Abstract
Three cases of feline atypical mycobacteriosis from different geographical regions in North America were characterized by large clusters of filamentous bacteria visible on hematoxylin-and-eosin-stained tissue sections. PCR amplification demonstrated the presence of Mycobacterium-specific nucleic acid in samples of skin lesions from these cases. PCR-assisted cloning and DNA sequence analysis of a 541-bp length of the Mycobacterium 16S rRNA gene generated DNA sequences which were >95% identical, suggesting that the three isolates were closely related. Two of the sequences were 99% identical and may represent the same species. Alignment with comparable 16S rRNA gene sequences from 66 Mycobacterium species and partially characterized isolates highlighted similarities (>94%) with Mycobacterium bohemicum, Mycobacterium haemophilum, Mycobacterium ulcerans, Mycobacterium avium subsp. avium, and isolate IWGMT 90242. Parsimony analysis of sequence data suggested relatedness to M. leprae. Significant molecular genetic and pathobiological differences between these three similar isolates and other known species of mycobacteria suggested that the organisms may not have been described previously and that these cases may represent a new form of mycobacterial disease in cats. We suggest the term “Mycobacterium visibilis” to describe the organism from which the two nearly identical sequences were obtained.
Cutaneous mycobacteriosis in cats is a rare group of diseases with worldwide distribution which manifests as three forms: (i) feline leprosy, (ii) atypical mycobacteriosis, and (iii) cutaneous tuberculosis. Each form is associated with infection by a different array of Mycobacterium species. Feline leprosy is a granulomatous nodular disease widely assumed to be caused by Mycobacterium lepraemurium or a closely related species and is characterized by single or multiple, firm to soft, intact nodules in the skin or subcutis which frequently ulcerate (9, 10, 12, 18). Nodules can be found anywhere on the body but are frequently located on the head, limbs, and trunk. Regional lymphadenopathy is common. Etiological diagnosis is frequently based on the lack of growth on routine culture medium for acid-fast organisms. In older literature, feline leprosy is sometimes called cutaneous atypical mycobacteriosis, but current usage now makes a clear distinction between the two syndromes (27). Atypical mycobacteriosis, also known as opportunistic mycobacterial granulomas, is characterized by chronic or recurrent fistulous tracts and ulcers, fasciitis, and nodules which ulcerate. Lesions are most frequently located on the ventral abdomen, and the majority of cases are caused by rapidly growing Mycobacterium species including M. fortuitum, M. phlei, M. smegmatis, and M. chelonae. In rare circumstances, slowly growing mycobacteria can also be associated with lesions resembling atypical mycobacteriosis. These include M. avium, M. chitae, M. xenopi, and M. ulcerans (4, 13, 19, 27). Culture of these organisms is relatively successful, thereby differentiating atypical mycobacteriosis infections from feline leprosy. The third form of disease, cutaneous tuberculosis, is associated with M. bovis, M. tuberculosis, or, rarely, M. avium or M. microti and is characterized by multiple ulcers, plaques, nodules, and abscesses that discharge a thick, yellow to green exudate. Lesions are microscopically described as pyogranulomatous dermatitis with extensive caseous necrosis; regional lymphadenopathy is common (10, 19).
Comparison studies have shown that traditional culture and biochemical schemes lack sensitivity and are limited by phenotypic variability (22). The use of molecular genetic techniques to identify infectious mycobacteria has been widely adopted. In particular, 16S rRNA gene sequences have been used to describe phylogenetic relationships between mycobacterial species (7, 16, 22, 24, 26). Similar tests that use sequences of the Mycobacterium-specific dnaJ gene, the 32-kDa protein gene, hsp 65, superoxide dismutase, and the recA, rpoV, and gyrB genes have been reported (2, 5, 15, 20, 21, 24, 25). The molecular genetic approach offers a fast and accurate means to identify Mycobacterium spp., from which information can be gained even if the isolate represents a new taxon or is nonculturable, nonviable, or archived only in formalin-fixed, paraffin-embedded tissue. A 16S rRNA sequence comparison was recently used to positively identify M. lepraemurium infection in three cats from New Zealand (4).
In this report, we describe a distinctive generalized granulomatous dermatitis, fasciitis, and multisystemic granulomatous disease in postmortem tissues from one cat and similar skin lesions in antemortem tissue samples from a second cat. Genetic characterization of Mycobacterium spp. from these two cats, as well as from archived postmortem material from a case of feline disseminated mycobacteriosis with similar histological lesions (11), is presented here. We have partially characterized mycobacterium 16S rRNA gene sequences isolated from each case and show that they are from the same or closely related previously undescribed species. We present data describing possible phylogenetic relationships to other mycobacteria.
