Abstract
A novel PCR-based restriction fragment length polymorphism analysis of the Brachyspira nox gene was developed. The restriction patterns for Brachyspira hyodysenteriae, B. pilosicoli, B. intermedia, B. murdochii, and B. innocens were highly distinct with two restriction endonucleases only. The assay proved to be user-friendly and robust.
The porcine intestinal tract is frequently colonized by different Brachyspira species. Brachyspira hyodysenteriae is the causative agent of swine dysentery, and B. pilosicoli causes intestinal spirochetosis (9). The other species, namely, B. intermedia, B. murdochii, and B. innocens, are considered nonpathogenic (9). Due to differences in pathogenicity (9) and the zoonotic potential of B. pilosicoli (15), rapid and user-friendly differentiation between species is important with respect to the economic use of medication, preharvest food safety, and animal trade.
Currently, porcine Brachyspira isolates are differentiated based on phenotypic criteria, including intensity of hemolysis, indole production, hippurate hydrolysis, and activities of α-galactosidase, α-glucosidase, and β-glucosidase (6, 14). In addition, a positive ring phenomenon is indicative of B. hyodysenteriae (11). Due to variable results in these tests and difficulty in correctly assessing the intensity of hemolysis, it seems desirable to replace phenotypic identification schemes with genotypic methods. The currently used sequencing analyses of 16S or 23S ribosomal DNA (rDNA), the nox gene, or unidentified DNA segments or PCR methods based on these data (1, 4, 5, 10, 12, 13) either are not practical for routine veterinary diagnostic purposes or are confined to the identification of selected species only. Therefore, the purpose of the present study was to develop a robust and user-friendly method for the identification of all porcine Brachyspira species and to evaluate its performance in routine diagnostic work.
Based on an analysis of sequencing data derived from GenBank for the nox genes of 15 Brachyspira strains (Table 1), we designed a nox gene-specific PCR with subsequent evaluation of restriction fragment length polymorphisms (RFLPs) of the PCR products. A forward primer [Bnoxf; 5′-TAG C(CT)T GCG GTA T(CT)G C(AT)C TTT GG-3′] and a reverse primer [Bnoxr; 5′-CTT CAG ACC A(CT)C CAG TAG AAG CC-3′] specific for the Brachyspira nox gene were designed to include a 939-bp fragment encompassing positions 345 to 1283 of the B. hyodysenteriae strain B204 nox gene. To perform the PCR, bacterial growth was removed from a 2-day-old pure culture on Columbia blood agar (Oxoid, Wesel, Germany), and a bacterial suspension was prepared with 1 mM Tris (pH 8.0)-0.1 mM EDTA. To achieve bacterial lysis, the suspension was boiled for 8 min in a microwave. Ten microliters of lysate was added to 40 μl of a PCR premixture to yield final concentrations of 1.5 mM MgCl2 (Gibco BRL, Karlsruhe, Germany), PCR buffer (Gibco BRL), a 0.2 mM concentration of each deoxynucleoside triphosphate (Roth, Karlsruhe, Germany), 0.5 μM Bnoxf, 0.5 μM Bnoxr, and 2.5 U of Taq polymerase (Gibco BRL). The mixture was subjected to 30 cycles of amplification in a Crocodile III cycler (Quantum Appligene, Illkirch, France). After initial denaturation at 94°C for 3 min, each cycle involved denaturation at 94°C for 30 s, annealing at 59°C for 40 s, and extension at 72°C for 54 s. The amplification was finished after a final extension step at 72°C for 10 min. An aliquot of 10 μl of the PCR product was visualized after separation by electrophoresis in a 1.5% agarose gel and staining with ethidium bromide.
TABLE 1.
GenBank accession numbers of nox gene sequences of Brachyspira spp.
