Abstract
Genetic investigations were carried out with 50 phenotypically selected strains of Pseudomonas aeruginosa from 18 patients attending an Australian cystic fibrosis (CF) center. The isolates were analyzed by restriction fragment length polymorphism (RFLP) analysis by pulsed-field gel electrophoresis (PFGE). Phylogenetic analysis of the macrorestriction patterns showed rates of genetic similarity ranging from 76 to 100%; 24 (48%) of the strains from 11 patients had greater than 90% similarity. A dominant strain emerged: 15 isolates from seven patients had identical PFGE patterns, and 4 other isolates were very closely related. The 50 isolates were grouped into 21 pulsotypes on the basis of visual delineation of a three-band difference. Ten of the 18 (56%) patients were infected with clonal or subclonal strains. Sequence analysis of PCR products derived from the mucA gene showed 20 mutations, with the number of mutations in individual isolates ranging from 1 to 4; 19 of these changes are reported here for the first time. Potentially functional changes were found in 22 (44%) isolates. Eight changes (five transversions and three single base deletions) led to premature stop codons, providing support for the presence of mucA mutations as one pathway to mucoidy. There was a trend toward an association between the dominant strain and lack of potentially functional mucA mutations (P = 0.09 by the χ2 test) but no relationship between genotype and phenotype. This is the first study of genetic variation in P. aeruginosa isolates from adult Australian CF patients. The findings highlight the need for further investigations on the transmissibility of P. aeruginosa in CF patients.
Chronic lung infection with Pseudomonas aeruginosa leading to respiratory failure is the major cause of both morbidity and mortality in patients with cystic fibrosis (CF) (6). Genetic analyses by techniques such as restriction fragment length polymorphism (RFLP) analysis by pulsed-field gel electrophoresis (PFGE) have indicated that most CF patients are colonized for long periods with single clones or subclones of P. aeruginosa (4, 7, 9, 17, 21, 23), and the appearance of new unrelated strains has often been associated with changes in antibiotic therapy (3). It has been believed that P. aeruginosa is generally environmentally acquired and that person-to-person spread occurs only rarely (3, 5, 9, 11, 16, 20, 21, 23). However, recent reports of the transmission of virulent multidrug-resistant mucoid strains of P. aeruginosa in a large Australian pediatric CF clinic (1) and two adult CF clinics in the United Kingdom (10, 13) and the person-to-person spread of P. aeruginosa among holiday campers with CF in Germany (9) have raised concerns about the effectiveness of standard infection control practices for P. aeruginosa. Nonetheless, confirmation of substantial cross infection with epidemic strains would be needed to support a policy of segregation like the one already practiced for Burkholderia cepacia.
Associated with the chronic colonization of P. aeruginosa in CF patients is a unique in vivo microbial adaptation process that involves transformation of the nonmucoid phenotype to the mucoid phenotype. Mucoidy contributes to the intractable and eventually fatal infection, promoting persistence by inhibiting phagocytosis and penetration by antibiotics. Investigations with derivatives of laboratory strains such as PAO-1 in vitro and in the mouse model have shown that frameshift or nonsense mutations in the muc gene cluster (algU, mucA, mucB, and mucD) of P. aeruginosa result in upregulation of the expression of the algU gene (2). The resultant overproduction of the exopolysaccharide alginate confers the mucoid phenotype. Evidence from in vitro studies suggests that the mucA gene is the preferential site for muc mutations. There is little information on the nature and frequency of mucA mutations in clinical isolates, apart from one survey that found mucA mutations in 84% of 55 mucoid strains from 53 CF patients but none in nonmucoid isolates (2). Strains of P. aeruginosa vary widely in the degree of mucoidy, but the relationship between particular mucA mutations and the mucoid phenotype has not been explored.
In the study described in this report, investigations were carried out with 50 P. aeruginosa isolates from the sputa of 18 patients treated at the CF Centre, Royal Prince Alfred Hospital, Sydney, Australia, between 2000 and 2001. The isolates were examined by PFGE to determine inter- and intrapatient variabilities, and direct sequence analysis of PCR products derived from the mucA gene was carried out to identify changes potentially leading to mucoidy. Associations between phenotype, genotype, and mucA mutations were sought.
MATERIALS AND METHODS
Bacterial strains.
