assay identified two extra positives and the other technologist reported five extra positives. Table 4 presents testing profiles of the seven discordant samples. The seven samples are from different sources, and all had been diluted 1:10 to 1:100 beyond each end point of detection. Two of the samples (PS32 and TNFαI 2I) were found negative by six replicates of testing by the in-house PCR assay of laboratory B, and the other five discordant samples were positive in that assay in one to three of the six replicates. Out of 60 specimens and using a positive result in at least one of the three runs as the criterion for detection of a positive specimen by each of the two technologists (data not shown) there were 28, 28, and 26 positives reported by the original LCx PCR-testing technologist, compared to 27, 29, and 31 positives reported by the other technologist, indicating no significant differences (Cochrane Q = 4.3 with 5 df; P = 0.50).
TABLE 4.
Testing profiles of seven specimens found discordant when tested by two technologists in replicates of three in the LCx C. pneumoniae RUO assay
| Identification no. (dilution) | No. of positives/total no. of replicates tested by laboratory B in-house PCR | Assay valuea for the indicated replicate tested by:
|
|||||
|---|---|---|---|---|---|---|---|
| Technologist 1
|
Technologist 2
|
||||||
| 1 | 2 | 3 | 1 | 2 | 3 | ||
| T2219 (10−7) | 3/6 | 354.0 (+) | 55.3 (−) | 10.7 (−) | 9.4 (−) | 9.5 (−) | 9.6 (−) |
| VR-1310 (10−7) | 2/6 | 482.7 (+) | 782.8 (+) | 710.6 (+) | 9.5 (−) | 9.9 (−) | 9.8 (−) |
| A03 (10−7) | 2/6 | 9.7 (−) | 10.3 (−) | 10.0 (−) | 239.4 (+) | 9.6 (−) | 9.7 (−) |
| 379 CBD (10−6) | 2/6 | 540.7 (+) | 10.0 (−) | 9.7 (−) | 10.7 (−) | 10.3 (−) | 10.1 (−) |
| TNFαI 3I (10−5) | 1/6 | 11.8 (−) | 9.5 (−) | 11.9 (−) | 10.8 (−) | 10.6 (−) | 521.1 (+) |
| PS32 (10−7) | 0/6 | 10.0 (−) | 273.2 (+) | 10.4 (−) | 10.0 (−) | 9.8 (−) | 9.6 (−) |
| TNFαI 2I (10−5) | 0/6 | 366.9 (+) | 10.4 (−) | 11.3 (−) | 9.8 (−) | 9.6 (−) | 9.7 (−) |
The cutoff for positivity in the LCx RUO PCR is 100.0, and positive (+) and negative (−) results are indicated by symbols in parentheses.
DISCUSSION
In order to evaluate the performance of the new LCx C. pneumoniae PCR assay developed for research use by Abbott Laboratories, we chose to compare it to three C. pneumoniae in-house PCR protocols from five different research laboratories in North America. These in-house assay types had already had some validation for sensitivity and specificity. All three had been compared in at least two other laboratories by using both calibrated artificial specimens and true clinical specimens. Each had been documented to be capable of detecting at least 1 inclusion-forming unit, and specificity had been documented against other chlamydia species and prokaryotic and eukaryotic DNA (6).
Our interlaboratory comparison of the detection of C. pneumoniae DNA in a panel of 60 preextracted specimens assayed in parallel by six PCR tests found that the LCx C. pneumoniae PCR assay was more sensitive than all of the in-house NAA tests, especially when the samples contained >1 copy of C. pneumoniae DNA per μl. Of the 24 samples that were diluted 1:100 past the end point, the five in-house tests and the LCx C. pneumoniae PCR RUO assay were in agreement that 8 were negative in panels A and B and 12 in panels C and D. Of these 20 samples, 6 were determined negative both times by consensus of all six assays. If these 12 (6 + 6) consensus negatives were moved from the positive group of specimens, the overall adjusted sensitivity for each test would increase (data not shown); that of the LCx C. pneumoniae PCR RUO assay would increase from 68 to 77% compared to those of the in-house PCR tests, which would range from 37 to 71%.
