Abstract
PCR assays have proved useful for detecting and characterizing Shiga toxin-producing Escherichia coli (STEC). Recent advances in PCR technology have facilitated the development of real-time fluorescence PCR assays with greatly reduced amplification times and improved methods for the detection of amplified target sequences. We developed and evaluated two such assays for the LightCycler instrument: one that simultaneously detects the genes for Shiga toxins 1 and 2 (stx1 and stx2) and another that simultaneously detects the genes for intimin (eae) and enterohemolysin (E-hly). Amplification and sequence-specific detection of the two target genes were completed within 60 min. Findings from the testing of 431 STEC isolates of human and animal origin, 73 isolates of E. coli negative for stx genes, and 118 isolates of other bacterial species with the LightCycler PCR (LC-PCR) assays were compared with those obtained by conventional block cycler PCR analysis. The sensitivities and specificities of the LC-PCR assays were each 100% for the stx1, eae, and E-hly genes and 96 and 100%, respectively, for the stx2 gene. No stx2 genes were detected from 10 stx2f-positive isolates because of significant nucleotide differences in their primer annealing regions. Melting curve analyses of the amplified Shiga toxin genes revealed sequence variation within each of the tested genes that correlated with described and novel gene variants. The performance characteristics of the LC-PCR assays, such as their speed, detection method, and the potential subtyping information available from melting curve analyses, make them attractive alternatives to block cycler PCR assays for detecting and characterizing STEC strains.
Shiga toxin-producing Escherichia coli (STEC) strains, also referred to as verocytotoxin-producing E. coli strains, are an important cause of food-borne disease worldwide. STEC strains produce one or more potent cytotoxins and are capable of causing watery diarrhea, bloody diarrhea, hemolytic uremic syndrome (HUS), thrombotic thrombocytopenic purpura, and death (6, 18, 19, 33, 38, 41). STEC strains constitute a serologically and biochemically heterogeneous group of organisms and are most reliably detected by methods that target the toxins they produce or the genes encoding these toxins. Among the methods that target the toxin genes, PCR assays have been used widely to detect small numbers of STEC present in stool specimens, enrichment broths, or primary fecal cultures (2, 10, 12, 16, 25, 27, 28, 31, 36). PCR assays have also been used extensively to characterize STEC strains with respect to other virulence and virulence-associated genes (1, 8, 14, 20, 21, 23, 26, 31). While conventional PCR assays are versatile from the standpoint of being able to detect and identify multiple target genes in one reaction, most first-generation PCR assays have cumbersome procedures for detecting the amplification products and many lack sequence-specific identification of the gene(s) being amplified. The extent of postamplification manipulation of samples generally performed for conventional PCR assays also carries significant risk of contaminating the work environment with amplification products. Recent advances in fluorescence-based, real-time PCR techniques have facilitated the development of assays that offer rapid, simultaneous amplification and sequence-specific detection of two target genes in a single reaction tube. In the present study, we developed and evaluated two duplex real-time fluorescence PCR assays for the LightCycler instrument for the detection and genotypic characterization of STEC.
MATERIALS AND METHODS
Bacterial strains.
E. coli strains included in the study were selected from the culture collections of our institutions and are described in Table 1. We studied 504 E. coli isolates, consisting of 431 STEC and 73 non-STEC, and 118 strains from the following bacterial genera: Treponema (1), Borrelia (3), Leptospira (1), Helicobacter (2), Legionella (5), Neisseria (2), Branhamella (1), Xanthomonas (1), Moraxella (2), Kingella (1), Acinetobacter (1), Sphingomonas (1), Pseudomonas (4), Brucella (2), Bordetella (4), Shigella (4; including Shigella dysenteriae type 1), Salmonella (2), Citrobacter (2), Klebsiella (2), Enterobacter (2), Serratia (2), Proteus (2), Morganella (1), Providencia (1), Yersinia (2), Vibrio (1), Aeromonas (1), Pasteurella (1), Haemophilus (3), Actinobacillus (1), Bacteroides (1), Campylobacter (3), Fusobacterium (1), Eikenella (1), Prevotella (1), Porphyromonas (1), Rickettsia (1), Ehrlichia (1), Coxiella (1), Bartonella (2), Chlamydia (3), Mycoplasma (2), Ureaplasma (1), Micrococcus (1), Staphylococcus (5), Streptococcus (5), Leuconostoc (1), Pediococcus (1), Peptostreptococcus (3), Bacillus (2), Clostridium (3), Lactobacillus (2), Listeria (1), Corynebacterium (5), Gardnerella (1), Propionibacterium (1), Actinomyces (2), Mycobacterium (4), Nocardia (2), Tsukamorella (1), Rhodococcus (1).
TABLE 1.
Characteristics of E. coli and other bacterial strains tested by conventional and LC-PCR
| Genotype | No. of strains | Serotypea | |||
|---|---|---|---|---|---|
| stx1 | stx2 | eae | E-hly | ||
| + | − | + | + | 135 | O26:H11/NM (70), O45:H2 (10), O46:H2 (1), O63:NM (1), O84:NM (1), O91:NM (1), O103:H2/H18/H25/NM (21), O111:H2/H8/NM (14), O125:NM (1), O145:NM (7), O153:H2 (1), O157:H7/NM (2), Orou:H2 (1), Orou:H11 (1), Orou:NM (1), Ound:NM (2) |
| − | + | + | + | 89 | O2:NM (1), O14:NM (1), O26:H11/NM (16), O103:H2/NM (5), O121:H19 (5), O144:NM (3), O145:NM (14), O157:H7/NM (41), O165:H25 (1), Orou:NM (1), Ound:NM (1) |
| + | + | + | + | 87 | O4:NM (1), O26:H11/NM (16), O75:NM (1), O111:H2/H8/NM (20), O145:NM (2), O120:NM (1), O145:NM (2), O157:H7/NM (43), Ound:NM (1) |
| − | + | − | − | 48 | O2:H5 (1), O8:H14/NM (2), O16:H32/H48 (2) O22:H5 (1), O23:NM (1), O28:H25 (1), O30:NM (1), O40:H6/NM (2), O60:H2/NM (4), O73:H17 (2), O79:H7 (1), O77:NM (1), O80:H42 (1), O92:NM (1), O101:H9 (1), O103:NM (1), O113:H21 (1), O121:H19 (1), O128:H45 (2), O140:NM (1), O146:NM (1), O162:NM (1), O171:H2 (1), OX3:H21 (1), Orou:NM (3), Ound:NM (13) |
| − | − | − | − | 159 | O1:H7 (1), O2:NM (1), O4:H5 (1), O6:H1/H10/H16/H31/NM (5), O8:H42 (1), O11:H18 (1), O16:H5/H7 (2), O18:H1/H7 (3), O22:NM (1), O25:H4 (1), O34:H10 (1), O49:NM (1), O50:H7 (1), O62:H30 (1), O63:H20 (1), O64:NM (1), O75:NM (3), O88:H7 (1), O128:H27 (1), O135:H4 (1), O146:H28 (1), O148:H28/NM (4), O167:H5/NM (2), O169:H41 (1), Orou:H16/NM (4), Ound:H32 (1), bacterial species other than E. coli (117) |
| − | − | + | − | 24 | O2ab:H2 (1), O6:H1 (1):O37:NM (1), O53:NM (1), O55:NM (1), O63:NM (2), O86:H34 (1), O88:H25 (1), O101:NM (1), O111:NM (2), O119:H2/H6: (2), O128:H2 (1), O130:NM (1), O132:H34 (1), O142:NM (1), O145:H34 (1), O163:H6 (1), O170:NM (1), Orou:NM (1), Ound:H45/NM (2) |
| + | + | − | + | 24 | O60:H19 (1), O8:NM (1), O22:H8 (1), O62:NM (1), O70:NM (1), O75:H21 (1), O88:25 (1), O96:NM (1), O111:NM (1), O113:NM (2), O128:H2/NM (5), O128:NM (1), O137:H41(1), Ound:H8/NM (2), Orou:H19/NM (2), Ound:NM (2) |
| − | + | − | + | 12 | O2:H8 (1), O5:H21(1), O6:NM (1), O8:NM (1), O26:H19 (1), O38:H21 (1), O91:H21 (1), O104:H21 (2), O110:H28 (1), O113:H21 (1), O112:NM (1) |
| − | + | + | − | 11 | O15:NM (1), O26:NM (1), O55:H7 (1), O63:H6 (1), O66:NM (1), O83:H19 (1), O128:NM (3), O135:NM (1), Ound:NM (1), Ound:NM (1) |
| + | − | − | − | 11 | O75:NM (1), O91:H14/NM (2), O112:NM (2), O117:H7 (1), O121:H7 (2), O146:H2 (1), Ound:H52 (1), S. dysenteriae type 1 (1) |
| − | − | + | + | 7 | O26:H4/H11 (2), O103:NM (1), O143:NM (1), O157:H7/NM (2), Orou:NM (1) |
| + | + | − | − | 6 | O28:H25 (1), O91:NM (1), O128:NM (1), O162:NM (1), Ound:NM (2) |
| + | − | + | − | 6 | O5:NM (1), O14:H11 (1), O26:H11 (1), O111:H8/NM (2), O119:H2 (1) |
| + | − | − | + | 2 | O112:NM (1), Orou:NM (1) |
| + | + | + | − | 1 | O83:H1 (1) |
| Total | 622 | ||||
Ound and Hund, O and H antigen, respectively, not determined with available typing sera; Orou, O antigen not determined because of incomplete or rough lipopolysaccharide; NM, nonmotile. Numbers in parentheses indicate number of strains with that serotype.