The 50 strains of P. aeruginosa investigated were isolated from single sputum samples from 18 patients attending the CF clinic at the Royal Prince Alfred Hospital on 15 occasions between June 2000 and May 2001. All phenotypically different strains from each patient except three nonmucoid strains (two from patient 6 and one from patient 12) were tested. The number of isolates from individual patients varied from one to four (mean, three). The group comprised 11 males and 7 females whose ages ranged from 11 to 33 years (median age, 23 years). The mean forced expiratory volume in 1 s for the group was 55.78% predicted (standard deviation, 17.03% predicted). Patient demographics are presented in Table 1. Nonmucoid strain P. aeruginosa PAO-1 (GenBank accession number U491151; kindly supplied by V. Krishnapillai, Monash University, Melbourne, Australia) served as the reference strain.
TABLE 1.
Patient demographics and genotypic characteristics of P. aeruginosa isolates from patients' sputaa
| Isolate | Sex | Age (yr) | FEV1 (% predicted) | Pulso- type | mucA mutations | Effect of mutation |
|---|---|---|---|---|---|---|
| PAO-1 | None | |||||
| 1A | M | 33 | 40 | XXI | A → G at 342b | S |
| 1B | XXI | A → G at 342, C → T at 351 | S, stop codon at 351 | |||
| 2A | F | 23 | 69 | IC | A → G at 342 | S |
| 2B | I | A → G at 342 | S | |||
| 2C | I | A → G at 342, C → T at 500 | S, Arg → Cys | |||
| 2D | ID | A → G at 342, C → T at 500 | S, Arg → Cys | |||
| 3A | M | 22 | 61 | XI | A → G at 342, deletion of GCAGGT at 434 | S, deletion of Glut + Val |
| 4A | M | 20 | 50 | VIA | A → G at 342, T → G at 381, T → G at 198, T → C at 126 | S, S, S, S |
| 4B | VI | A → G at 342, T → G at 381, T → G at 198, T → C at 126 | S, S, S, S | |||
| 5A | F | 19 | 63 | I | A → G at 342 | S |
| 5B | I | A → G at 342 | S | |||
| 6A | F | 24 | 48 | I | A → G at 342 | S |
| 6B | I | A → G at 342 | S | |||
| 7A | M | 22 | 93 | I | A → G at 342 | S |
| 7B | I | A → G at 342 | S | |||
| 7C | I | A → G at 342 | S | |||
| 8A | F | 20 | 45 | I | A → G at 342, T → G at 198, T → G at 381 | S, S, S |
| 8B | I | A → G at 342, T → G at 198, A → G at 243, T → G at 381 | S, S, S, S | |||
| 8C | I | A → G at 342, deletion of G at 425 | S, stop codon at 439 | |||
| 9A | M | 23 | 35 | IB | A → G at 342 | S |
| 9B | II | A → G at 342, deletion of GCAGGT at 434 | S, deletion of Glut + Val | |||
| 10A | F | 19 | 59 | XV | A → G at 342 | S |
| 10B | XA | A → G at 342 | S | |||
| 10C | X | A → G at 342, C → T at 349 | S, stop codon | |||
| 11A | M | 31 | 40 | XX | A → G at 342, deletion of G at 430 | S, stop codon at 439 |
| 11B | XVI | A → G at 342, T → C at 133, C → T at 156, deletion of C at 205 | S, S, S, stop codon | |||
| 11C | XVIII | A → G at 342, C → T at 435 | S, S, stop codon at 435 | |||
| 12A | M | 32 | 34 | XIV | A → G at 342 | S |
| 12B | IV | A → G at 342 | S | |||
| 13A | M | 21 | 62 | VIII | A → G at 342, T → C at 133, TC → GT at 381/2 | S, S, silent at 381, stop codon at 382 |
| 13B | VIII | A → G at 342, T → C at 133, TC → GT at 381/2 | S, S, silent at 381, stop codon at 382 | |||
| 13C | VIII | A → G at 342, T → C at 133, TC → GT at 381/2 | S, S, silent at 381, stop codon at 382 | |||
| 14A | M | 21 | 40 | V | A → G at 342 | S |
| 14B | VII | A → G at 342, T → C at 376, deletion of G at 430 | S, Leu → Pro, stop codon at 439 | |||
| 14C | XVII | A → G at 342, C → T at 358 | S, Thr → Ile | |||
| 14D | III | A → G at 342, T → C at 376, deletion of G at 430 | S, Leu → Pro, stop codon at 439 | |||
| 15A | M | 24 | 73 | IX | A → G at 342, C → T at 340 | S, stop codon at 340 |
| 15B | IX | A → G at 342, deletion of C at 364 | S, stop codon at 439 | |||
| 15C | IX | A → G at 342, deletion of C at 364 | S, stop codon at 439 | |||
| 15D | XII | A → G at 342 | S | |||
| 16A | F | 30 | 43 | XIII | A → G at 342 | S |