| Strain | GenBank accession no. |
|---|---|
| B. hyodysenteriae B204 | U19610 |
| B. hyodysenteriae R1 | AF060802 |
| B. hyodysenteriae B169 | AF060801 |
| B. hyodysenteriae B78 | AF060800 |
| B. pilosicoli WesB | AF060808 |
| B. pilosicoli 42167 | AF060809 |
| B. pilosicoli P43/6/78 | AF060807 |
| B. pilosicoli HRM7 | AF060806 |
| B. intermedia 2818.5 | AF060810 |
| B. intermedia PWS/A | AF060811 |
| B. intermedia 4482 | AF060812 |
| B. murdochii 155-20 | AF060803 |
| B. murdochii 56-150 | AF060813 |
| B. innocens B256 | AF060804 |
| B. innocens 4/71 | AF060805 |
The buffer composition of the remaining PCR product was modified to meet the conditions for restriction digestion with DpnII (New England Biolabs, Frankfurt, Germany) and BfmI (MBI Fermentas, St. Leon-Rot, Germany). We prepared 10× PCR restriction buffer (230 mM Tris acetate [pH 7.6], 41 mM potassium acetate, 9.25 mM magnesium acetate, 30 mM spermidine [Sigma, Taufkirchen, Germany], 1 mg of bovine serum albumin/ml). For restriction digestion, 12.5 μl of the PCR product, 2.5 μl of 10× PCR restriction buffer, and 2.5 μl of dithiothreitol (10 mM; Roth) were mixed; the volume was adjusted to 25 μl with distilled H2O; and 3 or 1.5 U of DpnII or BfmI, respectively, was added. Digestion was carried out at 37°C for 2 h. The restriction fragments were separated in a 2% agarose gel and visualized after staining with ethidium bromide.
The predicted fragment lengths (Table 2) were confirmed by using the reference strains B. hyodysenteriae B204 (kindly provided by D. L. Harris and S. C. Whipp, Department of Veterinary Microbiology and Preventive Medicine, Iowa State University, Ames) and B. pilosicoli P43/6/78, B. intermedia AN26:93, B. innocens C336, and B. murdochii C301 (kindly provided by C. Fellström, Department of Medicine and Surgery, Faculty of Veterinary Medicine, Swedish University of Agricultural Sciences, Uppsala, Sweden) (Fig. 1). In addition, 132 field strains isolated from clinical specimens from diseased and healthy pigs were identified conventionally as described by Stanton et al. (14) and Fellström et al. (6). Hemolysis was judged based on the appearance of growth on Trypticase soy agar (BD, Heidelberg, Germany) with 10% bovine blood (WDT, Hoyerhagen, Germany) and on the ring phenomenon occurring upon incision of the inoculated agar (11). Hippurate hydrolysis and α-galactosidase, α-glucosidase, and β-glucosidase activities were determined by using Rosco diagnostic tablets (Rosco Diagnostic, Taastrup, Denmark). To determine indole production, growth was collected with a swab from a 2-day-old pure culture on Columbia blood agar, and 1 or 2 drops of DMACA reagent (BD) was added; a blue-green color appearing within 5 min was considered a positive result.
TABLE 2.
Predicted fragment sizes after digestion of the nox-specific PCR product with DpnII and BfmI for the five porcine Brachyspira spp.
| Species | Predicted restriction fragments (bp) obtained with:
|
|
|---|---|---|
| DpnII | BfmI | |
| B. hyodysenteriae | 684, 209, 24 | 741, 197 |
| B. pilosicoli | 896, 24 | 741, 197 |
| B. intermedia | 684, 209, 24 | 504, 238, 197 |
| B. innocens | 684, 209, 24 | 504, 210, 197, 25 |
| B. murdochii | 684, 157, 24 | 504, 210, 197, 25 |
Values in bold are fragments not visualized in ethidium bromide-stained gels.
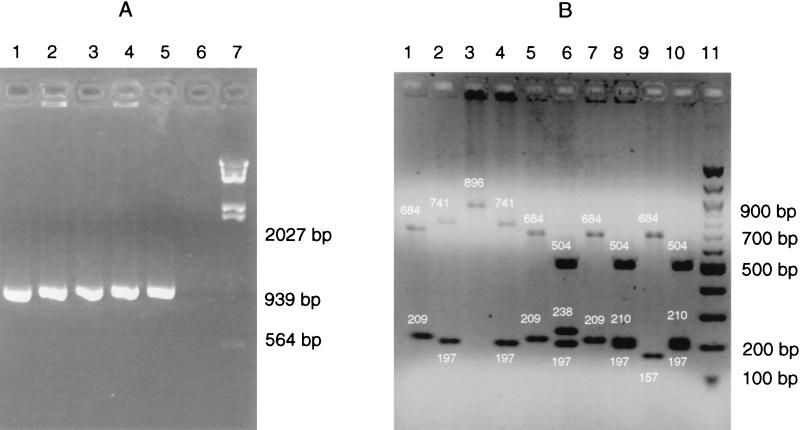
FIG. 1.
nox-specific PCR (A) and RFLP (B) analyses of the five Brachyspira reference strains. (A) Lanes 1 to 5, B. hyodysenteriae B204, B. pilosicoli P43/6/78, B. intermedia AN26:93, B. innocens C336, and B. murdochii C301; lane 6, distilled H2O as a template; lane 7, HindIII-digested λ DNA. (B) Lanes 1 and 2, B. hyodysenteriae B204; lanes 3 and 4, B. pilosicoli P43/6/78; lanes 5 and 6, B. intermedia AN26:93; lanes 7 and 8, B. innocens C336; lanes 9 and 10, B. murdochii C301. Digestion was done with DpnII (odd-numbered lanes) and BfmI (even-numbered lanes). Lane 11, 100-bp DNA ladder. The small white numbers are fragment sizes (in base pairs).
