Abstract
Background
Cyanobacteria have diversified through their long evolutionary history and occupy a wide range of environments on Earth. To advance our understanding of their adaptation mechanisms in extreme environments, we performed stress tolerance characterizations, whole genome sequencing, and comparative genomic analyses of a novel heat-tolerant and euryhaline strain of the unicellular cyanobacterium Cyanobacterium sp. Dongsha4 (DS4). This strain was isolated from a lagoon on Dongsha Island in the South China Sea, a habitat with fluctuations in temperature, salinity, light intensity, and nutrient supply.
Results
DS4 cells can tolerate long-term high-temperature up to 50 ℃ and salinity from 0 to 6.6%, which is similar to the results previously obtained for Cyanobacterium aponinum. In contrast, most mesophilic cyanobacteria cannot survive under these extreme conditions. Based on the 16S rRNA gene phylogeny, DS4 is most closely related to Cyanobacterium sp. NBRC102756 isolated from Iwojima Island, Japan, and Cyanobacterium sp. MCCB114 isolated from Vypeen Island, India. For comparison with strains that have genomic information available, DS4 is most similar to Cyanobacterium aponinum strain PCC10605 (PCC10605), sharing 81.7% of the genomic segments and 92.9% average nucleotide identity (ANI). Gene content comparisons identified multiple distinct features of DS4. Unlike related strains, DS4 possesses the genes necessary for nitrogen fixation. Other notable genes include those involved in photosynthesis, central metabolisms, cyanobacterial starch metabolisms, stress tolerances, and biosynthesis of novel secondary metabolites.
Conclusions
These findings promote our understanding of the physiology, ecology, evolution, and stress tolerance mechanisms of cyanobacteria. The information is valuable for future functional studies and biotechnology applications of heat-tolerant and euryhaline marine cyanobacteria.
Supplementary Information
The online version contains supplementary material available at 10.1186/s12866-025-03993-7.
Keywords: Cyanobacterium, Thermotolerance, Genome, Comparative genomics
Background
Cyanobacteria are great contributors to photosynthesis, biological nitrogen fixation, and the increase of oxygen in the modern atmosphere [1–3]. During their evolutionary history, cyanobacteria of different lineages have occupied and adapted to a wide range of environments on Earth, including hot desert soil crusts, nonacidic hot springs, and Arctic/Antarctic areas [1, 4]. Some of these cyanobacteria can survive stressful conditions related to light intensity, salinity, temperature, and water availability. However, their adaptation mechanisms to diverse extreme environments are still not fully understood [5]. These cyanobacteria and their natural bioproducts, including carotenoids, phycobilins and lantipeptides (lantibiotics), have been utilized in numerous biotechnology applications [6, 7]. Additionally, saltwater cyanobacteria hold significant potential for biotechnological and bioremediation applications, especially as freshwater resources become increasingly scarce due to the impacts of climate change [6].
Dongsha Atoll is a coral reef atoll located in the South China Sea [8]. Dongsha Island, situated on the western part, is the only above-water part of the atoll during high tide. This island has a small lagoon (at 20° 70’N and 116° 80’E) with an average depth of about one meter. The surface seawater temperature around Dongsha Atoll in a year is between 22 and 32 °C [9]. However, due to the effects of strong sunlight and poor water exchange, the seawater temperature in this lagoon is higher than 35 °C constantly during daytime and can rise over 38 °C in summer [9]. This unusually high seawater temperature makes this location a ground of natural selection for thermotolerance [10]. A new species of thermotolerant microalga, Tetraselmis sp. DS3, was isolated and characterized recently from this location [10]. The DS3 cells could grow at 40 °C, the highest temperature reported for marine microalgal growth. In addition, this small lagoon is subject to seasonal changes and daily fluctuations in temperature, salinity, light and nutrient supply. Therefore, a thermotolerant cyanobacterium isolated from this site may be also tolerant to other kinds of abiotic stresses. The new isolate might be an attractive biological system for cellular and biochemical study of these tolerance mechanisms, as well as for potential biotechnology applications.
In this work, we report the isolation and stress tolerance properties of a novel heat-tolerant, euryhaline, unicellular cyanobacterial strain Dongsha4 (DS4) from the lagoon of Dongsha Island. Phylogeny analysis based on 16 S rRNA genes showed that DS4 is likely a member of the genus Cyanobacterium [11, 12]. This new isolate could grow in media containing 3% salt at 50 °C. This stress tolerance is unusual among the reported mesophilic cyanobacteria but shared with the closely related thermotolerant species Cyanobacterium aponinum. To investigate the distinctive genetic features and underlying adaptation mechanisms of this new strain, we sequenced the whole genome and conducted comparative genome analysis of DS4 with related cyanobacteria that have genome sequence information available.
Methods
Isolation of DS4 from Dongsha lagoon
Seawater samples were collected from the small lagoon of Dongsha Atoll (20°70’N, 116°72’E) in the South China Sea. Cells in the samples were collected using centrifugation at 3000 x g for 10 min at room temperature, and spread onto 1.5% agar plates. These plates were prepared with modified f medium in which the levels of nitrate and phosphate were changed to 3.5 mM NaNO3 and 0.15 mM NaH2PO4, respectively, and Na2SiO3 was dropped [13]. These plates were incubated under continuous 80 µmol photons m-2s-1 white light at 28 °C for five days, and then 32 °C under the same illumination conditions to grow single colonies. The DS4 strain was isolated and chosen for this study.
Growth conditions
The DS4 strain was grown axenically at different temperatures, in BG11 or modified BG11 (mBG11, with 3.3% NaCl) liquid medium [14] according to the exact experiments as specified in the Results section. These cultures were bubbled with filtered air (pore size 0.2 μm, Millipore) in an orbital shaker (speed 150 rpm). The light intensity was 30 µmol photons m-2s-1.
Microscopy
The procedures for Scanning Electron Microscopy (SEM) and Transmission Electron Microscopy (TEM) were described previously [15].
Genome sequencing and analysis
Total genomic DNA was isolated from 150 mL of DS4 culture grown for 4–5 days. The DNeasy Plant Maxi Kit (QIAGEN, Germany) was used to extract the DNA that was used for Illumina MiSeq sequencing. To obtain high quality DNA for Oxford Nanopore Technologies (ONT) MinION sequencing, a modified CTAB method, and followed by further purification with DNeasy Blood & Tissue Kit (QIAGEN, Venlo, Netherland) was used as described in Chen et al. [16]. Quality and quantity of the purified genomic DNA samples were assessed by using the NanoDrop 2000 spectrophotometer (ThermoFisher, USA) and 1% agarose gel electrophoresis.
The procedures for genome sequencing and analysis were based on those described in our previous work [17]. All bioinformatics tools were used with the default settings unless stated otherwise. Further details of the methodology are provided below.
The raw reads from both platforms were combined for de novo assembly by using Unicycler v0.4.9b [18]. For validation, the Illumina and ONT raw reads were mapped to the assembly by using BWA v0.7.12 [19] and Minimap2 v2.15 [20], respectively. The results were programmatically checked by using SAMtools v1.2 [21] and manually inspected by using IGV v2.3.57 [22]. The finalized assembly was submitted to the National Center for Biotechnology Information (NCBI) and annotated by using their Prokaryotic Genome Annotation Pipeline (PGAP) v5.3 [23].