| 16B | I | A → G at 342, deletion of 24 bases 272 to 295 | S, UK | |||
| 16C | IA | A → G at 342, deletion of 24 bases 272 to 295 | S, UK | |||
| 17A | M | 32 | 64 | I | A → G at 342 | S |
| 17B | I | A → G at 342 | S | |||
| 17C | VA | A → G at 342, insertion of 24 bases at 279 | S, UK | |||
| 18A | F | 27 | 85 | XIX | A → G at 342 | S |
| 18B | XIXA | A → G at 342 | S | |||
| 18C | XIX | A → G at 342 | S | |||
| 18D | XIX | A → G at 342 | S |
Abbreviations: M, male; F, female; FEV1, forced expiratory volume in 1 s; UK, unknown; S, silent.
All isolates had an A → G change at nt 342 (silent mutation).
Media and growth conditions.
Sputa were plated onto blood, MacConkey, and cetrimide fucidin cephaloridine (CFC) agars (Oxoid, Melbourne, Australia) for incubation at 37°C for 48 h. All P. aeruginosa isolates were identified by colony morphology, mucoidy, pigment, growth on CFC agar, a positive oxidase test, growth at 42°C on Mueller-Hinton agar (Becton Dickinson, Sydney, Australia), and resistance to C390 (Rosco, Taastrup, Denmark; Dutec Diagnostics, Sydney, Australia). Isolates showing equivocal results following these investigations were tested with the API 20 NE system (bioMerieux Vitek Inc., Marcy-l'Etoile, France). Provisional strain differentiation was made on the basis of colony size, pigment (visual assessment), and degree of mucoidy (± to +++). Strains were stored in Protect bacterial preservers (Technical Service Consultants Ltd., Heywood, Lancashire, United Kingdom) at −70°C.
PFGE analysis.
RFLP analysis and PFGE were carried out by using modifications of the procedures published by Grothues and Tummler (8) and Barth and Pitt (1). Cells grown in nutrient broth were washed with PIV buffer (1 M NaC1, 10 mM Tris-HCl [pH 7.6]) and set into low-melting-point agarose (final concentration, 1.6%; Bio-Rad Laboratories, Hercules, Calif.) plugs. The plugs were incubated overnight with lysis buffer (6 mM Tris-HCl [pH 7.6], 1 M NaCI, 0.1 M EDTA [pH 8.0], 0.2% sodium deoxycholate, 0.5% sodium lauryl sarcosine, 0.5% Brij 58, 1 mg of lysozyme per ml) at 37°C in a shaking water bath and were then incubated with 1 ml of proteolysis buffer (0.5 M EDTA [pH 8.0], 1% sodium lauryl sarcosine, 1 mg of proteinase K per ml) overnight at 50°C in a shaking water bath. The plugs were washed with 10 mM Tris-1 mM EDTA (TE; pH 8) buffer containing 0.175 mg of phenylmethylsulfonyl fluoride per ml and then four times for 30 min each time with TE buffer alone. Genomic DNA was digested with SpeI (MBI Fermentas, Vilnius, Lithuania), and macrorestriction fragments were separated by PFGE on a CHEF-DR II apparatus (Bio-Rad Laboratories) in a 1.2% agarose gel run in 0.5× Tris-borate-EDTA buffer at 14°C. Pulse-field parameters were as follows: two linear ramps of 0.5 to 25 s for 20 h, followed by 30 to 60 s for 4 h at 6 V/cm. The MidRange 1 PFG marker (New England Biolabs, Beverly, Mass.) and DNA from the PAO-1 reference strain were run on every gel. The gels were stained with ethidium bromide, visualized in a transilluminator, and photographed. A TIFF image of each gel image was created and entered into a database in Gelcompar II software (version 2.5; Applied Maths, Sint-Martens-Latem, Belgium). Each image was normalized by using PAO-1. The percent relatedness was calculated by use of the Dice coefficient, and the unweighted pair group method with arithmetic averages was used for clustering to produce a dendrogram with band optimization settings of 4% and a band position tolerance of 1.5 to 2%. A visual analysis by the method of Tenover et al. (25) was also performed. Strains with up to three band differences were considered closely related, strains with four to six band differences were considered possibly related, and strains with greater than six band differences were considered unrelated. Results were based on at least two independent experiments.
