Abstract
Between December 1999 and December 2000, teams from the National Institute of Cholera and Enteric Diseases, Calcutta, India, examined eight outbreaks of cholera, which occurred in different parts of the country distant from each other. In two of these outbreaks each, only V. cholerae O1 biotype ElTor or V. cholerae O139 could be isolated, while in the remaining four outbreaks, both O1 and O139 were isolated. The interesting feature is the escalating association of V. cholerae O139 with outbreaks of cholera; two of the most recent outbreaks, one in Calcutta and one in Orissa, were caused exclusively by O139. The O139 strains from the six different outbreaks were genotypically closely related. These trends indicate a shift in the outbreak propensity of V. cholerae O139.
Vibrio cholerae O139 Bengal, the new causative strain of cholera, first emerged in September 1992 in the south Indian coastal city of Madras and then spread rapidly to different areas of cholera endemicity in India and its neighboring countries (M. J. Albert, A. K. Siddique, M. S. Islam, A. S. G. Faruque, M. Ansaruzzaman, S. M. Faruque, and R. B. Sack, Letter, Lancet 341:704, 1993; T. Ramamurthy, S. Garg, R. Sharma, S. K. Bhattacharya, G. B. Nair, T. Shimada, T. Takeda, T. Karasawa, H. Kurazono, A. Pal, and Y. Takeda, Letter, Lancet 341:703-704, 1993). This was an unprecedented event in the history of the disease cholera, and the genesis of serogroup O139 was, at that time, thought to be the beginning of the next, or eighth, pandemic of cholera. In the beginning, the new strain totally displaced the existing V. cholerae O1, the only serogroup responsible for epidemics and pandemics of cholera at that time in Calcutta (5). However, serogroup O1, biotype ElTor, reappeared in September 1993, and by February 1994 it had replaced serogroup O139 (3) in the Indian subcontinent. There was a feeling among cholera workers that the appearance of the new serogroup may have been a one-time event. But the resurgence of serogroup O139 in September 1996 in Calcutta (R. Mitra, A. Basu, D. Dutta, G. B. Nair, and Y. Takeda, Letter, Lancet 348:1181, 1996) and the coexistence of serogroups O1 and O139 in many of the areas of cholera endemicity in India and elsewhere suggested that serogroup O139 had come to stay. Serogroup O139 currently accounts for approximately 17% of laboratory-confirmed cholera cases in Asian countries in which cholera is endemic (6). No evidence is currently available to indicate whether or not this strain has become a new threat. However, we have recently begun to witness a conspicuous increase in the association of V. cholerae O139 with cholera outbreaks in India.
Since the National Institute of Cholera and Enteric Diseases is the national reference center for cholera in India, it periodically receives requests from the government to investigate outbreaks of cholera when they occur in that country. Stool samples or rectal swabs of patients hospitalized during the outbreaks investigated were directly plated on thiosulfate citrate bile salt sucrose agar (TCBS; Eiken, Tokyo, Japan) and incubated at 37°C for 16 to 18 h. V. cholerae was identified by previously published methods (7). Strains were serotyped by using O139 antiserum and polyvalent O1 and monospecific Inaba and Ogawa antisera.
V. cholerae strains isolated from patients affected in the outbreaks were examined for resistance to ampicillin (10 μg), chloramphenicol (30 μg), cotrimoxazole (25 μg), ciprofloxacin (5 μg), furazolidone (100 μg), gentamicin (10 μg), neomycin (30 μg), nalidixic acid (30 μg), norfloxacin (10 μg), streptomycin (10 μg), and tetracycline (30 μg) by using commercial disks (HiMedia, Bombay, India). Strains were characterized as susceptible, of reduced susceptibility, or resistant based on the size of the inhibition zone around each disk, according to the manufacturer's instructions, which matched the interpretive criteria recommended by the World Health Organization as followed in a previous study (3).
A method modified from the work of Murray and Thompson (4) was used for DNA extraction from the V. cholerae O139 strains examined in this study. Randomly amplified polymorphic DNA (RAPD) PCR fingerprinting was carried out with primer 1281 (5′-AACGCGCAAC) (1) in a 25-μl reaction mixture containing 2.5 μl of 10× PCR buffer, 20 ng of V. cholerae genomic DNA, 2.5 μl of 25 μM MgCl2, 20 pmol of the primer, 1 U of Amplitaq DNA polymerase, and 2.5 μl of 2.5 mM deoxynucleoside triphosphates. PCR was performed in an automated thermal cycler (Gene Amp 9700; Perkin-Elmer, Foster City, Calif.). The cycling program was 45 cycles of 94°C for 1 min, 36°C for 1 min, and 72°C for 2 min. After PCR, an 8-μl aliquot of the reaction product was electrophoresed in a 1% agarose gel containing 0.5 μg of ethidium bromide/ml and photographed under UV light by using Gel Doc 2000 (Bio-Rad, Richmond, Calif.). A 1-kb ladder (New England Biolabs, Beverly, Mass.) was used as a size marker in all gels.