According to these criteria, 80 isolates belonged to the species B. hyodysenteriae, 7 were B. pilosicoli, 4 were B. intermedia, 24 were B. innocens, and 17 were B. murdochii. By nox-specific RFLP typing, 121 of these isolates were classified accordingly, while 11 isolates showed different results (Table 3). Five isolates showed a banding pattern that appeared to be the result of a mixture of fragments from two Brachyspira spp. In order to confirm this result, a bacterial suspension was serially diluted and plated to obtain the growth of single colonies. Single colonies were retested and proven to be a pure culture of one of the formerly identified species. For another five isolates, the two methods yielded different identifications (Table 3). One indole-negative, apparently strongly hemolytic B. hyodysenteriae isolate with a weakly positive α-galactosidase reaction was identified as B. innocens by the nox-specific RFLP method. One hippurate-positive, weakly indole-positive, and α-galactosidase-negative B. pilosicoli isolate showed the banding pattern of B. intermedia. Two indole-negative, α-galactosidase-positive B. innocens isolates were identified as B. intermedia and B. murdochii. Finally, one weakly hemolytic isolate with only β-glucosidase activity (B. murdochii) showed the restriction fragment pattern of B. intermedia. Since there are known variations and even discrepancies in species descriptions based on biochemical reactions (6, 7, 14), the genotypic identification is to be considered the correct one. This conclusion is supported by the finding that only a single isolate biochemically classified as B. innocens could not be assigned to one of the five relevant species with the proposed nox-specific RFLP method.
TABLE 3.
Comparison of the results of identification by conventional testing and nox-specific RFLP typing
| Phenotypic identification (no. of isolates) | Identification by nox-specific RFLP typing
|
||
|---|---|---|---|
| No. of identical isolates | Different (n) | Mixed culture of two species (n) | |
| B. hyodysenteriae (80) | 77 | B. innocens (1) | B. hyodysenteriae + B. innocens (2) |
| B. pilosicoli (7) | 5 | B. intermedia (1) | B. pilosicoli + B. murdochii (1) |
| B. intermedia (4) | 3 | B. intermedia + B. murdochii (1) | |
| B. innocens (24) | 20 | B. intermedia (1) B. murdochii (1); no identification (1) | B. innocens + B. intermedia (1) |
| B. murdochii (17) | 16 | B. intermedia (1) | |
Comparable attempts to separate B. hyodysenteriae, B. pilosicoli, B. intermedia, B. murdochii, and B. innocens by DNA-based methods had been made previously, also with nox-specific RFLP or 23S rDNA-based RFLP analysis. Both methods established by Barcellos et al. (2, 3) did not allow the differentiation of all weakly hemolytic spirochetes due to overlapping banding patterns, and the nox-based method also showed strain variations of RFLP patterns for B. intermedia, B. pilosicoli, and B. innocens. A nox-based PCR recently developed by Ateyo et al. (1) required four different primers and a complex PCR protocol not easily established in a routine diagnostic laboratory. In addition, it also did not allow the differentiation of all five relevant Brachyspira species. Another method, based on Southern blotting analysis of EcoRV chromosomal DNA digests with a PCR-amplified and digoxigenin-labeled fragment of the flaA1 gene, was described by Fisher et al. (8). It corresponded well to the known classification of all five relevant Brachyspira species but is not a practical method for a routine diagnostic laboratory. The 23S rDNA-based PCR developed by Leser et al. (10) needed different sets of primers and different annealing temperatures and could not identify all biochemical groups.
To our knowledge, the nox-specific RFLP method described here is the first PCR-based method developed that facilitates the complete discrimination of all five porcine Brachyspira spp. of veterinary interest without the help of sequencing or hybridization technology. It is user-friendly and robust enough to be performed in clinical veterinary laboratories with basic equipment for molecular biology work.