The specificity of the LCx RUO PCR was 100%, and few false positives were observed in the other PCR tests. Negative specimens were purposely placed in sequence after strong positives, but the study results showed high specificity, with little or no contamination, for these assays performed by experienced laboratory technologists. These results may not be generalizable to all laboratory settings.
Apfalter et al. (1) conducted a nine-center comparison of 16 PCR test methods on identical sets of 15 experimental atheroma samples and 5 spiked controls. The majority of specimens with 1 inclusion body of tissue homogenate were identified by all of the participants, but only 19% of the test methods reported specimens with 0.01 of an inclusion body as positive. In the Apfalter study, 3 out of 16 negative samples were rated as positive, whereas in our study, a minimal number of false positives were reported by the in-house assays (overall specificity, 97.9%). All three of the false positives were reported by two laboratories, and the LCx PCR assay had no false positives. Regardless of previous experience, a high specificity needs to be continually demonstrated by instituting measures to minimize carryover contamination, including detection of contamination through an adequate number of negative controls in each run. The LCx C. pneumoniae PCR RUO assay kit has a nonnested automated format, with fewer steps where contamination might occur.
It was surprising that the overall sensitivity varied markedly for the six testing sites, because extraction was standardized, three laboratories used the same nested PCR primers, and the laboratory technologists performing the assays were highly experienced. The panel was intentionally designed to maximize differences between tests by diluting specimens beyond the end point of detection. Thus, a large number of samples had low DNA copy numbers. These differences in sensitivity may or may not reflect on how well these tests will perform with clinical specimens. With a high C. pneumoniae DNA copy number, all of these assays would be expected to perform well on clinical specimens. Based on our previous observations (21), the C. pneumoniae DNA copy number can be low in clinical specimens such as sputum, nasopharyngeal aspirates, and peripheral blood mononuclear cells, as shown by replicate testing and probit regression modeling. The observations in that study indirectly confirm the findings in this study, but multicenter comparisons need to be repeated with clinical specimens.
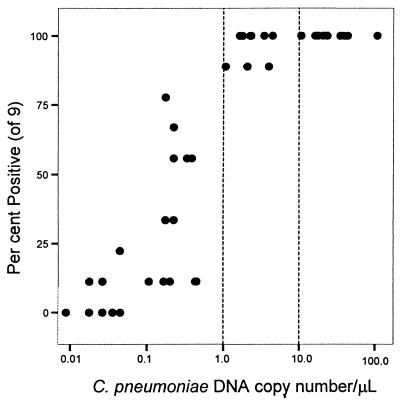
We demonstrated the critical dependence of test sensitivity on relative DNA copy number in each sample (Table 2 and Fig. 2). Figure 3 plots the percentage of positives in the LCx RUO PCR assay against the DNA copy number per microliter and illustrates the ability of this assay to reproducibly detect samples with lower copy numbers, based on testing nine replicates of 20 μl each and using the criterion that at least one of three replicates must test positive for the sample to be declared positive. A very high percentage of positives can be expected when the number of DNA copies is above 1.0 per 1 μl of sample. The percentage varies from 77 to 12% when the number of DNA copies ranges from 0.99 to 0.01 per μl. Reproducibility was also assessed by testing the same specimens a second time in a different random order. All laboratories showed high kappa agreement except for one. The LCx C. pneumoniae PCR assay had the highest kappa agreement at 0.77. This level of intraobserver agreement remained high as larger numbers of runs in the RUO assay were factored into the calculations. The reproducibility of the RUO assay between two different technologists performing three runs each (180 samples) was also excellent, and presumably when this assay becomes more universally used, similarly good agreement may be expected.
FIG. 3.
Relationship between the reproducibility of the Abbott LCx C. pneumoniae RUO PCR and the C. pneumoniae DNA copy number among 48 specimens assayed nine times each. Dashed lines indicate DNA copy numbers of 1 (left) and 10 (right)/μl.
Having access to an industry-produced assay in kit form such as this LCx C. pneumoniae RUO PCR from Abbott Laboratories should ensure consistent reagent production and performance, and provide research laboratories with data that lend themselves to cross-interpretation and comparative findings. A direct comparison between tests in large numbers of patients' specimens is needed to validate the equal or superior performance of this LCx RUO PCR assay for detection in clinical specimens.