The serotype and presence of the genes for Shiga toxins 1 and 2 (stx1 and stx2), enterohemolysin (E-hly), and intimin (eae) for the E. coli isolates are summarized in Table 1. Of the E. coli isolates included, all except 10 were from human sources; the 10 exceptions were positive for the stx2f genes by PCR (35) and were isolated from pigeons. E. coli strains used as controls included ATCC 43895, containing the genes for stx1, stx2, eae, and E-hly; ATCC 43889, containing the genes for stx2, eae, and E-hly; and ATCC 43890, containing the genes for stx1, eae, and E-hly. The following stx-negative diarrheagenic strains were used: H10407 (O78:H11), containing the genes for thermostable and thermolabile enterotoxins; TX1 (O78:H12), containing the gene for thermostable enterotoxin; B170 (O111:NM), containing the genes for eae, bfpA, and the EAF plasmid; EDL1284 (O124:NM), containing the genes for ipaH; and 3591-78 (O75:NM), containing the plasmid involved in mediating the enteroaggregate-type adherence to HEp-2 cells in vitro.
Bacterial isolates were stored frozen at −70°C in tryptic soy broth containing 10% glycerol and were subcultured prior to testing onto either MacConkey agar (Merck, Darmstadt, Germany) or tryptic soy agar containing 5% sheep blood.
Serologic characterization.
Serologic testing for O and H antigens was performed by standard methods using microtitration plates instead of tubes (7).
Preparation of template DNA.
To prepare template DNA from pure bacterial cultures, a 1-cm-long sweep of bacterial growth from the quadrant of the MacConkey or blood agar plate with heaviest growth was suspended in 300 μl of lysis buffer composed of 10 mM Tris-HCl (pH 8.0), 1% Triton X-100, 0.5% Tween 20, and 1 mM EDTA (37). The bacterial suspension was heated at 99.9°C for 10 min and then centrifuged at 1,000 × g for 2 min to sediment the debris. Aliquots (2 to 5 μl) of the clear DNA supernatant were then directly transferred to the PCRs.
Block cycler PCR assays.
Genes associated with STEC were detected by PCR by using previously described methods and the primers listed in Table 2. A 5-μl aliquot of supernatant from the boiled suspension of bacterial cells was used as the source of template DNA in 50-μl reaction mixtures consisting of 10 mM Tris-HCl (pH 8.3), 50 mM KCl, 1.5 mM MgCl2, 0.01% gelatin, a 200 μM concentration of each deoxynucleoside triphosphate, a 0.4 μM concentration of each primer, and 2 U of Taq DNA polymerase. Thermocycling conditions for the individual amplification reactions and the expected size of the amplicons are listed in Table 2.
TABLE 2.
Oligonucleotide primers used in the detection and characterization of STEC by conventional PCR
| Target gene | Primer | Primer sequence (5′-3′)a | Amplification conditionsb | Amplicon size (bp) | Reference |
|---|---|---|---|---|---|
| stxB1 | SLTI-1 | CAg TTA ATg Tgg Tgg CgA Ag | 35 × (95°C, 15 s; 60°C, 45 s; 72°C, 1.5 min) | 475 | 25 |
| SLTI-2 | CAC AgA CTg CgT CAg TgA gg | 25 | |||
| stxB2 | SLTII-1 | CTT Cgg TAT CCT ATT CCC gg | 35 × (95°C, 15 s; 60°C, 45 s; 72°C, 1.5 min) | 863 | 25 |
| SLTII-3 | CgC TgC AgC TgT ATT ACT TTC | 25 | |||
| stxB2/2c | GK3 | ATg AAg AAg ATg TTT ATg | 35 × (94°C, 30 s; 52°C, 1 min; 72°C, 40 s) | 260 | 17 |
| GK4 | TCA gTC ATT ATT AAA CTg | 17 | |||
| stxB2d | VT2-cm | AAg AAg ATA TTT gTA gCg g | 35 × (94°C, 30 s; 52°C, 1 min; 72°C, 1 min) | 256 | 35 |
| VT2-f | TAA ACT gCA CTT CAg CAA AT | 35 | |||
| stxB2e | FK1 | CC ggA TCC AAg AAg ATg TTT ATA g | 35 × (94°C, 30 s; 55°C, 1 min; 72°C, 40 s) | 280 | 9 |
| FK2 | CCC gAA TTC TCA gTT AAA CTT CAC C | 9 | |||
| stxA2f | 128-1 | AgA TTg ggC gTC ATT CAC Tgg TTg | 35 × (94°C, 30 s; 57°C, 1 min; 72°C, 1 min) | 428 | 40 |
| 128-2 | TAC TTT AAT ggC CgC CCT gTC TCC | 40 | |||
| stxA/B2d | SD-a | ATT TAC CAg gCT CgC TTT Tg | 30 × (94°C, 30 s; 53°C, 1 min; 72°C, 1.5 min) | 1,361 | Present study |
| SD-b | ACA TTg CTg CAC ACT ACg | ||||
| stxA/B2e | SE-a | gAg CAg ACg ACA CgA TAA CA | 30 × (94°C, 30 s; 53°C, 1 min; 72°C, 1.5 min) | 1,547 | Present study |
| SE-b | AAT CAg CAT CCA CAA CAC TA | ||||
| eae | C1 | TCg TCA CAg TTg CAg gCC Tgg T | 35 × (94°C, 20 s; 53°C, 45 s; 72°C, 1.5 min) | 1,110 | 21 |
| C2 | CgA AgT CTT ATC CgC CgT AAA gT | 21 | |||
| E-hly | MFS1F | ACg ATg Tgg TTT ATT CTg gA | 35 × (94°C, 20 s; 53°C, 45 s; 72°C, 1.5 min) | 166 | 10 |
| MFSIR | CTT CAC gTC ACC ATA CAT AT | 10 |
g = guanosine. A lowercase letter is used for this nucleotide to avoid confusion with C (cytosine) in the printed table.
Total cycle numbers, temperatures, and periods of denaturation, annealing, and elongation steps are given.
In addition to reactions performed with template DNA from known positive and negative control strains, a reaction mixture containing water instead of template was included in each experiment to detect possible reagent contamination. Amplified fragments were separated by agarose gel electrophoresis and visualized after ethidium bromide staining with UV illumination. PCR was also performed on 5 to 10 individual colonies to isolate STEC from mixed bacterial cultures that had initially tested positive for stx genes.
Subtyping of the stx2 genes was accomplished by restriction fragment analysis of amplicons generated by block cycler PCR restriction fragment length polymorphism assays (PCR-RFLP) or by toxin-specific block cycler PCR assays. To differentiate stx2 from stx2c, PCR-RFLP was performed with primers GK3 and GK4 (17) and amplicons were digested with the restriction endonucleases HaeIII and FokI (Roche Diagnostics, Mannheim, Germany) as described by Rüssmann et al. (39). Toxin-specific PCR assays were used to characterize stx2d, stx2e, and stx2f: stx2d was detected by using primers VT2-cm and VT2-f (35), stx2e was detected by using primers FK1 and FK2 (9), and stx2f was detected by using primers 128-1 and 128-2 (40). To amplify the complete A- and B-subunit genes for sequencing, primers SD-a and SD-b were used for stx2d and primers SE-a and SE-b were used for stx2e.
Oligonucleotide primers and fluorescent-labeled probes for the LightCycler PCR (LC-PCR) assays.