CASE REPORTS
Case 1.
An 8-year-old spayed female domestic shorthair cat was presented to a veterinarian in western Canada in February 1994 in an emaciated state with several open sores on the hocks and carpi; cutaneous swelling; extensive alopecia of the head, neck, forelimbs, and thoracic regions; and thickened subcutaneous tissues of the head and forelimbs. Histological examination of skin biopsy specimens showed severe granulomatous fasciitis, panniculitis, and deep dermatitis, associated with very large numbers of thin, filamentous organisms that were visible on hematoxylin-and-eosin-stained formalin-fixed tissue in foamy macrophages and in large, lipid-like vacuoles. The organisms were found to be acid fast (Fite's), and the cat was euthanized for postmortem examination.
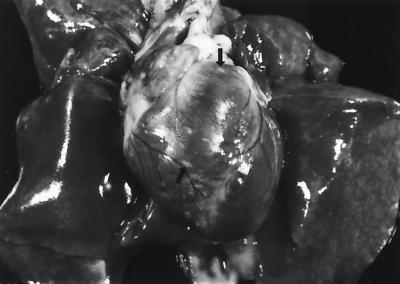
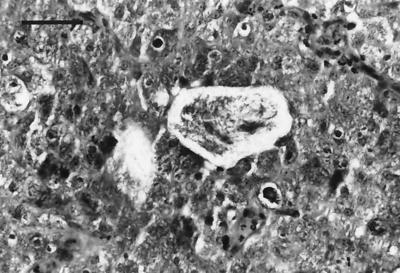
Necropsy revealed extensive alopecia and edema of the head, neck, and forelimbs. The edema fluid was viscous and contained numerous light-yellow, flat, miliary foci which were also present throughout all skeletal muscles of the body and in fascial planes. White to yellow foci (diameter, 1 to 2 mm), often elongated, were scattered over the epicardium, especially on the right side (Fig. 1). White flecks were visible floating in the anterior chambers of both eyes. All lymph nodes were enlarged and edematous. No bacteria were cultured from several tissues submitted. Cytological evaluation of imprints from skin and lymph nodes revealed epithelioid macrophages, lymphocytes, and foreign-body giant cells with unstained 10.4- by 2.6-μm filamentous structures in the intra- and extracellular space. Granulocytic hyperplasia and macrophages containing similar bacteria were visible in imprints of the bone marrow smears. There was marked proteinuria. On histopathological examination, all organs and tissues, except the liver and kidneys, showed extensive granulomatous cell infiltrates, accompanied by clear, lipid vacuole-like structures containing numerous mildly gram-positive, acid-fast filamentous bacteria (Fig. 2). The tissues most affected were the skin, skeletal muscle, interstitium, facial planes, and optic globes.
FIG. 1.
Photograph of heart and lung of cat 1. White 1- to 2-mm-diameter foci (small arrows) are visible on the epicardial surface.
FIG. 2.
Photomicrograph of dermis of cat 1 showing numerous thin, filamentous bacteria within a lipid-like vacuole, surrounded by large macrophages. Hematoxylin and eosin stain was used. Magnification, ×250. Bar, 50 μm.
Case 2.
Histological evaluation of skin biopsied (Portland, Oreg., 1999) from a 6-year-old spayed female domestic shorthair cat with a history of severe pruritis and ulcerative skin lesions revealed extensive ulceration with exudation of plasma proteins and degenerate granulocytes. The epidermis was variably ulcerated or severely hyperplastic. The dermis and subcutis had severe, diffuse to multinodular, densely cellular infiltrates composed of large macrophages with foamy or vacuolated cytoplasm. Numerous poorly stained filamentous bacilli consistent with Mycobacterium spp. were visible in larger cytoplasmic vacuoles. Fite's (acid-fast) staining of these organisms was positive. Biopsy material was frozen for subsequent molecular analysis. The Cryptococcus antigen titer was negative. The animal was treated with clindamycin and clofazimin and responded well to therapy, with only sporadic ulcerated areas on the trunk.
Case 3.