REFERENCES
- 1.Apfalter, P., F. Blasi, J. Boman, C. A. Gaydos, M. Kundi, M. Maass, A. Makristathis, A. Meijer, R. Nadrchal, K. Persson, M. L. Rotter, C. Y. W. Tong, G. Stanek, and A. M. Hirschl. 2001. Multicenter comparison trial of DNA extraction methods and PCR assays for detection of Chlamydia pneumoniae in endarterectomy specimens. J. Clin. Microbiol. 39:519-524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Black, C. M., P. I. Fields, T. O. Messmer, and B. P. Berdal. 1994. Detection of Chlamydia pneumoniae in clinical specimens by polymerase chain reaction using nested primers. Eur. J. Clin. Microbiol. Infect. Dis. 13:752-756. [DOI] [PubMed] [Google Scholar]
- 3.Boman, J., C. A. Gaydos, and T. C. Quinn. 1999. Molecular diagnosis of Chlamydia pneumoniae infection. J. Clin. Microbiol. 37:3791-3799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Campbell, L. A., E. R. O'Brien, A. L. Cappuccio, C. C. Kuo, S. P. Wang, D. L. Patton, D. Stewart, and J. T. Grayston. 1995. Detection of Chlamydia pneumoniae in atherectomy tissue from patients with symptomatic coronary artery disease. J. Infect. Dis. 172:585-588. [DOI] [PubMed] [Google Scholar]
- 5.Campbell, L. A., M. Perez Melgosa, D. J. Hamilton, C. C. Kuo, and J. T. Grayston. 1992. Detection of Chlamydia pneumoniae by polymerase chain reaction. J. Clin. Microbiol. 30:434-439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dowell, S. F., R. W. Peeling, J. Boman, G. M. Carlone, B. S. Fields, J. Guarner, M. R. Hammerschlag, L. A. Jackson, C. C. Kuo, M. Maass, T. O. Messmer, D. F. Talkington, M. L. Tondella, S. R. Zaki, et al. 2001. Standardizing Chlamydia pneumoniae assays: recommendations from the Centers for Disease Control and Prevention (USA) and the Laboratory Centre for Disease Control (Canada). Clin. Infect. Dis. 33:492-503. [DOI] [PubMed] [Google Scholar]
- 7.Gaydos, C. A., T. C. Quinn, and J. J. Eiden. 1992. Identification of Chlamydia pneumoniae by DNA amplification of the 16S rRNA gene. J. Clin. Microbiol. 30:796-800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Holland, S. M., C. A. Gaydos, and T. C. Quinn. 1990. Detection and differentiation of Chlamydia trachomatis, Chlamydia psittaci, and Chlamydia pneumoniae by DNA amplification. J. Infect. Dis. 162:984-987. [DOI] [PubMed] [Google Scholar]
- 9.Khan, M. A., and C. W. Potter. 1996. The nPCR detection of Chlamydia pneumoniae and Chlamydia trachomatis in children hospitalized for bronchiolitis. J. Infect. 33:173-175. [DOI] [PubMed] [Google Scholar]
- 10.Kubota, Y. 1996. A new primer pair for detection of Chlamydia pneumoniae by polymerase chain reaction. Microbiol. Immunol. 40:27-32. [DOI] [PubMed] [Google Scholar]
- 11.Kuo, C. C., A. Shor, L. A. Campbell, H. Fukushi, D. L. Patton, and J. T. Grayston. 1993. Demonstration of Chlamydia pneumoniae in atherosclerotic lesions of coronary arteries. J. Infect. Dis. 167:841-849. [DOI] [PubMed] [Google Scholar]
- 12.Maass, M., E. Krause, P. M. Engel, and S. Kruger. 1997. Endovascular presence of Chlamydia pneumoniae in patients with hemodynamically effective carotid artery stenosis. Angiology 48:699-706. [DOI] [PubMed] [Google Scholar]