For comparative analysis, the 16S rRNA gene of DS4 was used as the query to run BLASTN [24] against the NCBI Genbank 16S rRNA gene sequences and nucleotide collection (nt) databases [25]. Related strains with genome sequences available were identified (Table 1). To quantify genome similarity, FastANI v1.1 [26] was used to calculate the percentage of genomic segments mapped and the average nucleotide identity (ANI) based on these segments for each genome pair; only the chromosomes were included in this analysis and the plasmids were excluded. The homologous gene clusters (HGCs) among these genomes were inferred using OrthoMCL v1.3 [27]. For pairwise genome alignment, the NUCleotide MUMmer (NUCmer) program of the MUMmer package v3.23 [28] was used with the setting “–maxmatch–mincluster 2000.”. Additionally, MAUVE v2015-02-25 [29] was also used for chromosome level alignment.
Table 1.
Genome statistics of representative closely related cyanobacteria. The 16S rRNA gene sequence identity values were calculated based on comparisons with DS4
| Cyanobacterium sp. Dongsha4 | Cyanobacterium aponinum PCC 10605 | Geminocystis sp. NIES-3708 | Geminocystis sp. NIES-3709 | Cyanobacterium sp. HL-69 | Cyanobacterium stanieri PCC 7202 | Planktothrixagardhii NIES-204 | |
|---|---|---|---|---|---|---|---|
| Strain | DS4 | PCC10605 | NIES3708 | NIES3709 | HL69 | PCC7202 | NIES204 |
| Environment | marine lagoon | freshwater thermal spring | freshwater stream | freshwater stream | hypersaline lake | alkaline pond |
freshwater lake |
| GenBank Accession | CP084098-CP084102 | GCF_000317685.1 | GCF_001548105.1 | GCF_001548125.1 | GCA_002813905.1 | GCA_000317665.1 | GCA_003609765.1 |
| 16 S rRNA gene seq. id. (%) | - | 99.46 | 95.07 | 95.54 | 93.78 | 93.50 | 89.82 |
| Annotation Date | 24-Sep-21 | 7-Mar-21 | 15-Dec-21 | 15-Dec-21 | 16-Jan-18 | 17-Jul-13 | 3-Apr-18 |
| Genome Size (bp) | 4,320,404 | 4,176,973 | 4,042,078 | 4,426,059 | 3,296,945 | 3,163,381 | 5,084,134 |
| G + C Content (%) | 35 | 34 | 31 | 32 | 36 | 39 | 39 |
| Coding Density (%) | 83.8 | 80.2 | 83.5 | 81 | 83.4 | 85.9 | 85.8 |
| No. of Protein-coding Genes | 3663 | 3451 | 3495 | 3636 | 2947 | 2837 | 4492 |
| No. of Pseudogenes | 63 | 77 | 67 | 151 | 0 | 0 | 0 |
| No. of rRNA Genes | 9 | 9 | 9 | 9 | 8 | 9 | 11 |
| No. of tRNA Genes | 43 | 43 | 44 | 43 | 43 | 43 | 43 |
For phylogenetic inference, the multiple sequence alignment was conducted using MUSCLE v3.8.31 [30]. The maximum likelihood inference was conducted using PhyML v3.3 [31]. The proportion of invariable sites and the gamma distribution parameter were estimated using the dataset, and the number of substitute rate categories was set to four.
Results and discussion
Stress tolerance properties
The strain DS4 exhibited distinct heat tolerance properties when compared with other mesophilic cyanobacteria. The DS4 cells showed normal photosynthetic growth at 30 °C under both BG11 and mBG11 (with 3.3% NaCl) growth conditions and maintained apparent photosynthetic growth under mBG11 growth conditions up to 50 ℃ (Fig. 1A). In addition, 3 h of 50 °C thermal stress followed by 21 h of 35 °C recovery each day did not slow down the growth rate of the DS4 cells. Moreover, DS4 cells can tolerate high temperature stress at 70 ℃ for 3 h and the mortality rate of the cells was only 7.1 ± 1.8% (n = 3) (Fig. 1 and Supplementary Fig. 1). In comparison, previous studies reported that Cyanobacterium aponinum PCC10605 and other related strains can grow at temperature up to 45–50 ℃ [10, 32]. PCC10605 was isolated from Euganean thermal springs in Italy and is the type strain for C. aponinum [12]. C. aponinum is a thermotolerant cyanobacterial species found in different thermal springs and desert environments [12, 32–37]. Because of their thermotolerant properties, C. aponinum strains have been widely used for scientific research, including biotechnological applications [35–37]. In contrast, the model mesophilic cyanobacterium Synechocystis sp. PCC6803 cannot grow above 43 ℃ [38, 39] and can only tolerate short-term heat stress up to 50 ℃ [38, 39].
Fig. 1.
Stress tolerance properties of DS4 cells. (A) Photosynthetic growth of Cyanobacterium sp. DS4 at different temperatures (30, 45 and 50 °C) and different salinity [in BG11 or mBG11 (with 3.3% NaCl) liquid medium]; (B) Photosynthetic growth of DS4 cells in BG11 liquid medium supplemented with different levels of NaCl at 30 °C. All cultures were bubbled with filtered air in an orbital shaker (speed 150 rpm), except for “low-air mBG11” cultures in panel (A), which were not bubbled with air. The light intensity was 30 µmol photons m− 2s− 1. Data are the mean ± SD (n = 2)
Furthermore, DS4 cells were able to grow at 30 ℃ in mBG11 medium with salt levels from 0 to 6.6% NaCl (Fig. 1B). Thus, DS4 showed euryhaline (halo- and osmotic tolerant) properties. Moreover, the growth of DS4 cells at 50 ℃ required the saline growth medium (e.g., 3.3% NaCl or sea salts) and bubbling with air (Fig. 1A). Salinity therefore seemed to have a positive effect on thermotolerance of DS4. Similar effects of salinity on thermotolerance have been reported in some hot-spring cyanobacteria, including C. aponinum [32, 40]. These stress tolerant properties may help DS4 adapt and survive under harsh environmental conditions, such as fluctuations in temperature, salinity, and other environmental factors, in the small lagoon of Dongsha Island.
Cell morphology
The images of SEM, TEM, and light microscopy showed that the DS4 cells were typically round to ellipsoid in shape, about 2–3 μm long (Fig. 2 and Supplementary Fig. 1). DS4 cells at the stage of binary fission were observed. Especially at 45 ℃ and in BG11 liquid medium, the shapes of many DS4 cells were elongated. This is possibly due to impairments in cell fission (Fig. 2C and G). In addition, the SEM images showed that the surface of DS4 cells was covered with a thick cell wall, presumably an extracellular polysaccharide layer (Fig. 2A-D). The TEM images also showed the irregular thylakoid structures in the DS4 cells. Fimbriae-like protrusions are present on the surface of DS4 (Fig. 2E-H).
Fig. 2.