As previously reported by Matthews and Liggitt (11), an 11-year-old domestic longhair cat from a farm in northern Idaho was presented to a veterinary clinic in an emaciated state, showing hair loss and open skin lesions. Attempts to culture, isolate, and transmit the disease organism to laboratory animals were unsuccessful. One notable feature of hematoxylin-and-eosin-stained tissue sections was histiocytes with foamy to granular cytoplasm and cytoplasmic vacuoles containing eosinophilic material which, when subsequently stained with Ziehl-Neelsen acid-fast stain, revealed the presence of acid-fast bacteria in clumps. Archived paraffin-embedded formalin-fixed tissues from the cat were obtained from the College of Veterinary Medicine, Washington State University.
MATERIALS AND METHODS
Bacterial culture and histopathology.
A variety of standard Mycobacterium culture media were used in an attempt to culture acid-fast bacilli from tissues of cats 1 and 3 (culture was not attempted in case 2). Tissues from the three cats were prepared for histological examination by fixation in 10% buffered formalin and embedding in paraffin. Sections were stained with hematoxylin and eosin as well as with a variety of special stains including Brown and Brenn (gram stain for bacteria), modified Fite's (acid-fast), periodic acid-Schiff, and Grocott's methenamine silver stains.
Molecular genetic analysis.
Fresh tissue samples from cat 1 were sent to the National Reference Centre for Tuberculosis (Ottawa, Ontario, Canada) for identification using the AccuProbe Mycobacterium avium culture identification kit (Gen-Probe, San Diego, Calif.).
Fixed tissue was retrieved from 10 10-μm-thick paraffin-embedded sections (cases 1 and 3) by using a microtome with a sterile disposable blade. Paraffin was removed by xylene and 95% ethanol solvent treatment. The tissue pellet was dried thoroughly under a vacuum. The frozen biopsy specimen (case 2) was minced by using a sterile scalpel. Genomic DNA was extracted from these three prepared tissue samples by lysis buffer (100 mM NaCl, 500 mM Tris [pH 8], 10% sodium dodecyl sulfate) and complete proteinase K (0.2 mg/ml) digestion. Two solvent extractions with phenol-chloroform (1:1) and one with chloroform alone were performed. Nucleic acids were concentrated by precipitation in cold 95% ethanol.
PCR followed by restriction fragment length polymorphism (RFLP) analysis of the mycobacterial dnaJ gene was performed as described by Takewaki et al. (24). PCR was conducted using 10× buffer (Invitrogen, Burlington, Ontario, Canada), 1.5 mM Mg2+, primers 5′-GGGTGACGCGGCATGGCCCA-3′ and 5′-CGGGTTTCGTCGTACTCCTT-3′, and 1.25 U of Taq polymerase (Invitrogen) on a PTC200 thermal cycler (MJ Research, Watertown, Mass.; licensed by Perkin-Elmer Cetus, Norwalk, Conn.). Briefly, 40 cycles of PCR amplification were performed in a 50-μl volume with 30 s of denaturation at 94°C, 60 s of annealing at 65°C and 120 s of extension at 72°C. DNA extractions were controlled with a “no-tissue” extraction blank, and “no-DNA” samples were tested as negative PCR controls. The PCR product, prepared by ethanol precipitation and resuspension in 30 μl of sterile water, was digested with restriction enzymes FokI, HinfI, NaeI, and SmaI (Invitrogen; New England Biolabs, Beverly, Mass.) according to the manufacturers' instructions.
PCR amplification of approximately 577 bp of the16S rRNA gene, corresponding to bases 1 to 577 of the M. tuberculosis (GenBank accession no. AJ131120) 16S rRNA gene, was performed as previously described by Hughes et al. (4) by using primers pA (5′-AGAGTTTGATCCTGGCTCAG-3′) and MSHE (5′-GCGACAAACCACCTACGAG-3′). The PCR fragment generated by using primers pA and MSHE was “TA-cloned” into plasmid pCRII (TA-Cloning Kit; Invitrogen) according to the manufacturer's instructions. Transformed Escherichia coli (strain INVF′) colonies were selected on the basis of 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal)-mediated blue-white discrimination and ampicillin resistance. Individual bacterial colonies that contained a fragment of approximately 550 bp and supported PCR amplification (primers pA and MSHE) were prepared for sequencing. Bidirectional sequencing of plasmid and insert was performed by the DNA Services Laboratory, Plant Biotechnology Institute, National Research Council (Saskatoon, Saskatchewan, Canada). The resulting sequences were analyzed by using GenBank BLAST software (1) and ClustalV (Megalign, version 3.15; DNAStar, Madison, Wis.) alignments performed on 16S rRNA gene sequences from 66 different mycobacteria and 24 other members of the Actinomycetales. Phylogenetic analysis was performed using the PHYLIP maximum-likelihood algorithm (3).