Genomic DNAs of various strains of V. cholerae were prepared in agarose plugs as described previously (H. Kurazono, J. Okuda, Y. Takeda, G. B. Nair, M. J. Albert, R. B. Sack, M. Chong-nguan, and W. Chaicumpa, Letter, J. Infect. 29:109-110, 1994). Pulsed-field gel electrophoresis (PFGE) was performed by using the contour-clamped homogeneous electric field (CHEF) method on a CHEF Mapper system (Bio-Rad) with 1% PFGE grade agarose in 0.5× TBE buffer (44.5 mM Tris-HCl, 44.5 mM boric acid, 1.0 mM EDTA [pH 8.0]) for 40 h, 24 min. Run conditions were generated by the autoalgorithm mode of the CHEF Mapper PFGE system by using a size range of 20 to 300 kb for V. cholerae strains. Gels were stained with 1 μg of an ethidium bromide solution/ml for 30 min. Gels were then destained in water for 15 min, and photographs were taken under UV light by using the Gel Doc 2000 gel documentation system (Bio-Rad).
Between December 1999 and December 2000, teams from our institute examined eight outbreaks of cholera, which occurred in different parts of the country distant from each other, as shown in Table 1. Of these, in two outbreaks each, only V. cholerae O1 biotype ElTor (Kerala, January 2000, and Calcutta, April 2000) or V. cholerae O139 (Calcutta, October 2000, and Orissa, December 2000) could be isolated, while in the remaining four, both serogroup O1 and serogroup O139 strains were isolated (Table 1). Even though the numbers of strains isolated from Calcutta in April and October 2000 and from Orissa in December 2000 were low, the numbers of cholera cases recorded during the outbreaks were 71, 710, and 194, respectively. These strains, isolated from cholera patients at the time of investigation, served as representatives of the different hospitals and wards. It often happens that by the time the outbreak investigation begins, the outbreak is on the wane, and therefore only strains from the last few cases are isolated. The interesting feature here is the escalating association of V. cholerae O139 with outbreaks of cholera: two of the most recent outbreaks, one in Calcutta (October 2000) and one in Orissa (December 2000), were exclusively caused by O139 (Table 1). These trends indicate a shift in the outbreak propensity of V. cholerae O139. During 2001, the government requested our institute to investigate only one outbreak, which occurred in Madhya Pradesh (central India) between August and September of that year. This outbreak was exclusively caused by V. cholerae O1, biotype Ogawa. However, cholera caused by V. cholerae O139 was reported for the first time in the coastal Karnataka region of India (M. Ballal, B. Nandanan, and P. G. Shivananda, Letter, Indian J. Pathol. Microbiol. 44:177, 2001). Furthermore, among the 2,818 diarrhea cases examined between October and November 2001 in the Communicable Diseases Hospital in Madras, India, the incidences of V. cholerae serogroups O1 and O139 were 14 and 12%, respectively (P. Kunganathan, personal communication). While the data above do not pertain to outbreaks of cholera, they are indications that the escalating trend in the isolation of V. cholerae O139 is continuing.
TABLE 1.
V. cholerae serogroups associated with various outbreaks of cholera in India from December 1999 to December 2000
| Outbreak state (mo yr) | No. of strains examined | No. of strains in serogroup:
|
||
|---|---|---|---|---|
| O1 | O139 | Non-O1, non-O139 | ||
| Orissa (December 1999) | 72 | 61 | 11 | NDa |
| Kerala (January 2000) | 16 | 16 | ND | ND |
| Ahmedabad (January 2000) | 190 | 124 | 46 | 4 |
| Calcutta (April 2000) | 2 | 2 | ND | ND |
| Karnataka (May 2000) | 9 | 5 | 4 | ND |
| Hyderabad (July 2000) | 22 | 13 | 7 | 2 |
| Calcutta (October 2000) | 6 | ND | 6 | ND |
| Orissa (December 2000) | 14 | ND | 14 | ND |
ND, not detected.
The phenotypic and genotypic traits of the O139 strains associated with the outbreaks during 1999 to 2000 were examined. The antimicrobial resistance patterns of the O139 strains were similar; strains from six outbreaks were resistant to furazolidone, ampicillin, and streptomycin. With the exception of O139 strains from the Calcutta 2000 outbreak, which were sensitive to nalidixic acid, strains from the outbreaks were resistant to nalidixic acid.