Scanning electron micrograph (SEM) and transmission electron micrograph (TEM) of Cyanobacterium sp. DS4 cells grown under different temperatures and different salinity (BG11 and mBG11) conditions. All cultures were bubbled with filtered air in an orbital shaker (speed 150 rpm). (A–D) SEM images; (E–H) TEM images. (A) and (E), at 30 °C in the BG11 liquid medium; (B) and (F), at 30 °C in the mBG11 liquid medium; (C) and (G), at 45 °C in the BG11 liquid medium; (D) and (H), at 45 °C in the mBG11 liquid medium
Genome sequence
The whole-genome shotgun sequencing generated 1,865,196*2 pair-end Illumina reads and 43,869 ONT long reads (average length = 28,001 bp, N50 length = 38,678 bp, total length = 1,228,373,655 bp). These reads provided ∼260- and ~ 284-fold coverage based on the Illumina and ONT data set, respectively. The complete genome sequence of DS4 contains one circular chromosome that is 4,250,085 bp in size and has 34.8% G + C content. Additionally, four circular plasmids were found, which are 26,702, 20,643, 13,205, and 9,769 bp in size, respectively. For gene content, the annotation includes 43 tRNA genes, three complete set of 16S–23S-5S rRNA genes, and 3,663 intact protein-coding genes. These genomic properties are similar to those found in other related strains, particularly Cyanobacterium aponinum PCC10605 (Table 1).
Phylogenetic placement
Based on the 16S rRNA gene phylogeny (Fig. 3A), DS4 is most closely related to Cyanobacterium sp. NBRC 102756 (NBRC102756, previously named as MBIC10216 or Synechocystis aquatilis SI-2) isolated from Iwojima Island, Japan (99.9% identity) [11, 41] and to a few strains isolated from Vypeen Island, India (Cyanobacterium sp. MCCB114/115/238) (99.9% identity) [42, 43]. NBRC 102,756 is known to be able to accumulate amylopectin-like polysaccharides in cells, designated as cyanobacterial starch [11]. However, the genome information of NBRC 102756 and MCCB114 strains is unavailable. Among the cyanobacteria with genome sequences available, the DS4 has the highest sequence similarity with Cyanobacterium aponinum PCC 10,605 (99.5% identity).
Fig. 3.
Maximum likelihood phylogeny containing DS4 and related strains. Levels of bootstrap supports were estimated based on 1,000 replicates. (A) 16S rRNA gene phylogeny. Branches with bootstrap support lower than 70% are plotted with grey dashed lines. (B) Core genome phylogeny. All branches received 100% bootstrap support
Comparisons with other strains
Based on the 16S rRNA gene phylogeny (Fig. 3A), we selected the related strains that have genome sequences available for comparative analysis (Table 1). These include three strains classified as Cyanobacterium [44] and two strains classified as Geminocystis [45, 46]. In addition to these closely related strains in the order Chroococcales, Planktothrix agardhii NIES-204 from the order Oscillatoriales was included as the outgroup [47].
For overall genome similarity, C. aponinum PCC10605 is the most closely related strain of the DS4, sharing 81.7% of the genomic segments and 92.9% ANI (Fig. 4). This ANI value is below the 95% threshold suggested for delineation of bacterial species [26], indicating that DS4 may not be a member of C. aponinum. However, to reach a more solid conclusion regarding species assignment, comparisons with additional strains within this genus upon genome sequence availability is needed.
Fig. 4.

Average nucleotide identity (ANI) analysis of DS4 and related cyanobacterial strains with complete genome assemblies available. For the pairwise comparisons, the ANI value (%) and the proportion of genomic segments mapped (%) are indicated in the cells above and below the diagonal, respectively. The genome sizes (Mb) are indicated in the cells along the diagonal
The genome alignment between DS4 and PCC10605 revealed no notable synteny conservation (Fig. 5). Such lack of synteny conservation in chromosome organization has been reported previously for other cyanobacteria [15, 48, 49].
Fig. 5.
Genome alignments between DS4 and PCC10605. (A) Alignment inferred by MUMmer. Matched regions with the same and opposite orientations are indicated by red and blue dots, respectively. (B) Alignment inferred by MAUVE. The strain DS4 is used as the reference. Matched regions are indicated by the same color in both assemblies and linked by a line connecting the two regions. In PCC10605, matched regions with the same and opposition orientations with DS4 are plotted above and below the black line in the center, respectively
For gene content comparison, 1,610 homologous gene clusters (HGCs) were shared by all of the strains compared. Among these HGCs, 1,525 were conserved as single-copy genes in all strains. The concatenated alignment of these 1,525 HGCs contained 518,506 aligned amino acid sites and was used to infer a maximum-likelihood phylogeny that received 100% bootstrap support in branches (Fig. 3B). Consistent with the results from the 16S rRNA gene phylogeny and the ANI analysis, the DS4 is most closely related to PCC10605. The two Geminocystis strains NIES-3708 and NIES-3709 collected in Japan form the sister group, while the other two Cyanobacterium strains PCC7202 and HL-69 collected in the USA appeared to be more distantly related. In addition to the phylogenetic divergence, the two Cyanobacterium strains collected in the USA both have much smaller genome sizes (Fig. 4). These findings, together with the 16S rRNA gene phylogeny, indicated that the genus Cyanobacterium is a polyphyletic group that contains diverse lineages, and future re-classification of strains or taxonomic revisions is required to avoid confusion [50].
Overview of the notable genetic differentiations among DS4 and other strains
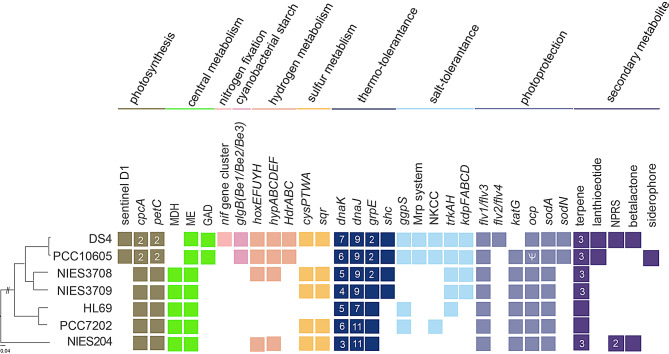
Notable genetic differentiations among the strains compared are summarized in Fig. 6. The strain DS4 has many unique genomic features which are different from those in the genome of PCC10605 and other closely related strains, including gene clusters involved in nitrogen fixation, photo-protection, and novel natural product biosynthesis. In addition, DS4 and PCC10605 shared many distinctive genomic features, including many distinct genes/gene clusters involved in cyanobacterial starch metabolism, central metabolism, hydrogen metabolism, thermotolerance, salt tolerance as well as lantipeptide/bactericide biosynthesis and export systems. More detailed information regarding the inferred function, distribution patterns, and molecular evolution of these genes are provided in the following sections.
Fig. 6.
Overview of notable gene content. Gene presence and absence are indicated by colored and white background, respectively. Multi-copy genes are indicated by a dark-colored background with the copy numbers labeled
Nitrogen fixation
One of major distinct features in the DS4 genome is the presence of a large gene cluster for nitrogen fixation (Dongsha4_16200–16345) (Fig. 6). This nitrogen-fixing (nif) gene cluster contained 29 predicted genes which included nifBSUHDKEN encoded for a Mo dependent nitrogenase system. This nif gene cluster was not found in PCC10605 and several other closely related strains (Fig. 6). The amino acid sequence of NifH and NifDK in DS4 are identical to those in Cyanobacterium. sp. NBRC10276 and show about 90% amino acid sequence identity to NifH and NifDK genes in baeocystous cyanobacteria, such as Xenococcus sp. PCC7305 and Chroococcidiopsis sp. PCC6712 (Supplementary Fig. 3) [51].