Nucleotide sequence accession numbers.
Sequences Mbv1, Mbv2, and Mbv3 have been assigned GenBank accession no. AY061984, AY061985, and AY061986, respectively.
RESULTS
Bacterial culture and histopathology.
Bacterial culture from cat 1 showed sparse growth of a mycobacterium-like organism after 8 weeks on malachite-green culture medium slants, but the organism did not maintain viability. Culture was not attempted on material from cat 2, and previous attempts using material from cat 3 were unsuccessful (11). Samples from all three cats were positive for acid-fast rods. The rods were consistent in size and shape with mycobacteria. Bacterial culture and other staining performed on material from cat 1 showed no evidence of mixed infection with other microorganisms.
Molecular analysis.
Gen-Probe's AccuProbe test specific for M. avium (which cross-reacts with M. gordonae and M. kansasii) was negative on cultured bacilli from cat 1. Amplification of the Mycobacterium dnaJ gene using nucleic acid isolated from formalin-fixed, paraffin-embedded cat 1 tissue resulted in production of a DNA fragment of approximately 240 bp. RFLP analysis showed that the fragment could be digested by FokI and HinfI but not by the NaeI or SmaI restriction enzyme. According to Takewaki et al. (24), RFLP analysis of the mycobacterial dnaJ gene can be used to differentiate mycobacteria into nine groups which correlate roughly with the Runyon groupings (17). Digestion patterns observed with amplified DNA from cat 1 did not match any of the 19 RFLP patterns (12).
Approximately 550 consecutive bases, spanning variable regions 1, 2, and 3 of the 16S rRNA gene (6), were cloned and sequenced from tissue from all three cats. Sequences Mbv1 and Mbv2 (GenBank accession no. AY061984 and AY061985, respectively) were found to be 99.4% similar to each other and 95.0 and 95.2% similar to Mbv3 (GenBank accession no. AY061986) over the entire length of available sequence. The three sequences were 96 to 94% similar to corresponding sequences in M. avium, M. bohemicum, M. ulcerans, and M. haemophilum. They were also 95% similar to the corresponding sequence in an M. avium-like isolate (IWGMT 90242) described by Wayne et al. (26).
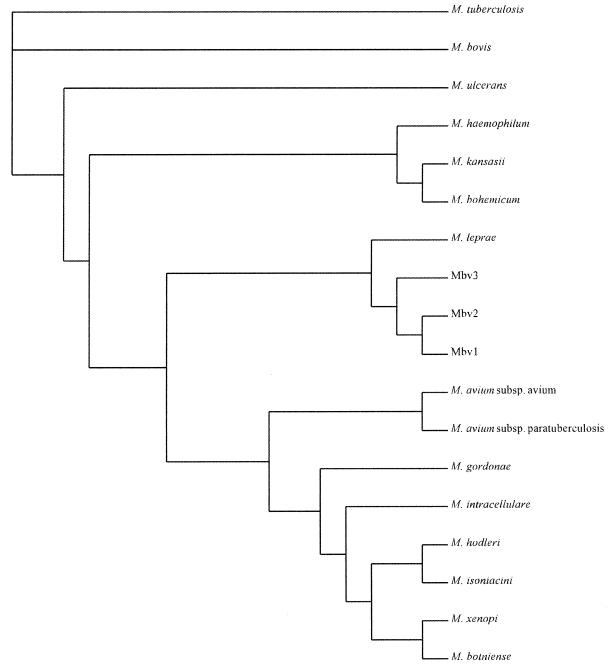
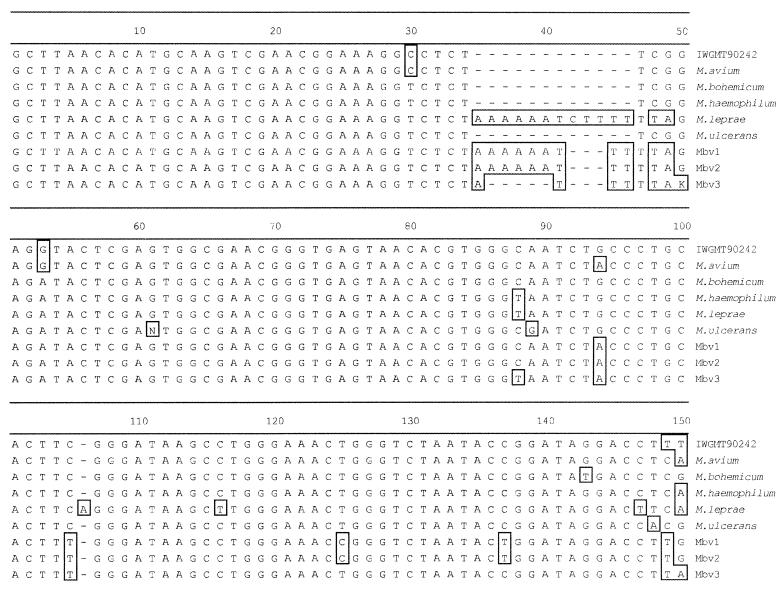