Analysis of mucA mutations by sequence analysis of PCR products.
Nucleic acids were extracted from bacteria by standard protocols with phenol. PCR performed with primers whose sequences have been published previously (12) amplified the 623 bases of the mucA gene (2), according to our optimized protocols (3.5 mM MgCl2, 0.25 μM primer, 250 μM deoxynucleoside triphosphates, and 50 ng of target per 25-μl reaction mixture). Cycling was carried out for 40 cycles at an annealing temperature of 64°C.
Sequence analysis was performed by our published procedures (18, 19) with pooled PCR products by using forward, reverse, and internal PCR primers. The PCR products were purified by polyethylene glycol precipitation. Cycle sequencing reaction mixtures containing the PCR products were assembled by using the Taq DyeDeoxy Terminator kit (ABI), and cycling was performed on a fast thermal sequencer Corbett Research, Sydney, Australia). Excess terminators were removed by phenol-chloroform extraction, the extension products were precipitated, and the products were analyzed on an ABI 378A DNA sequencer. The sequences were aligned with Clustal V software, available through Bionavigator, by using P. aeruginosa PAO-1 as the reference strain. Associations between the RFLP and PFGE profiles and mucA mutations and between the degree of mucoidy and mucA mutations were sought.
RESULTS
Phenotypic characteristics.
On the basis of phenotypic characteristics alone, 19 of the 20 patients were infected with multiple mucoid strains of P. aeruginosa.
PFGE analysis.
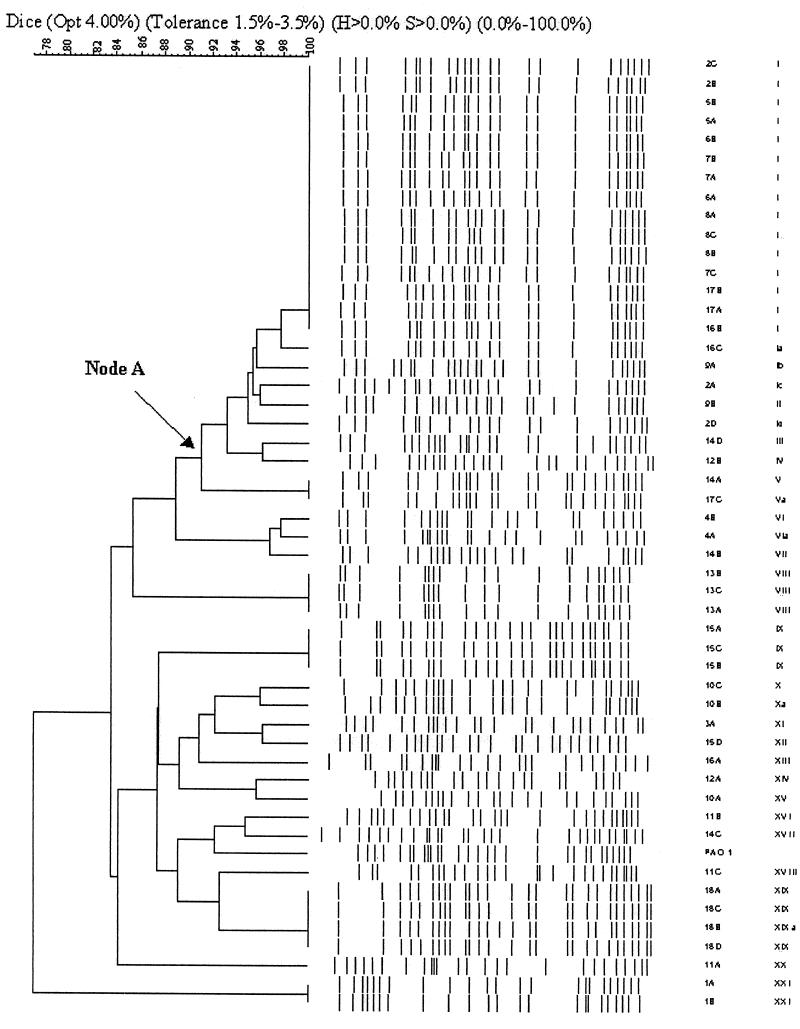
PFGE analysis carried out with 50 isolates from the 18 patients resulted in 16 to 23 bands. The dendrogram produced with Gelcompar II software (Fig. 1) showed rates of genomic similarity ranging from 76 to 100%. Twenty-four of the 50 (48%) isolates from 11 patients branched off from node A and had greater than 90% similarity. Fifteen identical isolates from seven patients (isolates 2B, C, 5A, B, 6A, B, 7A to 7C, 8A to 8C, 16B, 17A, and 17B) had the major band pattern for this cluster. No other major clusters had comparable percent similarities. The