Photosynthesis
The major structural components of photosynthesis genes are generally conserved in the DS4 genome. DS4 has phycobilisome genes for the allophycocyanin core and phycocyanin subunits, but not for phycoerythrin subunits. One of the distinctive features in DS4 (also in PCC10605) is the presence of the atypical psbA gene (Dongsha4_01220) (Fig. 6) which encoded a predicted sentinel D1 protein with several mutations on essential manganese ligands to the oxygen-evolution complex (Supplementary Fig. 2) [52, 53]. The expression of sentinel D1 protein during the dark period may protect and enable nitrogen fixation and hydrogenation reactions which are highly oxygen sensitive processes. In addition, DS4 and PCC10605 have two copies of cytochrome b6-F complex Rieske iron sulfur protein (petC) genes, whereas most cyanobacterial genomes have only one copy.
Central carbon metabolism
The cyanobacterial tricarboxylic acid (TCA) cycle does not use the typical α-ketoglutarate dehydrogenase but rather uses α-ketoglutarate decarboxylase (KGD), which sequentially converts α-ketoglutarate into succinic semialdehyde and succinate [54]. DS4 and PCC10605 have one distinct gene for glutamate decarboxylase (GAD) which is absent in most cyanobacterial genomes (Fig. 6). The GAD enzyme converts glutamate to γ-aminobutyric acid (GABA) [55]. Thus, DS4 and PCC10605 may be able to use the GABA shunt pathway to complete the TCA cycle independent of the KGD bypass (Fig. 7). In addition, DS4 and PCC10605 genomes do not have the gene for malate dehydrogenase (MDH), which catalyzes the NAD/NADH-dependent interconversion of malate and oxaloacetate. A recent study reported that malic enzyme (ME), not MDH, is mainly responsible for the oxidization of malate from the TCA cycle in cyanobacteria (Fig. 7) [56]. Thus, MDH may be dispensable and may have been lost during the evolution of DS4 and PCC10605 genomes. It would be interesting for future research to determine whether the GABA shunt and the absence of MDH may contribute to the stress tolerance mechanisms of DS4 and PCC10605.
Fig. 7.
Schematic model of the malic enzyme (ME)-dependent tricarboxylic acid (TCA) cycle and the GABA shunt in DS4 and PCC10605. GAD, glutamate decarboxylase; GDHA, glutamate dehydrogenase; GABA-AT, GABA transaminase. This scheme model is modified from Katayama et al., 2022
Cyanobacterial starch metabolism
While most cyanobacteria produce soluble glycogen, several species of unicellular nitrogen-fixing cyanobacteria, such as NBRC102756 and Cyanothece sp. ATCC51142, accumulated insoluble semi-amylopectin as the storage polysaccharides [11, 57]. DS4 has gene homologs for all three branching-enzyme (BE1, BE2, and BE3: Dongsha4_03540, 03220 and 06100) (Fig. 6) found in NBRC102756 and ATCC 51,142 which are required for biosynthesis of semi-amylopectin [57]. By contrast, most cyanobacteria, such as PCC 6803 and PCC7942, produced soluble glycogen as the major storage polysaccharides and contained only one branching enzyme gene (BE1) in their genomes [58].
Hydrogen metabolism
DS4 has a gene cluster for homologs of bidirectional NiFe hydrogenase complex (hoxEFUYH) [Dongsha4_03740 − 03725, 03710] and genes encoding for homologs of Hyp proteins (hypABCDEF) (Fig. 6) involved in the biosynthesis/maturation of these hydrogenases [59]. This NiFe hydrogenase complex may utilize the hydrogen production from nitrogen fixation processes to reduce the soluble electron carriers NADP or ferredoxin.
In addition, DS4 has a distinct gene cluster for homologs of heterodisulfide reductase complex (hdrABC) which is consisted of 5 heterodisulfide reductase genes arranged as hdrB-hdrC-hdrA-hdrB-hdrC (Dongsha4_08945–08965) (Fig. 6). HdrABC plays important functions in the energy metabolism of methanogenic archaea and were found in genomes of few species of cyanobacteria [60]. In most methanogenic archaea, HdrABC formed a complex with methyl-viologen reducing Ni/Fe hydrogenase (MvhAGD) and catalyzed reduction of the mixed disulfide (CoM–S–S–CoB) of the two coenzymes (M and B) and ferredoxin by H2 for methane formation [60]. The future study is required to identify reactions and substrates that were catalyzed by HdrABC in DS4.
Sulfur metabolism
The DS4 genome has a distinct gene cluster for sulfate ABC transport system (cysPTWA, Dongsha4_11405–11425) for sulfate uptake which was absent in PCC10605 (Fig. 6). In addition, DS4 and PCC10605 have the full set of genes (adenyl sulfate transferase, adenyl sulfate kinase, phospho-adenosine phosphor-sulfate reductase and sulfite reductase) involved in assimilatory sulfate reduction [61]. Furthermore, DS4 encodes a putative sulfide quinone oxidoreductase (SQR, Dongsha4_00975) which may oxidize sulfide to sulfur for anoxygenic photosynthesis or for detoxification of exogenous sulfide under low-O2 conditions [62].
Thermotolerance genes
The DS4 genome has high number of chaperone genes which may confer their thermos-tolerant properties (Fig. 7). For examples, DnaK interacts with its co-chaperones DnaJ and GrpE, and assist folding of denatured proteins under heat and other stress conditions [63]. DS4 and PCC10605 genomes have 7 and 6 copies of chaperone protein DnaK (Hsp70) genes, respectively, versus 3 copies in PCC6803. In addition, DS4 and PCC10605 have 9 copies of chaperone protein DnaJ (Hsp40) genes versus 4 copies in the PCC6803 genome. DS4 and PCC10605 also have 2 copies of chaperone protein GrpE genes versus a single copy in the PCC6803 genome.
In addition, the DS4 genome has the squalene-hopene cyclase (SHC) gene (Dongsha4_04150). The products of SHC, the hopanoids, condense lipid and provide stability of cell membranes under high temperatures and extreme acidity environments due to the rigid ring structures [64].
Salt and osmotic stress tolerance genes
DS4 exhibits euryhaline (high-salt and osmotic stress tolerant) properties (Fig. 1). The basic mechanism of salt acclimation in most cyanobacteria involves the accumulation of compatible solutes and the active extrusion of toxic inorganic ions [65, 66]. DS4 has the glucosylglycerol-phosphate synthase gene (ggpS, Dongsha4_01860) for the biosynthesis of the osmolyte glucosylglycerol as well as genes for the metabolism of glycerol-3-phosphate, a precursor of glucosylglycerol [glycerol-3-phosphate dehydrogenase (glpD, Dongsha4_00335)]. The accumulation of glucosylglycerol inside cells enabled cyanobacteria to tolerate salinities twice as high as that of seawater (about 6.6% NaCl) [65, 66].
Na+ is the main inorganic cation in oceans and in most other saline environments. DS4 has genes for NhaS3 type (Dongsha4_00020, 07485) and NhaP type (Dongsha4_05870, 01470) sodium/proton (or cation/proton) antiporters [67]. In addition, DS4 has genes (Dongsha4_01185 − 01165, 06870, 06875) for the multi-subunit cation/proton antiporter (Mrp system, composed of ABCDEFG subunits), which may function in osmotic stress tolerance under low salt conditions [68].
Potassium ion (K+) transport is also critical for acclimation of cyanobacterial cells to salt and osmotic stresses [67, 69]. DS4 has a distinct gene cluster (kdpDFABC) for the high affinity Kdp potassium uptake system (Dongsha4_15640–15655 and 15670) (Fig. 6) [69]. In addition, DS4 has the other type of potassium ion transporter system (TrkAH, Dongsha4_03140, 03145, 06635).