A complete phylogenetic relationship analysis was not possible because only one-third of the 16S rRNA gene was cloned and sequenced; however, maximum-likelihood analysis of parsimony suggested that sequences Mbv1, Mbv2, and Mbv3 were most closely related to sequences of M. leprae (Fig. 3) and were dissimilar to known disease-causing mycobacteria of cats (Table 1). Careful examination of sequence alignments revealed that Mbv1 and Mbv2 both had a 9-bp insert (AAAAAATTT) at position 34 of the cloned sequence, corresponding to variable region 1 (Fig. 4). This feature appeared to be unique among all mycobacteria examined (n = 66) (data not shown). The only other Mycobacterium species to possess similar extra nucleotides at this location was M. leprae, which had a 12-bp insert (AAAAAATCTTTT). The Mbv3 sequence at this locus was shorter, but still unique (ATTTT) among Mycobacterium spp. in 16S rRNA databases. Mbv1, Mbv2, and Mbv3 sequences were only 92.2, 92.1, and 92.6% similar to M. leprae, respectively, over the entire length of sequence available (554 bp), suggesting that they were not members of the M. leprae species.
FIG. 3.
Phylogenetic relationships based on approximately 550 bases of the 16S rRNA genes of selected mycobacteria and Mbv1, Mbv2, and Mbv3 sequences, calculated by the maximum-likelihood method.
TABLE 1.
Relationship between cloned Mbv1, Mbv2, and Mbv3 16S rRNA gene sequences and analogous genes in selected Mycobacterium species
| Mycobacterium species | % similaritya to:
|
||
|---|---|---|---|
| Mbv1 | Mbv2 | Mbv3 | |
| Mbv1 | 99.4 | 95.2 | |
| Mbv2 | 99.4 | 95.0 | |
| Mbv3 | 95.2 | 95.0 | |
| M. avium | 92.5 | 92.3 | 93.0 |
| M. bohemicum | 94.9 | 95.1 | 95.3 |
| M. bovis | 93.4 | 93.3 | 93.4 |
| M. chelonei | 86.8 | 84.1 | 86.4 |
| M. chitae | 83.3 | 82.9 | 86.6 |
| M. fortuitum | 88.2 | 87.8 | 83.6 |
| M. haemophilum | 94.4 | 94.6 | 96.8 |
| M. leprae | 92.2 | 92.1 | 92.6 |
| M. lepraemuriumb | 96.2 | 96.4 | 96.7 |
| M. microtib | 91.0 | 91.0 | 91.0 |
| M. phlei | 81.0 | 81.3 | 86.5 |
| M. smegmatis | 86.6 | 86.2 | 87.9 |
| M. tuberculosis | 93.6 | 93.4 | 93.8 |
| M. ulcerans | 95.1 | 94.9 | 94.4 |
| M. xenopi | 87.7 | 87.2 | 88.1 |
Sequence comparison made on the region equivalent to bases 39 to 572 of M. tuberculosis 16S rRNA gene (GenBank accession no. AJ131120).
Incomplete sequence; comparison made on only 430 bp.
FIG. 4.
Sequence alignment for a portion of the 16S rRNA gene from selected mycobacteria, showing sequence similarity between M. leprae and Mbv1, Mbv2, and Mbv3 at variable region 1. Boxed bases are different from the consensus sequence.