DNA oligonucleotide primers and hybridization probes were synthesized by TIB Molbiol, Berlin, Germany, and by the Biotechnology Core Facility, Centers for Disease Control, Atlanta, Ga. Hybridization probes were labeled with fluorescein, LC Red 640, or LC Red 705. The nucleotide sequences of primers and hybridization probes and their corresponding locations within the representative GenBank sequences are shown in Table 3. Oligonucleotide sequences for the hybridization probes were selected using Oligo (version 6.0; Molecular Biology Insights, Inc., Cascade, Colo.) to have comparable Tm values, moles percent G+C content within the range of 35 to 60%, and minimal secondary structure due to self-complementarity or palindromic sequences.
TABLE 3.
Oligonucleotide primers and LightCycler hybridization probes used in the PCR assay
| Oligo- nucleotide | Sequencea | Target gene(s) | Nucleotide position | GenBank accession no. | Reference |
|---|---|---|---|---|---|
| STEC-1 | gA(Ag) C(Ag)A AAT AAT TTA TAT gTg | stx1 and stx2 | 280-300 | AB015056 | Present study |
| STEC-2 | TgA TgA Tg(Ag) CAA TTC AgT AT | stx1 and stx2 | 800-781 | AB015056 | 28 |
| STEC-I HP-1 | TTT ACg TTT TCg gCA AAT ACA gAg ggg AT-[FL] | stx1 | 570-598 | AB015056 | Present study |
| STEC-I HP-2 | [Red 640]-TCg TAC AAC ACT ggA TgA TCT CAg Tgg g-Ph | stx1 | 601-627 | AB015056 | Present study |
| STEC-II HP-1 | TCA ggC ACT gTC TgA AAC TgC TCC TgT gTA-[FL] | stx2 | 778-807 | Z37725 | Present study |
| STEC-II HP-2 | [Red 705]-ACC ATg ACg CCg ggA gAC gTg gAC CT-Ph | stx2 | 809-834 | Z37725 | Present study |
| eaeAF | gAC CCg gCA CAA gCA TAA gC | eae | 24875-24894 | AF022236 | 31 |
| eaeAR | CCA CCT gCA gCA ACA AgA gg | eae | 25258-25239 | AF022236 | 31 |
| eae-HP-1 | ACA gTT CTg AAA gCg AAA TgA TgA Agg C-[FL] | eae | 25132-25159 | AF022236 | Present study |
| eae-HP-2 | [Red 640]-CCT ggT CAg CAg ATC ATT TTg CCA CT-Ph | eae | 25164-25189 | AF022236 | Present study |
| hlyAF | gCA TCA TCA AgC gTA CgT TCC | E-hly | 70-90 | X94129 | 31 |
| hlyAR | AAT gAg CCA AgC Tgg TTA AgC T | E-hly | 602-581 | X94129 | 31 |
| hlyA HP-1 | gCA Tgg CTC TTg ATg AAT TgC TgA gA-[FL] | E-hly | 434-459 | X94129 | Present study |
| hlyA HP-2 | [Red 705]-CAA Cgg gAA ggA gAg gAT ATA AgT CAg-Ph | E-hly | 464-489 | X94129 | Present study |
[FL], fluorescein; [Red 640], LC Red 640-N-hydroxy-succinimide ester; [Red 705], LC Red 705-phosphoramidite; [Ph], 3′-phosphate. g = guanosine. A lowercase letter is used for this nucleotide to avoid confusion with C (cytosine) in the printed table.
LC-PCR assay and product detection.
All LC-PCR assays were performed using a fluorescence temperature cycler (LightCycler; Roche Diagnostics). The amplification mixture consisted of 2 μl of 10× reaction mix (LightCycler-FastStart Master Hybridization Probes; Roche Diagnostics), 3 mM MgCl2, 0.5 μM concentrations of each oligonucleotide primer, 0.2 μM concentrations of each oligonucleotide probe, and 2 μl of template DNA in a final volume of 20 μl. Samples were amplified with an initial denaturation step at 95°C for 10 min to activate the FastStart Taq DNA polymerase and 50 cycles of denaturation at 95°C for 10 s, annealing at 50°C for 20 s, and an extension at 72°C for 30 s. The temperature transition rate was 20°C/s. The generation of target amplicons for each sample was monitored between the annealing and the elongation steps at 640 and 705 nm. Samples positive for target genes were identified by the instrument at the cycle number where the fluorescence attributable to the target sequences exceeded that measured for background. Those scored as positive by the instrument were confirmed by visual inspection of the graphical plot (cycle number versus fluorescence value) generated by the instrument.
Following the amplification phase, a melting curve analysis (starting at 40°C) was performed with a temperature transition rate of 0.2°C/s to determine the Tm values for the sequences targeted by the hybridization probes. Tm values were manually assigned from a plot generated by the instrument of the negative derivative of fluorescence versus temperature (−dF/dT) of the melting curve for amplification products measured at 640 and 705 nm.
Sensitivity of LC-PCR assays and protocol using LC-PCR assays for STEC detection in stool.
To assess the sensitivity of the PCR assays for the four target genes, E. coli ATCC 43895 was grown at 37°C in brain heart infusion broth to an optical density at 600 nm of 0.7 to 0.8 and diluted 10-fold in phosphate-buffered saline to yield 101 to 1010 CFU/ml as estimated by a standard plating procedure. DNA templates from a 300-μl aliquot of each dilution were prepared and 2-μl aliquots were tested in the LC-PCR assays and the block cycler PCR assays as described above to determine the minimum number of CFU per reaction mixture that could be detected by each method. In addition, 0.5-g portions of stool from a healthy volunteer were suspended in 0.5-ml aliquots of each bacterial dilution containing known numbers of CFU/ml to estimate the sensitivity of the assay for detecting the target genes in clinical specimens. For each spiked stool suspension (50% [wt/vol] stool suspension), a 5-μl aliquot was inoculated onto a MacConkey agar plate, streaked for isolated colonies, and incubated for 16 to 18 h at 35°C. The bacterial growth from the entire plate was harvested into 1.5 ml of lysis buffer as described above and processed for template DNA. A 2-μl aliquot of each sample was tested for STEC target genes by LC-PCR and block cycler PCR assays.
Nucleotide sequence analysis of PCR products.
Amplification products were purified using the HighPure PCR Product Purification kit (Roche Diagnostics), and cycle sequencing reactions of the stx1 and stx2 amplicons were performed as described in the PRISM Ready Reaction Dye Deoxy Terminator cycle sequencing kit protocol (Applied Biosystems, Weiterstadt, Germany). Both strands of amplicons originating from different amplification reactions were sequenced in duplicate to rule out the possibility of Taq DNA polymerase-induced errors. Optimal results were obtained using STEC-1 as the forward and STEC-2 as the reverse sequencing primer. The fluorescence-labeled reaction products were analyzed with an ABI PRISM 310 Genetic Analyzer (Applied Biosystems). To determine the complete sequence of the stx2d gene, amplification primers SD-a and SD-b (Table 2), which span subunit genes stxA2d and stxB2d, were used for cycle sequencing reactions. Likewise, amplification primers SE-a and SE-b (Table 2), which span subunit genes stxA2e and stxB2e, were used to sequence the complete stx2e gene.
Nucleotide sequence accession numbers.
For the sequence of the stx2 amplicon from strain 3143-97 with the 51°C melting point (described below), the sequence of a 1,324-bp region of the stx2 gene was determined and deposited in GenBank under accession no. AJ313015. For the nucleotide sequence of the stx2e gene from strain 3615-99 with the 63°C melting point (described below), a 1,509-bp region of its sequence was determined and deposited in GenBank under accession no. AJ313016.
RESULTS
Strategy for duplex LC-PCR assay design.