Furthermore, DS4 has a predicted gene for the Na-K-Cl cotransporter (NKCC) (Dongsha4_11575) (Fig. 6) which may catalyze the coordinated symport of chloride with sodium and potassium ions across the cell membrane in an electrically neutral manner [70].
Photoprotection
DS4 has four flavodiiron protein homologs (Flv1-4, Dongsha4_14815, 05170, 09665 and 05180) which play important photo-protective roles in photosynthesis of cyanobacteria [71]. Previous studies in PCC6803 showed that Flv1/Flv3 mediated an oxygen-dependent alternative electron flow under high CO2 condition, whereas Flv2/Flv4 was induced and mediated O2 photoreduction under CO2-limited conditions [71, 72]. Of note, Flv2/Flv4 was absent in the other closely related species (Fig. 6). By contrast, these closely related species all contained the catalase/peroxidase HPI gene which might substitute for the function of Flv2/Flv4.
DS4 also has a distinct gene operon (Dongsha4_07235, 07230) encoding for orange carotenoid binding protein (OCP) and fluorescence recovery protein (FRP) which involved in blue-green light induced photo-protection mechanism of cyanobacteria, known as non-photochemical fluorescence quenching [73]. Interestingly, the OCP gene was truncated in PCC10605 (Fig. 6).
Superoxide dismutases (SODs) remove superoxide free radicals produced from photosynthetic and respiratory electron transport chains in cyanobacteria. DS4 and PCC10605 have both iron-type (sodA) and nickel-type (sodN) superoxide dismutase genes (Dongsha4_10025 and 18425; CYAN10605_RS10260 and 11100) to detoxify harmful superoxide molecules generated in photosynthetic cells. By contrast, PCC7202, HL92 and NIES-3708 have iron-type (sodA) only (Fig. 6). Most cyanobacterial strains with sodN live in saltwater habitats and may have the advantage to grow under iron-limited marine environments [74].
Novel secondary metabolite genes
Marine cyanobacteria are important resources for pharmaceutically important natural products [75]. We identified the presence of secondary metabolite genes/gene clusters, including nonribosomal peptide (NRP), lantipeptide/bactericide, beta-lactone, and terpenes, in the DS4 genome by using antiSMASH v7.0.0 (see Fig. 6).
DS4 has one unique nonribosomal peptide synthetase (NRPS) gene cluster (Dongsha4_00260–00280) which are absent in PCC10605 and related strains (Fig. 6). This gene cluster is consisted of 2 amino acid adenylation domain-containing protein genes, as well as genes for phosphopantetheine-binding protein, AMP-binding protein and one hypothetical protein.
In addition, DS4 and PCC10605 genomes have a novel lantipeptide biosynthesis gene cluster (Dongsha4_03920–03950) (Fig. 8) [76]. This gene cluster is consisted of six copies of putative precursor peptide genes and one lantipeptide synthetase gene lanM (Supplementary Fig. 4). Precursor peptides contain an N-terminal leader sequence and a C-terminal core peptide. Typical precursor peptides of lantipeptides in cyanobacteria and bacteria are rich in Cys residues which are involved in cyclization of lantipeptides. However, the precursor peptides of DS4 and PCC10605 are rich in Ser/Thr residues and do not contain any Cys residue (Fig. 8). Thus, the products of gene clusters in DS4 and PCC10605 are expected to produce a novel type of linear D-amino acid containing lantiptides [77].
Fig. 8.
Type 2 lantipeptide synthetase and precursors. (A) Synteny plots of lantipeptide biosynthesis gene clusters. Chromosomal locations are labelled below strain name abreviations on the left. (B) Comparison of amino acid sequences of lantipeptide precursors in DS4 (Donsha4_03920–03950) with PCC10605. Precursor peptides contain an N-terminal leader sequence and a C-terminal core peptide. Two conserved glycine residues are labeled with red stars
Under iron-limiting conditions, many cyanobacteria secrete iron-chelating compounds (siderophores) to sequester the iron essential for growth from the environment [78]. One of the distinct differences between the DS4 and PCC10605 genomes is that PCC10605 has a unique NRPS-independent siderophore synthetase gene cluster that is involved in biosynthesis of the IucA/IucC family siderophores (Fig. 6) [78]. In addition, siderophore uptake across the outer membrane in cyanobacteria is usually transported by utilizing TonB-dependent receptors (TBDRs) in the outer membrane [79]. Of note, the PCC10605 genome has 10 TBDR genes, with none found in the DS4 genome. Furthermore, PCC10605 has a unique gene cluster for the ABC-type Fe3+-siderophore (the hydroxamate type) transport system. Thus, DS4 and PCC 10,605 have different types of mechanisms for iron uptake.
Conclusions
In this work, we reported distinctive genetic features and stress tolerance mechanisms of a thermotolerant and euryhaline strain of unicellular diazotrophic cyanobacterium DS4 isolated from a high-temperature lagoon at Dongsha Island. Unlike related model strains, DS4 has the capacity for nitrogen fixation, as demonstrated by comparative genomics. Additionally, DS4 possesses several distinct genetic features related to special metabolisms, stress tolerant mechanisms, and novel nature product biosynthesis. These findings deepen our understanding of the physiology, ecology, evolution, and stress tolerance mechanisms of cyanobacteria and provide valuable genomic resources for future functional studies and biotechnology applications of heat-tolerant and euryhaline marine cyanobacteria.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgements
The Illumina and Oxford Nanopore sequencing library preparation services were provided by the Genomic Technology Core (Institute of Plant and Microbial Biology, Academia Sinica). We thank Dr. Wen-Dar Lin in the Bioinformatics Core and Dr. Wann-Neng Jane in the Cell Biology Core (Institute of Plant and Microbial Biology, Academia Sinica) for technical assistance.
Abbreviations
- ANI
Average nucleotide identity
- BE
Branching-enzyme
- DS4
Cyanobacterium sp. Dongsha4
- FAAL
Fatty acyl-AMP ligase
- Flv
Flavodiiron protein
- FRP
Fluorescence recovery protein
- GABA
γ-aminobutyric acid
- GAD
Glutamate decarboxylase
- HdrABC
Heterodisulfide reductase complex
- KGD
α-ketoglutarate dehydrogenase
- MDH
Malate dehydrogenase
- mBG11
Modified BG11
- ME
Malic enzyme
- NKCC
Na-K-Cl cotransporter
- NRPS
Nonribosomal peptide
- OCP
Orange carotenoid binding protein
- PCC10605
Cyanobacterium aponinum strain PCC10605
- SEM
Scanning Electron Microscopy
- SHC
Squalene-hopene cyclase
- SOD
Superoxide dismutase
- SQR
Sulfide quinone oxidoreductase
- TBDR
TonB-dependent receptor
- TCA cycle
Tricarboxylic acid cycle
- TEM
Transmission Electron Microscopy
Author contributions
C.-N.C., C.-H.K., and H.-A.C. performed conceptualization, funding acquisition, project administration, and supervision. T.C.Y., K.-M.L., Y.-C.L., H.-Y.C. and Y.-F.C. performed investigation, validation, and visualization. C-NC: provided biological materials. TCY, K-ML, Y-CL, C-NC, C-HK, and H-AC: wrote the original draft. All authors review and editing the manuscript. All authors contributed to the article and approved the submitted version.