Mbv1, Mbv2, and Mbv3 were 96.1, 96.1, and 94.1% similar to partial M. lepraemurium sequences of 443 bp lacking variable region 1 (Table 1). Over the same span of bases, Mbv1 and Mbv2 were 100% identical to each other and 98% similar to Mbv3. The number of differences suggests that Mbv1, Mbv2, and Mbv3 isolates were also not members of the M. lepraemurium species.
In addition to the GenBank database, Mbv1, Mbv2, and Mbv3 sequences were submitted for comparison with sequences to two 16S RNA databases (Mycobacteriology Laboratory, Health Canada, and International Working Group on Mycobacterial Taxonomy, Medical School Hannover, Hannover, Germany), but no identical sequences were identified.
DISCUSSION
Characteristically, Mycobacterium spp. are not visible in tissue sections stained with hematoxylin and eosin, and acid-fast stains are used to detect their presence. The cats described in this report had granulomatous lesions associated with distinctive histologically visible mycobacteria.
The ability to identify Mycobacterium to the species level has generally been limited to those taxa which can be grown under laboratory conditions and are amenable to biochemical, serological, or structural analysis (22). Molecular biological techniques have improved the accuracy and reporting time, but comparisons are highly dependent on the completeness of semantide-based databases (6, 8, 14, 16). Two different techniques were used to identify the mycobacterium in each of the three cases. RFLP analysis of the Mycobacterium dnaJ gene was inconclusive because the pattern of digestion was unlike that of any of the 19 Mycobacterium species previously described (24).
Comparison of partial 16S rRNA gene sequences (Mbv1, Mbv2, and Mbv3) from cats 1, 2, and 3 showed >96% identity with each other over 541 bp, including three regions known to be variable among different mycobacterial species (6, 26). The high degree of sequence similarity between Mbv1 and Mbv2 would suggest that the two cats were infected with the same mycobacterial species and that Mbv3 sequences were from a closely related isolate with similar virulence factors and pathogenic properties. In our analysis, sequence similarity among isolates of the same species ranged from 98% to 100% but as low as 96% for comparisons between members of the M. avium-complex, while sequence similarity between differing species was <96% (data not shown). Rogall et al. (16) suggested that in Mycobacterium16S rRNA genes, overall sequence similarity was >94% across the entire genus, but their analysis differed by including more than 1,430 bp and excluding regions of alignment uncertainty.
Comparison of Mbv1, Mbv2, and Mbv3 with 16S rRNA sequences in GenBank and two other Mycobacterium-specific databases did not identify the organism as M. lepraemurium or any of the other species of mycobacteria associated with feline mycobacteriosis. Hughes et al. recently reported identifying M. lepraemurium in several cats from New Zealand (4). A relatively high sequence similarity between the three new sequences and M. lepraemurium sequences was observed but may be misleading, because this analysis was made over a short span of nucleotides (430 bp) and the available M. lepraemurium 16S rRNA gene sequence lacked data for variable region 1. More-complete sequences for the M. lepraemurium 16S rRNA gene are not currently available.
Comparisons of Mbv1, Mbv2, and Mbv3 16S rRNA gene sequences with those from 66 Mycobacterium species showed that the mycobacteria identified in these three cases were distinct from M. avium complex species and other species known to cross-react on the Gen-Probe test. This analysis is consistent with the findings that the Gen-Probe test for M. avium was negative and the dnaJ gene RFLP results did not support the identification of M. avium.
Mbv1, Mbv2, and Mbv3 all form secondary structures (helix 18, variable region 3 [6]) which include an extended region between bp 440 and 480. Extensions in this region are correlated with fastidious, slow-growing Mycobacterium species (16, 23). This finding is consistent with the observation that colonies of acid-fast bacilli became apparent only after 8 weeks in culture (case1) and with the lack of successful culturing experienced by Matthews and Liggitt (case 3) (11).
The 16S rRNA gene sequences identified in these three cases seem to be from Mycobacterium taxa not previously described nor sequenced within the slow-growing cluster of Mycobacterium spp. Until the identity of the apparently novel Mycobacterium sp. pathogen described here can be fully elucidated, we suggest the term “Mycobacterium visibilis,” reflecting its distinctive feature of being visible on hematoxylin-and-eosin-stained tissue sections. Based on the collective features of three feline cases sharing very similar histopathology which was distinct from those of feline leprosy, atypical mycobacteriosis, and cutaneous tuberculosis, the term feline multisystemic granulomatous mycobacteriosis was coined for this apparently novel syndrome.