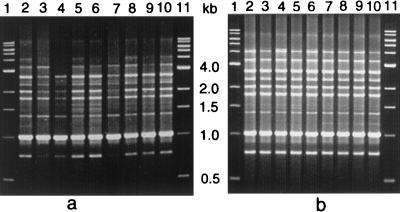
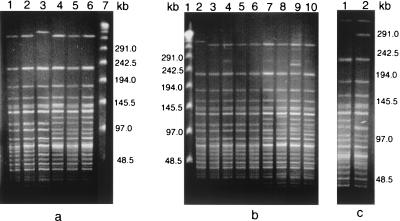
The RAPD patterns of 17 representative O139 strains from these outbreaks exhibited banding patterns similar to each other and to that of the O139 reference strain which was originally isolated from the Madras outbreak of O139 in 1992 (ATCC 51394; MO45) (Fig. 1) (Ramamurthy et al., letter). The PFGE patterns of the strains from five of the V. cholerae O139 outbreaks were also not very different from the ATCC 51394 Madras outbreak strain, and strains from the Ahmedabad outbreak exhibited a banding pattern identical to that of the Madras O139 strain (Fig. 2). Over the 10 years since its genesis, a variety of clones of O139 have been known to exist (2). It appears that the initial clone of O139 has stronger epidemic potential than later clones. These results indicate that O139 clones with minor variations are in circulation. The O139 strains from six different outbreaks were genotypically closely related. Therefore, the threat of O139 V. cholerae outbreaks appears to be escalating again, and continuous monitoring needs to be maintained in areas where V. cholerae O139 has established itself.
FIG. 1.
RAPD profiles of outbreak strains using primer 1281. (a) Lanes 1 and 11, 1-kb ladder; lanes 2 and 3, Orissa, 1999; lanes 4 and 5, Ahmedabad, 2000; lanes 6 and 7, Karnataka, 2000; lanes 8, 9, and 10, Hyderabad, 2000. (b) Lanes 1 and 11, 1-kb ladder; lane 2, O139 reference strain ATCC 51394 (originally designated MO45); lanes 3, 4, 5, and 6, Orissa, 2000; lanes 7, 8, 9, and 10, Calcutta, 2000.
FIG. 2.
PFGE profiles of NotI-digested chromosomal DNAs of various V. cholerae O139 outbreak strains. (a) Lanes 1 and 2, Orissa, 1999; lane 3, Ahmedabad, 2000; lanes 4, 5, and 6, Hyderabad, 2000; lane 7, λ ladder. (b) Lane 1, λ ladder; lane 2, O139 reference strain ATCC 51394; lanes 3, 4, 5, and 6, Calcutta, 2000; lanes 7, 8, 9, and 10, Orissa, 2000. (c) Karnataka, 2000 (both lanes).
Acknowledgments
This work was supported in part by the Council of Scientific and Industrial Research [grant 37(1019)/99EMR II] and the Japan International Co-operation Agency (JICA/NICED project 054-1061-E-O).
REFERENCES
- 1.Akopyanz, N., N. O. Bukenov, T. U. Westblam, S. Kresovich, and D. E. Berg. 1992. DNA diversity among clinical isolates of Helicobacter pylori detected by PCR-based RAPD fingerprinting. Nucleic Acids Res. 20:5137-5142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Faruque, S. M., M. N. Saha, Asadulghani, P. K. Bag, R. K. Bhadra, S. K. Bhattacharya, R. B. Sack, Y. Takeda, and G. B. Nair. 2000. Genomic diversity among Vibrio cholerae O139 strains isolated in Bangladesh and India between 1992 and 1998. FEMS Microbiol. Lett. 184:279-284. [DOI] [PubMed] [Google Scholar]
- 3.Mukhopadhyay, A. K., S. Garg, R. Mitra, A. Basu, K. Rajendran, D. Dutta, S. K. Bhattacharya, T. Shimada, T. Takeda, Y. Takeda, and G. B. Nair. 1996. Temporal shifts in traits of Vibrio cholerae strains isolated from hospitalized patients in Calcutta: a 3-year (1993 to 1995) analysis. J. Clin. Microbiol. 34:2537-2543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Murray, M. G., and W. F. Thompson. 1980. Rapid isolation of high molecular weight plant DNA. Nucleic Acids Res. 8:4321-4325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nair, G. B., T. Ramamurthy, S. K. Bhattacharya, A. K. Mukhopadhyay, S. Garg, M. K. Bhattacharya, T. Takeda, T. Shimada, Y. Takeda, and B. C. Deb. 1994. Spread of Vibrio cholerae O139 Bengal in India. J. Infect. Dis. 169:1029-1034. [DOI] [PubMed] [Google Scholar]
- 6.World Health Organization. 2000. Cholera, 1999. Wkly. Epidemiol. Rec. 75:249-256. [PubMed] [Google Scholar]
- 7.World Health Organization. 1993. Guidelines for cholera control. World Health Organization, Geneva, Switzerland.