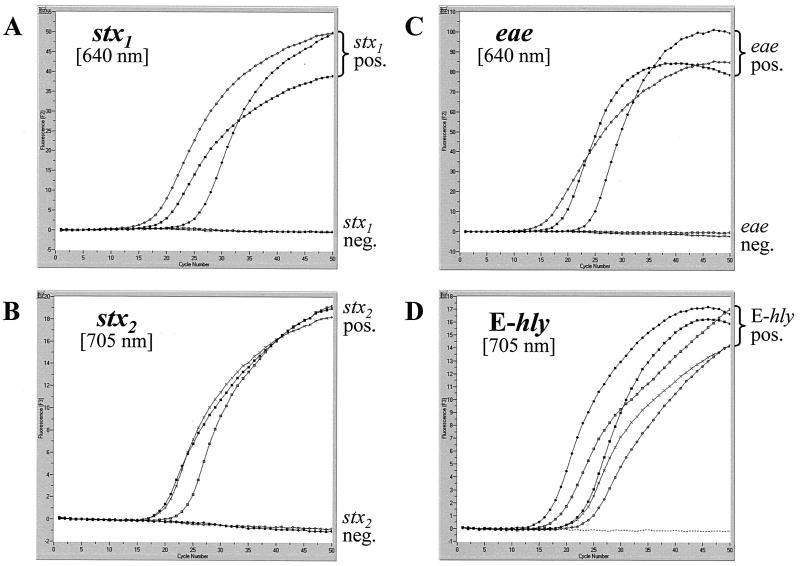
The two duplex real-time fluorescence PCR assays for the LightCycler simultaneously detect the most common virulence and virulence-associated genes present in STEC. We designed the assays to have identical amplification conditions so that both could be run at the same time on the instrument. By measuring the fluorescence values at 640 and 705 nm, we were able to detect stx1 and stx2, respectively, in one reaction capillary and eae and E-hly, respectively, in another reaction capillary. We used degenerate primer sequences from published primers (28) targeting conserved regions within the A subunit genes of stx1 and stx2 to give 512-bp and 518-bp amplicons, respectively, and we designed sets of hybridization probes, consisting of a “sensor” probe and an “anchor” probe, against sequences internal to the amplified regions of the target genes. We designed the sensor hybridization probes STEC-I HP-2 and STEC-II HP-2 to have a lower theoretical melting temperature than their anchor counterparts STEC-I HP-1 and STEC-II HP-1. We followed a similar strategy for comparably sized amplicons for the design of primers and hybridization probes targeting the genes encoding intimin (eae) and enterohemolysin (E-hly) (31). Graphs depicting the accumulation of fluorescence throughout the PCR for stx1, stx2, eae, and E-hly are shown in Fig. 1 for a panel of representative strains.
FIG. 1.
Amplification of stx1 and stx2 (A and B) and eae and E-hly (C and D) in duplex LC-PCR assays. Graphs depict fluorescence at 640 and 705 nm versus cycle number for the following E. coli strains with varied combinations of target genes: ▪, O157:H7 (stx1 positive; stx2 positive; eae positive; E-hly positive); •, O45:H2 (stx1 positive; stx2 negative; eae positive; E-hly positive); ○, O26:H11 (stx1 positive; stx2 negative; eae positive; E-hly positive); □, O110:H28 (stx1 negative; stx2 positive; eae negative; E-hly positive); x, O113:H21 (stx1 negative; stx2 positive; eae negative; E-hly positive); and - - - - , negative control.
Sensitivity and specificity of duplex LC-PCR assays for detecting target genes in pure bacterial culture.
To assess the ability of the LC-PCR assays to detect the four target genes in a serologically diverse collection of bacteria, we tested 504 isolates of E. coli and 118 isolates of other bacterial species with the LC-PCR assays and compared these findings with those obtained by block cycler PCR assays. The correlation of duplex LC-PCR and block cycler PCR findings and the sensitivities and specificities of the LC-PCR assays for detecting stx1, stx2, eae, and E-hly genes in pure bacterial cultures are summarized in Table 4. With the exception of the 10 stx2f-positive isolates from pigeons, the findings from the duplex LC-PCR assays and block cycler PCR assays were in complete agreement for all four target genes. The failure of the LC-PCR assay to detect stx2f was attributed to substantial nucleotide sequence differences between the stx primers and the primer annealing regions of the stx2f gene. Among the strains tested, the positive predictive values of the LC-PCR assays for stx1, stx2, eae, and E-hly were each 100%, and the negative predictive values for these genes were 100, 97, 100, and 100%, respectively.
TABLE 4.
Correlation of conventional PCR and LC-PCR results for 622 bacterial isolates
| Target gene | Results with conventional/LC PCR
|
Sensitivity (%) | Specificity (%) | |||
|---|---|---|---|---|---|---|
| +/+ | −/+ | +/− | −/− | |||
| stx1 | 272 | 0 | 0 | 350 | 100 | 100 |
| stx2 | 268 | 0 | 10a | 344 | 96 | 100 |
| eae | 360 | 0 | 0 | 262 | 100 | 100 |
| E-hly | 356 | 0 | 0 | 266 | 100 | 100 |
Ten of 10 stx2f isolates were not detected by the LC-PCR assay.
Time requirements for LC-PCR and block cycler PCR assays.
We performed 50 amplification cycles for the LC-PCR assays, which typically took 60 min to accomplish. For most samples, however, we could discriminate between PCR-positive and -negative samples after only 30 cycles, which took approximately 30 min to accomplish. Block cycler PCR assays with agarose gel detection of amplicons typically took 4.5 to 5 h to complete.
Differentiation of stx variants, eae, and E-hly by melting curve analysis.
Since considerable sequence heterogeneity exists among the described prototype and variant stx genes, we positioned the stx1- and stx2-specific sets of hybridization probes over hypervariable regions within the amplicon to distinguish gene variants by subsequent melting curve analyses. Sharp and clearly interpretable melting curves (plotted as the negative derivative of fluorescence versus temperature) were obtained with all tested stx1- and/or stx2-positive STEC isolates.
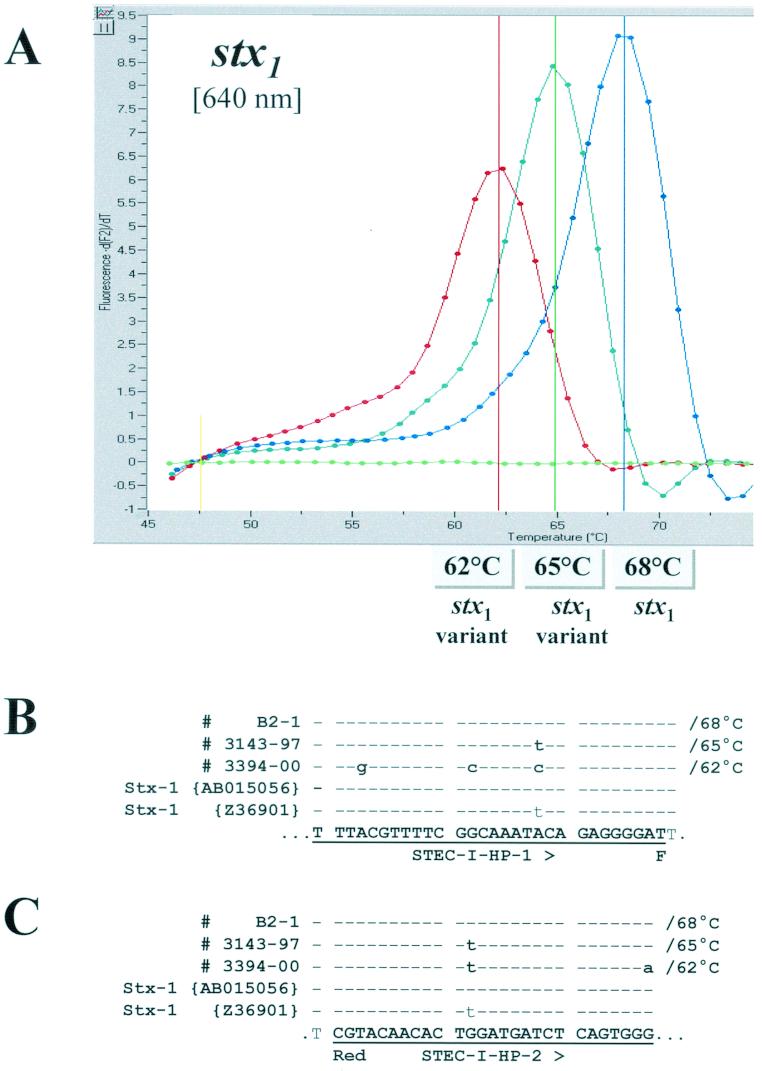
With the majority of stx1-positive strains, we observed a melting point of 68°C. This corresponds to a perfect match of hybridization probes STEC-I HP-1 and STEC-I HP-2 with the stx1 sequence AB015056 that has been deposited in GenBank (15). We noted reduced melting points for two stx1-positive E. coli strains: 62°C for strain 3394-00 (serotype O121:H7) and 65°C for strain 3143-97 (serotype O22:H8) (Fig. 2A). Findings from comparative nucleotide sequence analyses of the stx1 amplicons from strains 3394-00 and 3143-97 revealed partial and complete homologies, respectively, between these genes and the sequence of a variant stx1 gene from a strain described by Paton et al. (E. coli serotype OX3:H8; GenBank sequence Z36901) (30). We observed five and two nucleotide mismatches between the sequences of 3394-00 and 3143-97, respectively, and the sequences of the hybridization probes, which account for the reduced melting points of these isolates compared with those obtained for strains whose stx1 genes are completely homologous to the probes. Melting curve analyses of stx1 amplification products from representative strains with the 68, 62, and 65°C melting points and their corresponding sequence alignments are shown in Fig. 2.
FIG.2.