With the identification of more cases, the hypothesis that “M. visibilis” may be causally linked with multisystemic granulomatous mycobacteriosis in cats can be tested. The availability of nucleotide sequence data from this organism makes design of PCR primers and a rapid diagnostic test possible.
Acknowledgments
We are indebted to S. Myers (Prairie Diagnostic Services Inc.) and D. Haines (University of Saskatchewan) for critical reading of the manuscript, to R. Franklin (Idexx Veterinary Services) and E. Walder (Venice, Calif.) for recognition and submission of case 2, to C. Leathers (Washington State University) for archived material from case 3, and to C. Turenne (National Reference Centre for Mycobacteriology, Health Canada), P. Kirschner (Medizinische Hochschule Hannover), and E. Böttger (Medizinische Hochschule Hannover) for assistance with sequence data acquisition from nonpublic databases.
This study was supported in part by the Agriculture Development Fund of the Government of Saskatchewan and the Companion Animal Health Fund, Western College of Veterinary Medicine, University of Saskatchewan.
REFERENCES
- 1.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 17:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Comicini, S., D. Barbarini, S. Telecco, L. Bonno, and P. Marone. 1998. Rapid identification of Mycobacterium tuberculosis and Mycobacterium avium by polymerase chain reaction and restriction enzyme analysis within sigma factor regions. New Microbiol. 21:391-395. [PubMed] [Google Scholar]
- 3.Felsenstein, J. 1989. PHYLIP—Phylogeny Inference Package (version 3.2). Cladistics 5:164-166. [Google Scholar]
- 4.Hughes, M. S., N. W. Ball, L. A. Beck, G. W. de Lisle, R. A. Skuce, and S. D. Neill. 1997. Determination of the etiology of presumptive feline leprosy by 16S rRNA gene analysis. J. Clin. Microbiol. 35:2464-2471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kasai, H., T. Ezaki, and S. Harayama. 2000. Differentiation of phylogenetically related slowly growing mycobacteria by their gyrB sequences. J. Clin. Microbiol. 38:301-308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kempsell, K. E., Y. Ji, I. C. E. Estrada, M. J. Colston, and R. A. Cox. 1992. The nucleotide sequence of the promoter, 16S rRNA and spacer region of the ribosomal RNA operon of Mycobacterium tuberculosis and comparison with Mycobacterium leprae precursor rRNA. J. Gen. Microbiol. 138:1717-1727. [DOI] [PubMed] [Google Scholar]
- 7.Kirschner, P., and E. C. Böttger. 1998. Species identification of mycobacteria using rDNA sequencing. Methods Mol. Biol. 101:349-361. [DOI] [PubMed] [Google Scholar]
- 8.Kirschner, P., B. Springer, U. Vogel, A. Meier, A. Wrede, M. Kiekenbeck, F. C. Bange, and E. C. Böttger. 1993. Genotypic identification of mycobacteria by nucleic acid sequence determination: report of a 2-year experience in a clinical laboratory. J. Clin. Microbiol. 31:2882-2889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lawrence, W. E. 1963. Cat leprosy: infection by a bacillus resembling Mycobacterium lepraemurium. Aust. Vet. J. 39:390-393. [Google Scholar]
- 10.Lemarie, S. L. 1999. Mycobacterial dermatitis. Vet. Clin. N. Am. Small Anim. Pract. 29:1291-1301. [DOI] [PubMed] [Google Scholar]
- 11.Matthews, J. A., and H. D. Liggitt. 1983. Disseminated mycobacteriosis in a cat. J. Am. Vet. Med. Assoc. 183:701-702. [PubMed] [Google Scholar]
- 12.McIntosh, D. 1982. Feline leprosy: a review of forty-four cases from western Canada. Can. Vet. J. 23:291-295. [PMC free article] [PubMed] [Google Scholar]
- 13.Morfitt, D. C., J. A. Matthews, C. O. Thoen, and J. P. Kluge. 1989. Disseminated Mycobacterium avium serotype 1 infection in a seven-month-old cat. J. Vet. Diagn. Investig. 1:354-356. [DOI] [PubMed] [Google Scholar]