Funding
Open access funding provided by Academia Sinica
This work was supported in part by a grant MOST 110-2621-B-110-002-MY2 to C.-N. N. C. from the National Science and Technology Council of Taiwan and by funding from Academia Sinica to C-HK and H-AC.
Data availability
The complete genome sequence of Cyanobacterium sp. DS4 has been deposited in the NCBI GenBank under the accessions CP084098- CP084102. The raw reads have been deposited at the NCBI Sequence Read Archive under the accession code PRJNA765482.
Declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Ching-Nen Nathan Chen, Keng-Min Lin and Yu-Chen Lin contributed equally to this work.
Contributor Information
Chih-Horng Kuo, Email: chk@gate.sinica.edu.tw.
Hsiu-An Chu, Email: chuha@gate.sinica.edu.tw.
References
- 1.Whitton BA. Ecology of cyanobacteria II: their diversity in space and time. Springer Science & Business Media; 2012.
- 2.Sanchez-Baracaldo P, Cardona T. On the origin of oxygenic photosynthesis and Cyanobacteria. New Phytol. 2020;225:1440–6. 10.1111/nph.16249. [DOI] [PubMed] [Google Scholar]
- 3.Sanchez-Baracaldo P, Bianchini G, Wilson JD, Knoll AH. Cyanobacteria and biogeochemical cycles through Earth history. Trends Microbiol. 2022;30:143–57. 10.1016/j.tim.2021.05.008. [DOI] [PubMed] [Google Scholar]
- 4.Pathak J, Singh PR, Sinha RP, Rastogi RP. Evolution and distribution of Cyanobacteria. In: Rastogi RP, editor. Ecophysiology and biochemistry of Cyanobacteria. Singapore: Springer; 2021. pp.–1.
- 5.Cassier-Chauvat C, Blanc-Garin V, Chauvat F, Genetic. Genomics, and responses to stresses in cyanobacteria: biotechnological implications. Genes-Basel. 2021;12. 10.3390/genes12040500. [DOI] [PMC free article] [PubMed]
- 6.Selao TT. Exploring cyanobacterial diversity for sustainable biotechnology. J Exp Bot. 2022;73:3057–71. 10.1093/jxb/erac053. [DOI] [PubMed] [Google Scholar]
- 7.Patel A, Matsakas L, Rova U, Christakopoulos P. A perspective on biotechnological applications of thermophilic microalgae and cyanobacteria. Bioresour Technol. 2019;278:424–34. 10.1016/j.biortech.2019.01.063. [DOI] [PubMed] [Google Scholar]
- 8.Liu SYV, Green J, Briggs D, Hastings R, Jondelius Y, Kensinger S, et al. Dongsha Atoll is an important stepping-stone that promotes regional genetic connectivity in the South China sea. PeerJ. 2021;9:e12063. 10.7717/peerj.12063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dongsha Atoll Research Station. https://dongsha-mr.nsysu.edu.tw/ in the digital data section. https://www.marine.gov.tw/filesys/dlarea/536/file2.pdf. p. 141, and personal communication with the marine station staff.
- 10..10, Tsai HP, Chuang LT, Chen CN. Production of long chain omega-3 fatty acids and carotenoids in tropical areas by a new heat-tolerant microalga Tetraselmis Sp. DS3. Food Chem. 2016;192:682–90. 10.1016/j.foodchem.2015.07.071. [DOI] [PubMed] [Google Scholar]
- 11.Nakamura Y, Takahashi J-i, Sakurai A, Inaba Y, Suzuki E, Nihei S, et al. Some cyanobacteria synthesize semi-amylopectin type α-polyglucans instead of glycogen. Plant Cell Physiol. 2005;46:539–45. 10.1093/pcp/pci045. [DOI] [PubMed] [Google Scholar]
- 12.Moro I, Rascio N, La Rocca N, Di Bella M, Andreoli C. Cyanobacterium aponinum, a new cyanoprokaryote from the microbial mat of Euganean thermal springs (Padua, Italy). Algol Stud. 2007;123:1–15. 10.1127/1864-1318/2007/0123-0001. [Google Scholar]
- 13.Andersen RA. Algal culturing techniques. New York: Elsevier Academic; 2005. p. 507. [Google Scholar]
- 14.Rippka R, Deruelles J, Waterbury JB, Herdman M, Stanier RY. Generic assignments, strain histories and properties of pure cultures of cyanobacteria. J Gen Microbiol. 1979;111:1–61. 10.1099/00221287-111-1-1. [Google Scholar]
- 15.Cheng Y-I, Lin Y-C, Leu J-Y, Kuo C-H, Chu H-A. Comparative analysis reveals distinctive genomic features of Taiwan hot-spring Cyanobacterium Thermosynechococcus Sp. TA-1. Front Microb. 2022;13:932840. 10.3389/fmicb.2022.932840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen AL, Liu CY, Chen CH, Wang JF, Liao YC, Chang CH, et al. Reassessment of QTLs for late blight resistance in the tomato accession L3708 using a restriction site associated DNA (RAD) linkage map and highly aggressive isolates of Phytophthora infestans. PLoS ONE. 2014;9:e96417. 10.1371/journal.pone.0096417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Weng L-W, Lin Y-C, Su C-C, Huang C-T, Cho S-T, Chen A-P, et al. Complete genome sequence of Xylella taiwanensis and comparative analysis of virulence gene content with Xylella fastidiosa. Front Microb. 2021;12:684092. 10.3389/fmicb.2021.684092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wick RR, Judd LM, Gorrie CL, Holt KE. Unicycler: resolving bacterial genome assemblies from short and long sequencing reads. PLoS Comput Biol. 2017;13:e1005595. 10.1371/journal.pcbi.1005595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li H, Durbin R. Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics. 2009;25:1754–60. 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li H. Minimap2: pairwise alignment for nucleotide sequences. Bioinformatics. 2018;34(18):3094–100. 10.1093/bioinformatics/bty191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, et al. The sequence alignment/map format and samtools. Bioinformatics. 2009;25:2078–9. 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Robinson JT, Thorvaldsdóttir H, Winckler W, Guttman M, Lander ES, Getz G, et al. Integrative genomics viewer. Nat Biotechnol. 2011;29:24–6. 10.1038/nbt.1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tatusova T, DiCuccio M, Badretdin A, Chetvernin V, Nawrocki EP, Zaslavsky L, et al. NCBI prokaryotic genome annotation pipeline. Nucleic Acids Res. 2016;44:6614–24. 10.1093/nar/gkw569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Camacho C, Coulouris G, Avagyan V, Ma N, Papadopoulos J, Bealer K, et al. BLAST+: architecture and applications. BMC Bioinf. 2009;10:421. 10.1186/1471-2105-10-421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Benson DA, Cavanaugh M, Clark K, Karsch-Mizrachi I, Ostell J, Pruitt KD, et al. GenBank Nucleic Acids Res. 2018;46:D41–7. 10.1093/nar/gkx1094. [DOI] [PMC free article] [PubMed]