(A) Melting curve analysis performed on amplification products of three different stx1-positive isolates. Strain B2-1 represents a typical stx1-positive isolate (E. coli O111:H8), strains 3143-97 (E. coli O22:H8) and 3394-00 (E. coli O125:H7) are STEC isolates harboring variant stx1 sequences. (B and C) Sequence alignments with amplicons of the three different stx1 genotypes, GenBank AB015056, Z36901, anchor hybridization probe STEC-I HP-1 (B), and sensor hybridization probe STEC-I HP-2 (C). Sequence identity is indicated by dashes. The observed melting temperatures are indicated in the figure and next to the corresponding sequences in the alignments.
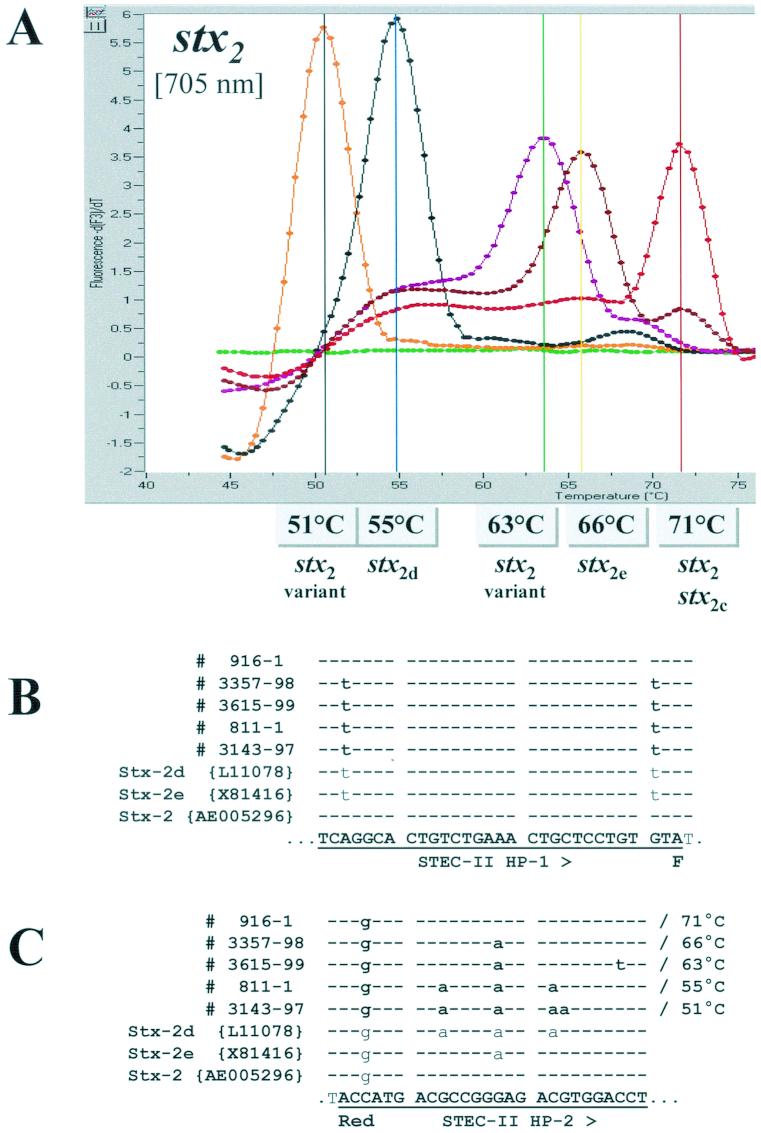
Because several subtypes of stx2 have been described, we examined the strains in our collection for which toxin subtype information was available by block cycler PCR or PCR-RFLP analysis for possible correlations between the melting points of the stx2 hybridization probes and the type of stx2 gene possessed. Findings from this comparison are summarized in Table 5, and melting curve profiles for representative strains of the five different melting temperatures observed are shown in Fig. 3A . Among the 117 strains included in this comparison, we observed melting points of 71°C for all 51 strains whose stx2 genes were characterized as either the prototype stx2 (n = 27), stx2c (n = 18), or a combination of these genes (n = 6). This melting point reflects a perfect match and a 1-nucleotide mismatch between the hybridization probes STEC-II HP-1 and STEC-II HP-2, respectively, and the prototype stx2 sequence AE005296 deposited in GenBank (32).
TABLE 5.
Melting peak temperatures of the stx2 LightCycler hybridization probes for 117 STEC strains whose stx2 genes were previously characterized by block cycler PCR and PCR-RFLP
| stx2 gene(s) present | No. of strains | Melting temp. (°C) | Serotype(s)a |
|---|---|---|---|
| stx2 | 27 | 71 | O6:NM (1), O26:H11/NM (7), O103:H2 (1), O111:NM (3), O145:NM (1), O157:H7/NM (13), Ound:NM (1) |
| stx2c | 18 | 71 | O23:NM (1), O30:NM (1), O77:NM (1), O92:NM (1), O120:NM (1), O145:NM (2), O157:H7/NM (10), Orou:NM (1) |
| stx2 and stx2c | 6 | 71 | O157:H7 (6) |
| stx2d | 39 | 55 | O8:NM (1), O16:H32/H48 (2), O40:H6/NM (2), O62:NM (1), O70:NM (1), O75:H21 (1), O91:NM (1), O96:NM (1), O103:NM (1), O113:NM (2), O128:H2/NM (8), O140:NM (1), O146:NM (1), O162:NM (1), Orou:H19/NM (4), Ound:H8 (4), Ound:NM (7) |
| 1b | 51 | O22:H8 (1) | |
| stx2e | 15 | 66 | O2:H5 (1), O8:Hund (1), O60:NM (2), O60:H2/NM (4), O101:H9 (1), Ound:NM (6) |
| 1c | 63 | Ound:NM (1) | |
| stx2f | 10 | No melting | O15:NM (1), O26:NM (1), O66:NM (1), O83:H19 (1), O128:NM (3), O135:NM (1), Ound:NM (1), Ound:NM (1) |
| Total | 117 |
Ound and Hund, O and H antigen, respectively, not determined with available typing sera; Orou, O antigen not determined because of incomplete or rough lipopolysaccharide; NM, nonmotile. Numbers in parentheses indicate number of strains with that serotype.
Nucleotide sequence variant of stx2d.
Nucleotide sequence variant of stx2e.
FIG.3.
(A) Melting curve analyses performed on amplification products of five different stx2-positive isolates representative of the different stx2 genotypes. Strain 916-1 is a typical stx2-positive isolate (E. coli O157:H7), strain 3357-98 is a stx2e-positive isolate (E. coli O60:NM), strain 3615-99 is a STEC isolate harboring a variant stx2 sequence (E. coli Ound:NM), strain 811-1 is a stx2d-positive isolate (E. coli O128:H2), and strain 3143-97 (E. coli O22:H8) is a STEC isolate harboring a variant stx2 sequence. (B and C) Sequence alignments with amplicons of the five different stx2 genotypes, GenBank L11078 (stx2d), X81416 (stx2e), AE005296 (stx2), anchor hybridization probe STEC-II HP-1 (B) and sensor hybridization probe STEC-II HP-2 (C). Sequence identity is indicated by dashes. The observed melting temperatures are indicated in the figure and next to the corresponding sequences in the alignments.
Among the 40 STEC strains that were positive for stx2d, we observed a melting point of 55°C for 39 strains and a melting point of 51°C for one strain (3143-97, serotype O22:H8) (Table 5). Findings from the comparative nucleotide sequence analysis of the stx2 amplicon from a representative strain with the 55°C melting point, 811-1 (serotype O128:H2) versus those stored in GenBank showed complete homology between it and the sequence of the stx2d gene deposited under GenBank L11078 (27) (Fig. 3B and C). In contrast, the sequence of the stx2 amplicon from strain 3143-97 with the 51°C melting point differed from those in GenBank, which prompted us to determine the sequence of a 1,324-bp region of its stx2 gene and deposit it in GenBank under accession no. AJ313015. Alignment of the stx2d sequences from strains 811-1 and 3143-97 with those of the hybridization probes revealed four and five mismatches, respectively, between the target genes and hybridization probe STEC-II-HP-2. Three and four of these mismatches account for the reduced melting points of the stx2d strains relative to those observed for the prototype stx2 and stx2c genes.