- 14.Pitulle, C., M. Dorsch, J. Kazda, J. Wolters, and E. Stackebrandt. 1992. Phylogeny of rapidly growing members of the genus Mycobacterium. Int. J. Syst. Bacteriol. 42:337-343. [DOI] [PubMed] [Google Scholar]
- 15.Plikaytis, B. B., B. D. Plikaytis, M. A. Yakrus, W. R. Butler, C. L. Woodley, V. A. Silcox, and T. M. Shinnick. 1992. Differentiation of slowly growing Mycobacterium species, including Mycobacterium tuberculosis, by gene amplification and restriction fragment length polymorphism analysis. J. Clin. Microbiol. 30:1815-1822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rogall, T., J. Wolters, T. Flohr, and E. C. Böttger. 1990. Towards a phylogeny and definition of species at the molecular level within the genus Mycobacterium. Int. J. Syst. Bacteriol. 40:323-330. [DOI] [PubMed] [Google Scholar]
- 17.Runyon, E. H. 1959. Anonymous mycobacteria in pulmonary disease. Med. Clin. N. Am. 43:273-290. [DOI] [PubMed] [Google Scholar]
- 18.Schiefer, B., B. R. Gee, and G. E. Ward. 1974. A disease resembling feline leprosy in Western Canada. J. Am. Vet. Med. Assoc. 165:1085-1087. [PubMed] [Google Scholar]
- 19.Scott, D. W. 1980. Mycobacteriosis. J. Am. Anim. Hosp. Assoc. 16:346-348. [Google Scholar]
- 20.Shivannavar, C. T., V. M. Katoch, V. D. Sharma, M. A. Patil, K. Katoch, V. P. Bharadwaj, R. K. Sharma, A. S. Bhatia, and B. M. Agrawal. 1996. Determination of mycobacterial phylogeny on the basis of immunological relatedness of superoxide dismutases. Int. J. Syst. Bacteriol. 46:1164-1169. [DOI] [PubMed] [Google Scholar]
- 21.Soini, H., E. C. Böttger, and M. K. Viljanen. 1994. Identification of mycobacteria by PCR-based sequence determination of the 32-kilodalton protein gene. J. Clin. Microbiol. 32:2944-2947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Springer, B., L. Stockman, K. Teschner, G. D. Roberts, and E. C. Böttger. 1996. Two-laboratory collaborative study on identification of mycobacteria: molecular versus phenotypic methods. J. Clin. Microbiol. 34:296-303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Stahl, D. A., and J. W. Urbance. 1990. The division between fast- and slow-growing species corresponds to natural relationships among the mycobacteria. J. Bacteriol. 172:116-124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Takewaki, S., K. Okuzumi, I. Manabe, M. Tanimura, K. Miyamura, K. Nakahara, Y. Yazaki, A. Ohkubo, and R. Nagai. 1994. Nucleotide sequence comparison of the mycobacterial dnaJ gene and PCR-restriction fragment length polymorphism analysis for identification of mycobacterial species. Int. J. Syst. Bacteriol. 44:159-166. [DOI] [PubMed] [Google Scholar]
- 25.van Soolingen, D., T. Hoogenboezem, P. E. de Haas, P. W. Hermans, M. A. Koedam, K. S. Teppema, P. J. Brennan, G. S. Besra, F. Portaels, J. Top, L. M. Schouls, and J. D. van Embden. 1997. A novel pathogenic taxon of the Mycobacterium tuberculosis complex, Canetti: characterization of an exceptional isolate from Africa. Int. J. Syst. Bacteriol. 47:1236-1245. [DOI] [PubMed] [Google Scholar]
- 26.Wayne, L. G., R. C. Good, E. C. Böttger, R. Butler, M. Dorsch, T. Ezaki, W. Gross, V. Jonas, J. Kilburn, P. Kirschner, M. I. Krichevsky, M. Ridell, T. M. Shinnick, B. Springer, E. Stackebrandt, I. Tarnok, H. Tasaka, V. Vincent, N. G. Warren, C. A. Knott, and R. Johnson. 1996. Semantide- and chemotaxonomy-based analysis of some problematic phenotypic clusters of slowly growing mycobacteria, a cooperative study of the International Working Group on Mycobacterial Taxonomy. Int. J. Syst. Bacteriol. 46:280-297. [DOI] [PubMed] [Google Scholar]
- 27.White, S. D., P. J. Ihrke, A. A. Stannard, C. Cadmus, C. Griffin, S. A. Kruth, E. J. Rosser, Jr., S. I. Reinke, and S. Jang. 1983. Cutaneous atypical mycobacteriosis in cats. J. Am. Vet. Med. Assoc. 182:1218-1222. [PubMed] [Google Scholar]