- 26.Jain C, Rodriguez-R LM, Phillippy AM, Konstantinidis KT, Aluru S. High throughput ANI analysis of 90K prokaryotic genomes reveals clear species boundaries. Nat Commun. 2018;9:5114. 10.1038/s41467-018-07641-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li L, Stoeckert CJ Jr., Roos DS. OrthoMCL: identification of ortholog groups for eukaryotic genomes. Genome Res. 2003;13:2178–89. 10.1101/gr.1224503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kurtz S, Phillippy A, Delcher AL, Smoot M, Shumway M, Antonescu C, et al. Versatile and open software for comparing large genomes. Genome Biol. 2004;5:R12. 10.1186/gb-2004-5-2-r12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Darling AC, Mau B, Blattner FR, Perna NT. Mauve: multiple alignment of conserved genomic sequence with rearrangements. Genome Res. 2004;14:1394–403. 10.1101/gr.2289704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–7. 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Guindon S, Gascuel O. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst Biol. 2003;52:696–704. 10.1080/10635150390235520. [DOI] [PubMed] [Google Scholar]
- 32.Adar O, Kaplan-Levy R, Schonhölz M, Ronen B, Banet G. Optimal temperature and salinity for growth of two Cyanobacteria from hammat Gader hot springs. Negev Dead Sea Arava Stud. 2017;9:128–36. https://www.adssc.org/wp-content/uploads/2018/10/Adar9-4.pdf. [Google Scholar]
- 33.Dobson Z, Ahad S, Vanlandingham J, Toporik H, Vaughn N, Vaughn M, et al. The structure of photosystem I from a high-light-tolerant cyanobacteria. Elife. 2021;10:e67518. 10.7554/eLife.67518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schubert MG, Tang T, Goodchild-Michelman I, Ryon KA, Henriksen JR, Chavkin T. Cyanobacteria newly isolated from marine volcanic seeps display rapid sinking and robust, high density growth. Appl Environ Microbiol. 2024;90:e0084124. 10.1128/aem.00841-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Karatay SE, Dönmez G. Microbial oil production from thermophile cyanobacteria for biodiesel production. Appl Energy. 2011;88:3632–5. 10.1016/j.apenergy.2011.04.010. [Google Scholar]
- 36.Winckelmann D, Bleeke F, Bergmann P, Klöck G. Growth of Cyanobacterium aponinum influenced by increasing salt concentrations and temperature. 3 Biotech. 2015;5:253–60. 10.1007/s13205-014-0224-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gris B, Sforza E, Morosinotto T, Bertucco A, La Rocca N. Influence of light and temperature on growth and high-value molecules productivity from Cyanobacterium aponinum. J Appl Phycol. 2017;29:1781–90. 10.1007/s10811-017-1133-3. [Google Scholar]
- 38.Inoue N, Taira Y, Emi T, Yamane Y, Kashino Y, Koike H, et al. Acclimation to the growth temperature and the high-temperature effects on photosystem II and plasma membranes in a mesophilic cyanobacterium, Synechocystis Sp. PCC6803. Plant Cell Physiol. 2001;42:1140–8. 10.1093/pcp/pce147. [DOI] [PubMed] [Google Scholar]
- 39.Tuominen I, Pollari M, Von Wobeser EA, Tyystjärvi E, Ibelings BW, Matthijs HC, et al. Sigma factor SigC is required for heat acclimation of the Cyanobacterium Synechocystis Sp. strain PCC 6803. FEBS Lett. 2008;582:346–50. 10.1016/j.febslet.2007.12.030. [DOI] [PubMed] [Google Scholar]
- 40.Banerjee M, Everroad RC, Castenholz RW. An unusual Cyanobacterium from saline thermal waters with relatives from unexpected habitats. Extremophiles. 2009;13:707–16. 10.1007/s00792-009-0258-y. [DOI] [PubMed] [Google Scholar]
- 41.Zhang K, Kurano N, Miyachi S. Outdoor culture of a Cyanobacterium with a vertical flat-plate photobioreactor: effects on productivity of the reactor orientation, distance setting between the plates, and culture temperature. Appl Microbiol Biotechnol. 1999;52:781–6. 10.1007/s002530051591. [Google Scholar]
- 42.Preetha R, Jayaprakash N, Singh IB, Synechocystis. MCCB 114 and 115 as putative probionts for Penaeus monodon post-larvae. Dis Aquat Org. 2007;74:243–7. 10.3354/dao074243. [DOI] [PubMed]
- 43.Deepa G, Mohandas A. Putative probiotic Cyanobacterium spp from the coastal waters of Cochin polyphasic taxonomy fate of axenic cultures and heterotrophic bacterial associates. In.: Cochin University of Science and Technology; 2019.
- 44.Mobberley J, Romine M, Cole J, Maezato Y, Lindemann S, Nelson W. Draft genome sequence of Cyanobacterium Sp. strain HL-69, isolated from a benthic microbial mat from a magnesium sulfate-dominated hypersaline lake. Genome Announc. 2018;6:e01583–17. 10.1128/genomea.01583-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hirose Y, Katayama M, Ohtsubo Y, Misawa N, Iioka E, Suda W, et al. Complete genome sequence of Cyanobacterium Geminocystis Sp. strain NIES-3708, which performs type II complementary chromatic acclimation. Genome Announc. 2015;3:e00357–15. 10.1128/genomea.00357-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hirose Y, Katayama M, Ohtsubo Y, Misawa N, Iioka E, Suda W, et al. Complete genome sequence of Cyanobacterium Geminocystis Sp. strain NIES-3709, which harbors a phycoerythrin-rich phycobilisome. Genome Announc. 2015;3:e00385–15. 10.1128/genomea.00385-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Shimura Y, Fujisawa T, Hirose Y, Misawa N, Kanesaki Y, Nakamura Y, et al. Complete sequence and structure of the genome of the harmful algal bloom-forming Cyanobacterium Planktothrix agardhii NIES-204T and detailed analysis of secondary metabolite gene clusters. Harmful Algae. 2021;101:101942. 10.1016/j.hal.2020.101942. [DOI] [PubMed] [Google Scholar]
- 48.Cheng Y-I, Chou L, Chiu Y-F, Hsueh H-T, Kuo C-H, Chu H-A. Comparative genomic analysis of a novel strain of Taiwan Hot-Spring Cyanobacterium Thermosynechococcus Sp. CL-1. Front Microb. 2020;11:82. 10.3389/fmicb.2020.00082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Frangeul L, Quillardet P, Castets A-M, Humbert J-F, Matthijs HC, Cortez D, et al. Highly plastic genome of Microcystis aeruginosa PCC 7806, a ubiquitous toxic freshwater Cyanobacterium. BMC Genomics. 2008;9:1–20. 10.1186/1471-2164-9-274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Oren A, Mareš J, Rippka R. Validation of the names Cyanobacterium and Cyanobacterium stanieri, and proposal of cyanobacteriota phyl. Nov. Int J Syst Evol Microbiol. 2022;72:005528. 10.1099/ijsem.0.005528. [DOI] [PubMed] [Google Scholar]
- 51.Jung P, D’Agostino PM, Brust K, Büdel B, Lakatos M. Final destination?? Pinpointing Hyella disjuncta Sp. Nov. PCC 6712 (Cyanobacteria) based on taxonomic aspects, multicellularity, nitrogen fixation and biosynthetic gene clusters. Life. 2021;11:916. 10.3390/life11090916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wegener KM, Nagarajan A, Pakrasi HB. An atypical PsbA gene encodes a Sentinel D1 protein to form a physiologically relevant inactive photosystem II complex in cyanobacteria. J Biol Chem. 2015;290:3764–74. 10.1074/jbc.M114.604124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Murray JW. Sequence variation at the oxygen-evolving centre of photosystem II: a new class of ‘rogue’cyanobacterial D1 proteins. Photosynth Res. 2012;110:177–84. 10.1007/s11120-011-9714-5. [DOI] [PubMed] [Google Scholar]