Among the 16 strains that were positive for stx2e, we detected melting points of 66°C for 15 strains and a melting point of 63°C for one strain (3615-99, serotype Ound:NM) (Table 5). Findings from the comparative nucleotide sequence analysis of the stx2e amplicon from a representative strain with the 66°C melting point (strain 3357-98, serotype O60:NM) revealed a perfect match between it and the stx2e GenBank sequence X81416 (9). Alignment of the sequence from this strain with the sequences of the hybridization probes revealed two nucleotide mismatches between it and hybridization probe STEC-II-HP-2, one of which accounts for the reduction in melting points observed for stx2e-positive strains compared with those obtained for prototype stx2- and stx2c-positive strains (Fig. 3B and C). The nucleotide sequence of the stx2e gene from strain 3615-99 with the 63°C melting point differed from other stx2 sequences deposited to date in GenBank, and a 1,509-bp region of its sequence was determined and deposited in GenBank under accession no. AJ313016. A similar alignment of its sequence with those of the hybridization probes showed three mismatches between its sequence and the sequence of hybridization probe STEC-II-HP-2 (Fig. 3B and C); two of these account for the reduced melting point temperature observed for this strain compared with prototype stx2- and stx2c-positive strains.
We observed a large number of different melting temperatures ranging from 55 to 67°C for E. coli isolates testing positive for eae and E-hly in the LC-PCR assay (data not shown). No subtype information for these genes was available on the strains included in this study for examination of correlations between the type of gene and melting point temperatures of the hybridization probes.
Analytical sensitivity of duplex LC-PCR assays.
To determine the lower limit of detection for the duplex LC-PCR assays, we performed these assays on DNA preparations from serial dilutions of ATCC 43895, a control strain that is positive for stx1, stx2, eae, and E-hly. In five separate experiments, we detected 5 × 102 CFU per reaction in three and 5 × 103 CFU per reaction in two of the experiments performed for stx1 and 5 × 102 CFU per reaction for stx2 in all five experiments performed (data not shown). For the simultaneous amplification and detection of eae and E-hly, we obtained a detection limit of approximately 5 × 102 CFU per reaction for both genes in the same number of experiments (data not shown). We observed no difference in the lower limits of detection for the four target genes when tested in the duplex PCR format or individually, demonstrating that the duplex assays were not significantly hampered by the formation of primer dimers, secondary structures, or other cross-reactions between oligonucleotides present in the reaction mixture (data not shown).
Sensitivity of duplex LC-PCR assays for detecting target genes in fecal specimens.
To assess the sensitivity of the LC-PCR assays for detecting STEC in fecal specimens, we compared the abilities of these assays with those of block cycler PCR assays to detect stx1, stx2, eae, and E-hly genes in mixed bacterial cultures harvested from primary isolation plates inoculated with spiked stool specimens. We observed the same lower limit of direct detection for both the LC-PCR and block cycler PCR assays for all four target genes: 5 × 103 CFU/ml of a 50% (wt/vol) stool suspension. For these four genes, findings from LC-PCR and block cycler PCR assays performed on overnight bacterial growth from primary isolation plates inoculated with 2.5 CFU were both negative, while plates inoculated with stool suspension containing 25 or more CFU were strongly positive with both assays (data not shown).
DISCUSSION
Our set of duplex assays covers the major variants of stx1 and stx2 currently associated with human and animal disease, as well as the genes encoding intimin (eae) and enterohemolysin (E-hly) (Tables 4 and 5). To detect and discriminate between the stx1 and stx2 genes, we designed a pair of degenerate primers for the amplification of both genes and used specific sets of hybridization probes to discriminate between and within the different families of toxin genes. We constructed the stx primers to hybridize to conserved regions within the published sequences of the A subunit genes of Shiga toxin from S. dysenteriae type 1 (stx), prototype stx1, a sequence variant of stx1, prototype stx2, stx2c, stx2d, and stx2e.
Because of insufficient homology between the nucleotide sequences of the A subunit genes of stx2f and the other stx genes (63.4% nucleotide sequence homology between stx2f and prototype stx2), we were not able to design specific primers to amplify this toxin variant with the other stx genes. The diagnostic importance of a false-negative reaction for this toxin with our assay is not clear. With the exception of one isolate from a human infant with diarrhea (strain H.I.8), the stx2f genes to date have been detected only in E. coli strains from apparently healthy feral pigeons (13, 22, 40). It is worth noting that stx2f genes, while not found in combination with other stx genes (11), were commonly present with the eae gene (22). Based on these findings, a strategy of subsequently testing isolates that are positive for only the eae gene in the LC-PCR assays with the stx2f-specific PCR assay described by Schmidt et al. (40) could be used to identify most stx2f-positive isolates until a specific LC-PCR assay for these strains is developed. Studies to assess the pathogenic potential of these and other stx variant strains for humans and animals are needed to help guide the development of diagnostic assays for clinically important STEC strains.
Excluding the stx2f-positive strains, our findings with 494 human E. coli isolates of more than 100 different serotypes and 118 other bacterial species show that the stx LC-PCR assay was 100% sensitive and specific for detecting and appropriately characterizing the toxin genes among the tested isolates. All 421 STEC strains positive for stx1, stx2, stx2c, stx2d, and stx2e, as well as a strain of S. dysenteriae type 1, were positive for the appropriate stx gene(s), and the respective stx gene variants were clearly identified by their characteristic hybridization probe melting points. Although we did not include a strain that was positive for the newly described sequence variant of stx2 (Stx2-NV206) reported by Bertin et al. (3), a comparison of the nucleotide sequence of this toxin with those of the primers and probes used in the LC-PCR assay showed that there would be sufficient homology between these sequences to allow detection of this stx2 variant gene by the LC-PCR assay with a predicted Tm of 65°C.
Regarding the design of the hybridization probes, it is worth mentioning that the 1-nucleotide mismatch between the sequence of hybridization probe STEC-II HP-2 and the sequence of prototype stx2 (GenBank AE005296) was intentional. In the course of evaluation experiments, hybridization probe STEC-II HP-2, which was designed complementary to a stx2 variant (GenBank Z37725) (29), revealed greater Tm differences between all the analyzed stx2 variants than a candidate hybridization probe with a perfect match to GenBank AE005296.
By melting curve analysis, we detected three Tms among the 271 stx1-positive isolates for the region of the stx1 gene targeted by the hybridization probes (Tms of 62, 65 and 68°C) (Fig. 2). With two exceptions, the stx1-positive isolates had a Tm of 68°C. One isolate had a Tm of 62°C (3394-00, serotype O121:H7), and another isolate (3143-97, serotype O22:H8) had a Tm of 65°C. The nucleotide sequence of the stx1 gene from 3143-97 was identical in the region of the hybridization probes to that reported by Paton et al. (30) for an OX3:H8 strain (GenBank Z36901). Although heterogeneity in the nucleotide and amino acid sequences of stx1 genes has been observed (30), no subtypes of stx1 meeting the criteria for designation specified by O'Brien et al. (24) are currently recognized. As would be expected, the S. dysenteriae type 1 isolate, which is positive for the Shiga toxin gene, was positive in the LC-PCR and block cycler PCR assays.
By melting curve analysis, we detected five Tm values (51, 55, 63, 66, and 71°C) among 278 stx2-positive isolates for the region of the stx2 gene targeted by the hybridization probes (Fig. 3). The majority of these (174) had a melting point of 71°C for their stx2 genes. Among a subset of 117 stx2-positive strains for which toxin subtype information was available by conventional PCR or PCR-RFLP tests, there was good correlation between the Tm values measured and the toxin subtype genes they possessed: strains positive for stx2 and/or stx2c had a Tm of 71°C; strains positive for stx2d, with one exception, had a Tm of 55°C; and strains positive for stx2e, with one exception, had a Tm of 66°C (Table 5).
For the two exceptional strains, 3143-97 (serotype O22:H8) and 3615-99 (serotype Ound:NM), Tms of 51 and 63°C, respectively, were obtained. Findings from the comparative nucleotide sequence analyses of the complete stx2 genes from these two isolates suggest that these isolates possess sequence variants of stx2d and stx2e, respectively. Degrees of similarity between the sequences of 3143-97 and stx2d (29 mismatches per 1,470 bp with respect to stx2d of GenBank L11078), and between the sequences of 3615-99 and stx2e (3 mismatches per 1,236 bp with respect to stx2e of GenBank X81416) were observed. Interestingly, the G-to-A mutation in 3143-97 at position 652 of GenBank AJ313015 introduces an additional start codon within the open reading frame of the stx2 gene. The effect of this start codon on the level of protein translation is not known.