Acknowledgments
A. Rothkamp is a fellow of Niedersächsische Tierseuchenkasse.
REFERENCES
- 1.Ateyo, R. F., T. B. Stanton, N. S. Jensen, D. S. Suriyaarachichi, and D. J. Hampson. 1999. Differentiation of Serpulina species by NADH oxidase gene (nox) sequence comparison and nox-based polymerase chain reaction tests. Vet. Microbiol. 67:47-60. [DOI] [PubMed]
- 2.Barcellos, D. E. S. N., N. Ikuta, V. R. Lunge, A. Fonseca, E. K. Marques, M. de Uzeda, and G. Duhamel. 2000. Identification and typing of porcine intestinal spirochetes by PCR-restriction fragment length polymorphism analysis of the nox gene, p. 47. In C. Cargill and S. McOrist (ed.), Proceedings of the 16th IPVS Congress. Causal Productions Pty Ltd, Rundle Mall, Australia.
- 3.Barcellos, D. E. S. N., M. de Uzeda, N. Ikuta, V. R. Lunge, A. S. K. Fonseca, I. I. T. A. Kader, and G. E. Duhamel. 2000. Identification of porcine intestinal spirochetes by PCR-restriction fragment length polymorphism analysis of ribosomal DNA encoding 23S rRNA. Vet. Microbiol. 75:189-198. [DOI] [PubMed] [Google Scholar]
- 4.Barcellos, D. E. S. N., M. R. Mathiesen, M. de Uzeda, I. I. T. A. Kader, and G. E. Duhamel. 2000. Prevalence of Brachyspira species isolated from diarrhoeic pigs in Brazil. Vet. Rec. 146:398-403. [DOI] [PubMed] [Google Scholar]
- 5.Elder, R. O., G. E. Duhamel, R. W. Schaffer, M. R. Mathiesen, and R. Ramanathan. 1994. Rapid detection of Serpulina hyodysenteriae in diagnostic specimens by PCR. J. Clin. Microbiol. 32:1497-1502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fellström, C., B. Pettersson, J. Thomson, A. Gunnarsson, M. Persson, and K.-E. Johansson. 1997. Identification of Serpulina species associated with porcine colitis by biochemical analysis and PCR. J. Clin. Microbiol. 35:462-467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Feltrup, C., J. Verspohl, and G. Amtsberg. 1999. Diagnosis of swine dysentery and spirochaetal diarrhoea. Part I. Cultural and biochemical differentiation of intestinal Serpulina in laboratory diagnosis. Dtsch. Tieraerztl. Wochenschr. 106:200-207. [PubMed] [Google Scholar]
- 8.Fisher, L. N., M. R. Mathiesen, and G. E. Duhamel. 1997. Restriction length polymorphism of the periplasmatic flagellar flaA1 gene of Serpulina species. Clin. Diagn. Lab. Immunol. 4:681-686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hampson, D. J. 2000. The Serpulina story, p. 1-5. In C. Cargill and S. McOrist (ed.), Proceedings of the 16th IPVS Congress. Causal Productions Pty Ltd, Rundle Mall, Australia.
- 10.Leser, T. D., K. Moller, T. K. Jensen, and S. E. Jorsal. 1997. Specific detection of Serpulina hyodysenteriae and potentially pathogenic weakly β-hemolytic porcine intestinal spirochetes by polymerase chain reaction targeting the 23S rDNA. Mol. Cell. Probes 11:363-372. [DOI] [PubMed] [Google Scholar]
- 11.Olson, L. D., and W. H. Fales. 1983. Comparison of stained smears and culturing for identification of Treponema hyodysenteriae. J. Clin. Microbiol. 18:950-955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Park, N. Y., C. Y. Chung, A. J. McLaren, R. F. Ateyo, and D. J. Hampson. 1995. Polymerase chain reaction for identification of human and porcine spirochaetes recovered from cases of intestinal spirochaetosis. FEMS Microbiol. Lett. 125:225-230. [DOI] [PubMed] [Google Scholar]
- 13.Petterson, B., C. Fellström, A. Andersson, M. Uhlén, A. Gunnarsson, and K.-E. Johansson. 1996. The phylogeny of intestinal porcine spirochetes (Serpulina species) based on sequence analysis of the 16S rRNA gene. J. Bacteriol. 178:4189-4199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Stanton, T. B., E. Fournié-Amazouz, D. Postic, D. J. Trott, P. A. D. Grimont, G. Baranton, D. J. Hampson, and I. Saint Girons. 1997. Identification of two new species of intestinal spirochetes: Serpulina intermedia sp. nov. and Serpulina murdochii sp. nov. Int. J. Syst. Bacteriol. 47:1007-1012. [DOI] [PubMed] [Google Scholar]
- 15.Trott, D. J., C. R. Huxtable, and D. J. Hampson. 1996. Experimental infection of newly weaned pigs with human and porcine strains of Serpulina pilosicoli. Infect. Immun. 64:4648-4654. [DOI] [PMC free article] [PubMed] [Google Scholar]