- 54.Zhang S, Bryant DA. The Tricarboxylic acid cycle in cyanobacteria. Science. 2011;334:1551–3. 10.1126/science.1210858. [DOI] [PubMed] [Google Scholar]
- 55.Xiong W, Brune D, Vermaas WF. The γ-aminobutyric acid shunt contributes to closing the Tricarboxylic acid cycle in Synechocystis Sp. PCC 6803. Mol Microbiol. 2014;93:786–96. 10.1111/mmi.12699. [DOI] [PubMed] [Google Scholar]
- 56.Katayama N, Iwazumi K, Suzuki H, Osanai T, Ito S. Malic enzyme, not malate dehydrogenase, mainly oxidizes malate that originates from the Tricarboxylic acid cycle in Cyanobacteria. Mbio. 2022;13:e02187–22. 10.1128/mbio.02187-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Suzuki R, Koide K, Hayashi M, Suzuki T, Sawada T, Ohdan T, et al. Functional characterization of three (GH13) branching enzymes involved in cyanobacterial starch biosynthesis from Cyanobacterium Sp. NBRC 102756. Biochim biophys acta. Proteins Proteom. 2015;1854:476–84. 10.1016/j.bbapap.2015.02.012. [DOI] [PubMed] [Google Scholar]
- 58.Welkie DG, Lee B-H, Sherman LA. Altering the structure of carbohydrate storage granules in the Cyanobacterium Synechocystis Sp. strain PCC 6803 through branching-enzyme truncations. J Bacteriol. 2016;198:701–10. 10.1128/jb.00830-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bothe H, Schmitz O, Yates MG, Newton WE. Nitrogen fixation and hydrogen metabolism in cyanobacteria. Microbiol Mol Biol Rev. 2010;74:529–51. 10.1128/mmbr.00033-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wagner T, Koch J, Ermler U, Shima S. Methanogenic heterodisulfide reductase (HdrABC-MvhAGD) uses two noncubane [4Fe-4S] clusters for reduction. Science. 2017;357:699–703. 10.1126/science.aan0425. [DOI] [PubMed] [Google Scholar]
- 61.Hamilton TL, Klatt JM, De Beer D, Macalady JL. Cyanobacterial photosynthesis under sulfidic conditions: insights from the isolate Leptolyngbya Sp. strain Hensonii. ISME J. 2018;12:568–84. 10.1038/ismej.2017.193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Liu D, Zhang J, Lü C, Xia Y, Liu H, Jiao N, et al. Synechococcus Sp. strain PCC7002 uses sulfide: Quinone oxidoreductase to detoxify exogenous sulfide and to convert endogenous sulfide to cellular sulfane sulfur. Mbio. 2020;11:e03420–19. 10.1128/mbio.03420-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Suzuki I, Simon WJ, Slabas AR. The heat shock response of Synechocystis Sp. PCC 6803 analysed by transcriptomics and proteomics. J Exp Bot. 2006;57:1573–8. 10.1093/jxb/erj148. [DOI] [PubMed] [Google Scholar]
- 64.Siedenburg G, Jendrossek D. Squalene-hopene cyclases. Appl Environ Microbiol. 2011;77:3905–15. 10.1128/AEM.00300-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hagemann M. Molecular biology of cyanobacterial salt acclimation. FEMS Microbiol Rev. 2011;35:87–123. 10.1111/j.1574-6976.2010.00234.x. [DOI] [PubMed] [Google Scholar]
- 66.Kirsch F, Klähn S, Hagemann M. Salt-regulated accumulation of the compatible solutes sucrose and glucosylglycerol in cyanobacteria and its biotechnological potential. Front Microb. 2019;10:2139. 10.3389/fmicb.2019.02139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Klähn S, Mikkat S, Riediger M, Georg J, Hess WR, Hagemann M. Integrative analysis of the salt stress response in cyanobacteria. Biol Direct. 2021;16:26. 10.1186/s13062-021-00316-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Fukaya F, Promden W, Hibino T, Tanaka Y, Nakamura T, Takabe T. An Mrp-like cluster in the halotolerant Cyanobacterium Aphanothece halophytica functions as a Na+/H + antiporter. Appl Environ Microbiol. 2009;75:6626–9. 10.1128/AEM.01387-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Nanatani K, Shijuku T, Takano Y, Zulkifli L, Yamazaki T, Tominaga A, et al. Comparative analysis of Kdp and Ktr mutants reveals distinct roles of the potassium transporters in the model Cyanobacterium Synechocystis Sp. strain PCC 6803. J Bacteriol. 2015;197:676–87. 10.1128/jb.02276-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Park J, Saier J. Phylogenetic, structural and functional characteristics of the Na-K-Cl cotransporter family. J Membr Biol. 1996;149:161–8. 10.1007/s002329900016. [DOI] [PubMed] [Google Scholar]
- 71.Santana-Sanchez A, Solymosi D, Mustila H, Bersanini L, Aro E-M, Allahverdiyeva Y. Flavodiiron proteins 1–to-4 function in versatile combinations in O2 photoreduction in cyanobacteria. Elife. 2019;8:e45766. 10.7554/eLife.45766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Shimakawa G, Shaku K, Nishi A, Hayashi R, Yamamoto H, Sakamoto K, et al. FLAVODIIRON2 and FLAVODIIRON4 proteins mediate an oxygen-dependent alternative electron flow in Synechocystis Sp. PCC 6803 under CO2-limited conditions. Plant Physiol. 2015;167:472–80. 10.1104/pp.114.249987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Kirilovsky D, Kerfeld CA. Cyanobacterial photoprotection by the orange carotenoid protein. Nat Plants. 2016;2:1–7. 10.1038/nplants.2016.180. [DOI] [PubMed] [Google Scholar]
- 74.Boden JS, Konhauser KO, Robbins LJ, Sánchez-Baracaldo P. Timing the evolution of antioxidant enzymes in cyanobacteria. Nat Commun. 2021;12:4742. 10.1038/s41467-021-24396-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Shah SAA, Akhter N, Auckloo BN, Khan I, Lu Y, Wang K, et al. Structural diversity, biological properties and applications of natural products from cyanobacteria. A review. Mar Drugs. 2017;15:354. 10.3390/md15110354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Haft DH, Basu MK, Mitchell DA. Expansion of ribosomally produced natural products: a nitrile hydratase-and Nif11-related precursor family. BMC Biol. 2010;8:1–15. 10.1186/1741-7007-8-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Yang X, Van Der Donk WA. Post-translational introduction of D-alanine into ribosomally synthesized peptides by the dehydroalanine reductase NpnJ. J Am Chem Soc. 2015;137:12426–9. 10.1021/jacs.5b05207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Årstøl E, Hohmann-Marriott MF. Cyanobacterial siderophores—physiology, structure, biosynthesis, and applications. Mar Drugs. 2019;17:281. 10.3390/md17050281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Krewulak KD, Vogel HJ. TonB or not TonB: is that the question? Biochem Cell Biol. 2011;89:87–97. 10.1139/O10-141. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The complete genome sequence of Cyanobacterium sp. DS4 has been deposited in the NCBI GenBank under the accessions CP084098- CP084102. The raw reads have been deposited at the NCBI Sequence Read Archive under the accession code PRJNA765482.