- 13.Madico, G., T. C. Quinn, J. Boman, and C. A. Gaydos. 2000. Touchdown enzyme time release-PCR for detection and identification of Chlamydia trachomatis, C. pneumoniae, and C. psittaci using the 16S-23S spacer rRNA genes. J. Clin. Microbiol. 38:1085-1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mahony, J. B., S. Chong, B. K. Coombes, M. Smieja, and A. Petrich. 2000. Analytical sensitivity, reproducibility of results, and clinical performance of five PCR assays for detecting Chlamydia pneumoniae DNA in peripheral blood mononuclear cells. J. Clin. Microbiol. 38:2622-2627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mahony, J. B., K. E. Luinstra, D. Jang, J. Sellors, and M. A. Chernesky. 1992. Chlamydia trachomatis confirmatory testing of PCR-positive genitourinary specimens using a second set of plasmid primers. Mol. Cell. Probes 6:381-388. [DOI] [PubMed] [Google Scholar]
- 16.Messmer, T. O., S. K. Skelton, J. F. Moroney, H. Daugharty, and B. S. Fields. 1997. Application of a nested, multiplex PCR to psittacosis outbreaks. J. Clin. Microbiol. 35:2043-2046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nystrõm-Rosander, C., S. Thelin, E. Hjelm, O. Lindquist, C. Páhlson, and G. Friman. 1997. High incidence of Chlamydia pneumoniae in sclerotic heart valves of patients undergoing aortic valve replacement. Scand. J. Infect. Dis. 29:361-365. [DOI] [PubMed] [Google Scholar]
- 18.Petitjean, J., F. Vincent, M. Fretigny, A. Vabret, J. D. Poveda, J. Brun, and F. Freymuth. 1998. Comparison of two serological methods and a polymerase chain reaction-enzyme immunoassay for the diagnosis of acute respiratory infections with Chlamydia pneumoniae in adults. J. Med. Microbiol. 47:615-621. [DOI] [PubMed] [Google Scholar]
- 19.Rasmussen, S. J., F. P. Douglas, and P. Timms. 1992. PCR detection and differentiation of Chlamydia pneumoniae, Chlamydia psittaci and Chlamydia trachomatis. Mol. Cell. Probes 6:389-394. [DOI] [PubMed] [Google Scholar]
- 20.Smieja, M., S. Chong, M. Natarajan, A. Petrich, L. Rainen, and J. B. Mahony. 2001. Circulating nucleic acids of Chlamydia pneumoniae and cytomegalovirus in patients undergoing coronary angiography. J. Clin. Microbiol. 39:596-600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Smieja, M., J. B. Mahony, C. H. Goldsmith, S. Chong, A. Petrich, and M. Chernesky. 2001. Replicate PCR testing and probit analysis for detection and quantitation of Chlamydia pneumoniae in clinical specimens. J. Clin. Microbiol. 39:1796-1801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tjhie, H. T. J., R. Roosendaal, J. M. M. Walboomers, J. J. H. Theunissen, R. R. M. Tjon Lim Sang, C. J. L. M. Meijer, D. M. MacLaren, and A. J. C. van den Brule. 1993. Detection of Chlamydia pneumoniae using a general Chlamydia polymerase chain reaction with species differentiation after hybridization. J. Microbiol. Methods 18:137-150. [Google Scholar]
- 23.Tong, C. Y., and M. Sillis. 1993. Detection of Chlamydia pneumoniae and Chlamydia psittaci in sputum samples by PCR. J. Clin. Pathol. 46:313-317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.van Kuppeveld, F. J. M., K. E. Johansson, J. M. D. Galama, J. Kissing, G. Bölske, J. T. M. van der Logt, and W. J. G. Melchers. 1994. Detection of mycoplasma contamination in cell cultures by a mycoplasma group-specific PCR. Appl. Environ. Microbiol. 60:149-152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Watson, M. W., P. R. Lambden, and I. N. Clark. 1991. Genetic diversity and identification of human infection by amplification of the chlamydial 60-kilodalton cysteine-rich outer membrane protein gene. J. Clin. Microbiol. 29:1188-1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wilson, P. A., J. Phipps, D. Samuel, and N. A. Saunders. 1996. Development of a simplified polymerase chain reaction-enzyme immunoassay for the detection of Chlamydia pneumoniae. J. Appl. Bacteriol. 80:431-438. [DOI] [PubMed] [Google Scholar]