The correlation of Tm values from the melting curve analyses with toxin subtypes was an unanticipated finding from our study. More than 99% (545 of 549) of the stx genes we examined by melting point analysis yielded Tm values that fell within 1°C of one of the four Tm values detected for the toxin subtype control strains. As more strains are tested, it is possible that polymorphisms in the region of the hybridization probes will be detected that may obscure the correlation between Tm value and toxin subtype. Testing additional strains, particularly nucleotide sequence variants of the recognized toxin subtypes, is needed to define the range of Tm values for each toxin subtype and to determine if the observed correlation between Tm value and toxin subtype remains. It is worth noting that during the course of these experiments we noticed that Tm values were influenced by the concentration of magnesium chloride in the reaction mixture; increasing the magnesium chloride concentration resulted in a higher observed Tm value than expected. We did not vary each component of the reaction mixture for its effect on Tm; however, because subtle changes may occur in the constituents of a reaction mixture that can influence the interaction of the hybridization probes with each other or their targets, we strongly advise running a set of control strains with each assay when characterizing toxin genes by melting curve analysis.
For the eae gene, we chose the PCR primers described by Paton et al. (31) because they amplify a segment at the 5′ end of eae that is conserved among STEC and enteropathogenic E. coli (EPEC) strains, which would suggest that these primers be used for the detection of both categories of diarrheagenic E. coli. In contrast, primers for E-hly were complementary to an area with minimal homology to the gene encoding E. coli alpha-hemolysin, which shares about 70% overall DNA sequence homology with E-hly. Hybridization probes complementary to the region of the genes bounded by these sets of primers were designed to provide good gene detection and as much discrimination as possible between the different alleles of each gene. LC-PCR assay findings obtained for 360 eae-positive and 356 E-hly-positive E. coli strains, as well as strains of E. coli and 116 isolates of other bacterial species lacking these genes, demonstrate the sensitivity and specificity of the eae and E-hly primer and hybridization probe sequences; all the eae- and E-hly-positive organisms were positive in the LC-PCR assays, while none of the isolates negative for these genes were positive. As expected, the EPEC control strain was positive for eae and negative for E-hly in the LC-PCR assay. Tms ranging from 55 to 67°C were observed by melting curve analyses for eae and E-hly among the strains tested, demonstrating sequence variability in the region of the eae and E-hly genes targeted by the hybridization probes and the potential for additional strain discrimination.
Although the detection of eae and E-hly among STEC isolates has been done for epidemiologic purposes for nearly a decade, the use of these genes diagnostically is still in its early stages. Findings from several studies have shown that the majority of STEC associated with human illness also are positive for eae and E-hly (4, 5, 34). While a positive finding of these genes among STEC strains causing human disease appears significant, STEC isolates lacking one or both of these genes, for example serotypes O113:H21, O91:H21, and O104:H21, have also been recovered from persons with clinically significant disease, and the role of these strains in causing disease needs further study. The initial testing of samples (mixed bacterial cultures from primary isolation plates or enrichment broths) for all four genes will allow the identification not only of STEC associated with more significant disease but also eae-negative and E-hly-negative STEC in subsequent studies.
Sensitivity testing experiments with serial dilutions of STEC in culture broth or saline revealed a conservative lower limit of detection for both the duplex LC-PCR and block cycler PCR assays of 5 × 103 organisms per reaction for the respective target genes. When stool samples were spiked with known numbers of STEC, the sensitivities of the LC-PCR and block cycler PCR assays for detecting the four target genes in overnight mixed bacterial cultures from primary isolation plates also compared favorably, with both assays easily detecting 5 × 103 CFU/ml of a 50% (wt/vol) stool suspension. DNA samples prepared from primary isolation plates inoculated with 2.5 CFU of STEC in the presence of competing normal fecal bacteria were negative, while samples prepared from plates inoculated with 25 CFU or more of STEC were strongly positive.
The duplex LC-PCR assays described in this report offer rapid cycling (1 h or less) with real-time, sequence-specific detection of amplicons. The fluorescence-labeled hybridization probes not only provide confidence in the identification of target genes but also reduce the risk of experiencing product contamination of the laboratory, because the amplification reaction, detection of PCR products, and their melting curve analyses are conducted in a single capillary tube. Although our current protocol for processing stool specimens for STEC requires an overnight enrichment step to achieve adequate sensitivity, the rapid amplification and reliable detection of stx1, stx2, eae, and E-hly genes in samples should facilitate the diagnosis and management of patients infected with STEC.
While the diagnosis and management of individual patients infected with STEC do not require an isolate, the isolation of STEC from patient samples is critical for public health purposes. The association of the STEC target genes with a particular bacterium requires testing isolates, and isolates are still needed for molecular subtyping (DNA fingerprinting), serologic characterization, and antimicrobial susceptibility testing for surveillance purposes. For these reasons, clinical laboratories are strongly encouraged to consult with their public health authorities for guidance on forwarding positive samples for isolation of STEC. The speed at which bacterial colonies can be screened for target genes and have these characterized with LC-PCR assays should facilitate the isolation of STEC and provide epidemiologically useful information for the identification and investigation of STEC outbreaks. Although they are currently more expensive to perform than block cycler PCR assays, the speed, greater information, and reliability of the results make the LC-PCR assays attractive alternatives to conventional block cycler PCR assays for the detection and characterization of STEC.
Acknowledgments
We thank Stefan Lukas and Barbara Plaschke for their active support and gratefully acknowledge the excellent technical assistance of Markus Bollwein, Michaela Hien, and Birgit Leppmeier during the study.
This work has been supported in part by the Arab Fund for Economic and Social Development, Kuwait. M.T.Y. was a recipient of the Arab Fund's Distinguished Scholar Award.
REFERENCES
- 1.Bastian, S. N., I. Carle, and F. Grimont. 1998. Comparison of 14 PCR systems for the detection and subtyping of stx genes in Shiga-toxin-producing Escherichia coli. Res. Microbiol. 149:457-472. [DOI] [PubMed] [Google Scholar]
- 2.Bellin, T., M. Pulz, A. Matussek, H.-G. Hempen, and F. Gunzer. 2001. Rapid detection of enterohemorrhagic Escherichia coli by real-time PCR with fluorescent hybridization probes. J. Clin. Microbiol. 39:370-374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bertin, Y., K. Boukhours, N. Pradel, V. Librelli, and C. Martin. 2001. Stx2 subtyping of Shiga toxin-producing Escherichia coli isolated from cattle in France: detection of a new Stx2 subtype and correlation with additional virulence factors. J. Clin. Microbiol. 39:3060-3065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Beutin, L., S. Aleksic, S. Zimmermann, and K. Gleier. 1994. Virulence factors and phenotypic traits of verotoxigenic strains of Escherichia coli isolated from human patients in Germany. Med. Microbiol. Immunol. 183:13-21. [DOI] [PubMed] [Google Scholar]
- 5.Boerlin, P., S. A. McEwen, F. Boerlin-Petzold, J. B. Wilson, R. P. Johnson, and C. L. Gyles. 1999. Associations between virulence factors of Shiga toxin-producing Escherichia coli and disease in humans. J. Clin. Microbiol. 37:497-503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Centers for Disease Control and Prevention. 1993. Update: multistate outbreak of Escherichia coli O157:H7 infections from hamburgers—western United States, 1993. Morb. Mortal. Wkly. Rep. 42:257-263. [PubMed] [Google Scholar]
- 7.Ewing, W. H. 1986. The genus Escherichia, p. 93-134. In W. H. Ewing and P. R. Edward (ed.), Edwards and Ewing's identification of Enterobacteriaceae, 4th ed. Elsevier Science B.V., New York, N.Y.
- 8.Feng, P., and S. R. Monday. 2000. Multiplex PCR for detection of trait and virulence factors in enterohemorrhagic Escherichia coli serotypes. Mol. Cell. Probes 14:333-337. [DOI] [PubMed] [Google Scholar]
- 9.Franke, S., D. Harmsen, A. Caprioli, D. Pierard, L. H. Wieler, and H. Karch. 1995. Clonal relatedness of Shiga-like toxin-producing Escherichia coli O101 strains of human and porcine origin. J. Clin. Microbiol. 33:3174-3178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fratamico, P. M., S. K. Sackitey, M. Weidmann, and M. Yi Deng. 1995. Detection of Escherichia coli O157:H7 by multiplex PCR. J. Clin. Microbiol. 33:2188-2191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Friedrich, A. W., M. Bielaszewska, W.-L. Zhang, M. Pulz, T. Kuczius, A. Ammon, and H. Karch. 2002. Escherichia coli harboring Shiga toxin 2 gene variants: frequency and association with clinical syndrome. J. Infect. Dis. 185:74-84. [DOI] [PubMed] [Google Scholar]
- 12.Gannon, V. P., R. K. King, J. Y. Kim, and E. J. Thomas. 1992. Rapid and sensitive method for detection of Shiga-like toxin-producing Escherichia coli in ground beef using the polymerase chain reaction. Appl. Environ. Microbiol. 58:3809-3815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gannon, V. P., C. Teerling, S. A. Masri, and C. L. Gyles. 1990. Molecular cloning and nucleotide sequence of another variant of the Escherichia coli Shiga-like toxin II family. J. Gen. Microbiol. 136:1125-1135. [DOI] [PubMed] [Google Scholar]
- 14.Gannon, V. P., M. Rashed, R. K. King, and E. J. Thomas. 1993. Detection and characterization of the eae gene of Shiga-like toxin-producing Escherichia coli using polymerase chain reaction. J. Clin. Microbiol. 31:1268-1274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Iwasa, M., S. Makino, H. Asakura, H. Kobori, and Y. Morimoto. 1999. Detection of Escherichia coli O157:H7 from Musca domestica (Diptera: Muscidae) at a cattle farm in Japan. J. Med. Entomol. 36:108-112. [DOI] [PubMed] [Google Scholar]
- 16.Karch, H., C. Janetzki-Mittmann, S. Aleksic, and M. Datz. 1996. Isolation of enterohemorrhagic Escherichia coli O157 strains from patients with hemolytic-uremic syndrome by using immunomagnetic separation, DNA-based methods and direct culture. J. Clin. Microbiol. 34:516-519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Karch, H., H. I Huppertz, J. Bockemühl, H. Schmidt, A. Schwarzkopf, and R. Lissner. 1997. Shiga toxin-producing Escherichia coli infections in Germany. J. Food Prot. 11:1454-1457. [DOI] [PubMed] [Google Scholar]
- 18.Karmali, M. A. 1989. Infections by verocytotoxin-producing Escherichia coli. Clin. Microbiol. Rev. 2:15-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Karmali, M. A., B. T. Steele, and L. C. Petric. 1983.. Sporadic cases of hemolytic uremic syndrome associated with fecal cytotoxin and cytotoxin-producing Escherichia coli. Lancet i:619-620. [DOI] [PubMed] [Google Scholar]
- 20.Lin, Z., H. Kurazono, S. Yamasaki, and Y. Takeda. 1993. Detection of various variant verotoxin genes in Escherichia coli by polymerase chain reaction. Microbiol. Immunol. 37:543-548. [DOI] [PubMed] [Google Scholar]
- 21.Louie, M., J. De Azavedo, R. Clarke, A. Borczyk, H. Lior, and M. Richter. 1994. Sequence heterogeneity of the eae gene and detection of verotoxin producing Escherichia coli using serotype-specific primers. Epidemiol. Infect. 112:449-461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Morabito, S. G., U. Dell'Omo, H. Agrimi, H. Schmidt, T. Karch, T. Cheasty, and A. Caprioli. 2001. Detection and characterization of Shiga toxin-producing Escherichia coli in feral pigeons. Vet. Microbiol. 82:275-283. [DOI] [PubMed] [Google Scholar]
- 23.Nataro, J. P., and J. B. Kaper. 1998. Diarrheagenic Escherichia coli. Clin. Microbiol. Rev. 11:142-201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.O'Brien, A. D., M. A. Karmali, and S. M. Scotland. 1994. A proposal for rationalizing the nomenclature of the Escherichia coli cytotoxins, p. 147-149. In M. A. Karmali and A. G. Golgio (ed.), Recent advances in verocytotoxin-producing Escherichia coli infections. Elsevier Science B.V., New York, N.Y.
- 25.Ølsvik, Ø., E. Rimstad, E. Hornes, N. Strockbine, Y. Wasteson, A. Lund, and K. Wachsmuth. 1991. A nested PCR followed by magnetic separation of amplified fragments for detection of Escherichia coli Shiga-like toxin genes. Mol. Cell. Probes 5:429-435. [DOI] [PubMed] [Google Scholar]
- 26.Oswald, E., H. Schmidt, S. Morabito, H. Karch, O. Marches, and A. Caprioli. 2000. Typing of intimin genes in human and animal enterohemorrhagic and enteropathogenic Escherichia coli: characterization of a new intimin variant. Infect. Immun. 68:64-71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Paton, A. W., J. C. Paton, and P. A. Manning. 1993. Polymerase chain reaction amplification, cloning and sequencing of variant Escherichia coli Shiga-like toxin type II operons. Microb. Pathog. 15:77-82. [DOI] [PubMed] [Google Scholar]
- 28.Paton, A. W., J. C. Paton, P. N. Goldwater, and P. A. Manning. 1993. Direct detection of Escherichia coli Shiga-like toxin genes in primary fecal cultures by polymerase chain reaction. J. Clin. Microbiol. 31:3063-3067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Paton, A. W., A. J. Bourne, P. A. Manning, and J. C. Paton. 1995. Comparative toxicity and virulence of Escherichia coli clones expressing variant and chimeric Shiga-like toxin type II operons. Infect. Immun. 63:2450-2458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Paton, A. W., L. Beutin, and J. C. Paton. 1995. Heterogeneity of the amino-acid sequences of Escherichia coli Shiga-like toxin type I operons. Gene 153:71-74. [DOI] [PubMed] [Google Scholar]
- 31.Paton, A. W., and J. C. Paton. 1998. Detection and characterization of Shiga toxigenic Escherichia coli by using multiplex PCR assays for stx1, stx2, eaeA, enterohemorrhagic E. coli hlyA, rfbO111, and rfbO157. J. Clin. Microbiol. 36:598-602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Perna, N. T., G. Plunkett III, V. Burland, B. Mau, J. D. Glasner, D. J. Rose, G. F. Mayhew, P. S. Evans, J. Gregor, H. A. Kirkpatrick, G. Posfai, J. Hackett, S. Klink, A. Boutin, Y. Shao, L. Miller, E. J. Grotbeck, N. W. Davis, A. Lim, E. Dimalanta, K. Potamousis, J. Apodaca, T. S. Anantharaman, J. Lin, G. Yen, D. C. Schwartz, R. A. Welch, and F. R. Blattner. 2001. Genome sequence of enterohemorrhagic Escherichia coli O157:H7. Nature 409:529-533. [DOI] [PubMed] [Google Scholar]
- 33.Pickering, L. K., T. G. Obrig, and F. B. Stapleton. 1994. Hemolytic-uremic syndrome and enterohemorrhagic Escherichia coli. Pediatr. Infect. Dis. J. 13:459-476. [DOI] [PubMed] [Google Scholar]
- 34.Piérard, D., D. Stevens, L. Moriau, H. Lior, and S. Lauwers. 1997. Isolation and virulence factors of verocytotoxin-producing Escherichia coli in human stool samples. Clin. Microbiol. Infect. 3:531-540. [DOI] [PubMed] [Google Scholar]
- 35.Piérard, D., G. Muyldermans, L. Moriau, D. Stevens, and S. Lauwers. 1998. Identification of new verocytotoxin type 2 variant B-subunit genes in human and animal Escherichia coli isolates. J. Clin. Microbiol. 36:3317-3322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ramotar, K., B. Waldhart, D. Church, R. Szumski, and T. J. Louie. 1995. Direct detection of verotoxin-producing Escherichia coli in stool samples by PCR. J. Clin. Microbiol. 33:519-524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Reischl, U., M. Pulz, W. Ehret, and H. Wolf. 1994. PCR-based detection of mycobacteria in sputum samples using a simple and reliable DNA extraction protocol. BioTechniques 17:844-845. [PubMed] [Google Scholar]
- 38.Riley, L. Q., R. S. Remis, S. D. Helgerson, H. B. McGee, J. G. Wells, B. R. Davis, R. J. Hebert, E. S. Olcott, L. M. Johnson, N. T. Hargrett, P. A. Blake, and M. L. Cohen. 1983. Hemorrhagic colitis associated with a rare Escherichia coli serotype. N. Engl. J. Med. 308:681-685. [DOI] [PubMed] [Google Scholar]
- 39.Rüssmann, H., H. Schmidt, J. Heesemann, A. Caprioli, and H. Karch. 1994. Variants of Shiga-like toxin II constitute a major toxin component in Escherichia coli O157 strains from patients with haemolytic uraemic syndrome. J. Med. Microbiol. 40:338-343. [DOI] [PubMed] [Google Scholar]
- 40.Schmidt, H., J. Scheef, S. Morabito, A. Caprioli, L. H. Wieler, and H. Karch. 2000. A new Shiga toxin variant (stx2f) from Escherichia coli isolated from pigeons. Appl. Environ. Microbiol. 66:1205-1208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Smith, H. R., and S. M. Scotland. 1988. Vero cytotoxin-producing strains of Escherichia coli. J. Med. Microbiol. 26:77-85. [DOI] [PubMed] [Google Scholar]