algorithmic analysis correlated well with the visual appraisal when the criteria of Tenover et al. (25) were applied. Twenty-one pulsotypes were identified: the pattern represented by the 15 identical isolates served as the reference pattern and was designated pulsotype I. Four isolates from three patients (isolates 2A, D, 9A, and 16C) showed one to three band differences from this pattern and were designated subtypes of pulsotype I. Taken together, these 19 isolates represented the dominant strain. An examination of intrapatient relationships showed that 10 of the 18 (56%) patients carried clonal or subclonal strains; five of these patients (patients 2, 5, 6, 7, and 8) carried strains belonging only to pulsotype I or its subtypes. Eight patients (patients 9, 10, 11, 12, 14, 15, 16, and 17) carried strains of different pulsotypes.
FIG. 1.
Dendrogram of P. aeruginosa isolates showing percent similarities of patterns and the pulsotypes of the individual isolates.
Sequence analysis of mucA.
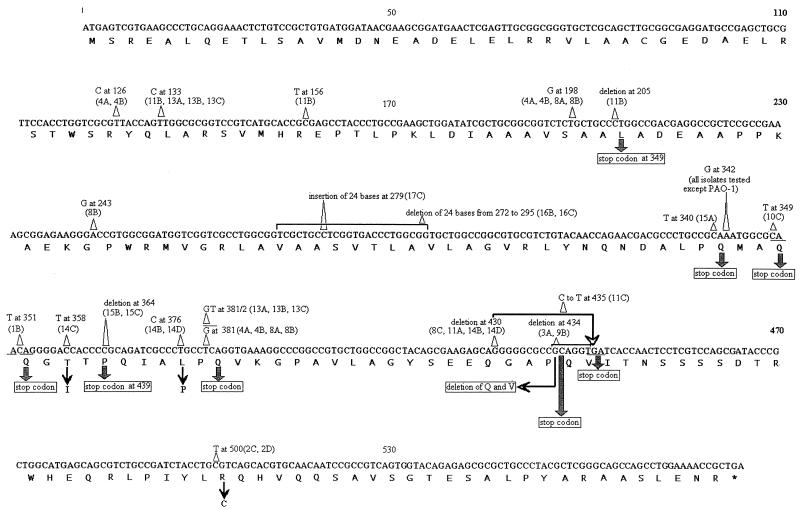
Twenty mutations were identified among the 50 isolates, with the number in individual isolates ranging from one to four (Table 1; Fig. 2). All contained a silent A-to-G transition at nucleotide (nt) 342, while the next most common change was a silent T-to-G mutation at nt 381 which affected seven isolates from three patients. Of five isolates from four patients containing large-scale changes, two contained a deletion of 24 bases at nt 272, two contained a 6-base deletion at nt 434, and one contained a 24-base insertion at nt 279. Of eight changes that led to a stop codon, five were transitions and three were single base deletions. Overall, 22 of the 50 (44%) isolates contained mucA mutations that may have accounted for their conversion to mucoidy. It was not possible to assess the relationship between the degree of mucoidy and any specific mucA mutations because of the large number of different mucA mutations identified.
FIG. 2.
Mutations in the mucA genes of P. aeruginosa clinical isolates. The mutations on the sequence of the mucA gene of nonmucoid reference strain PAO-1 are indicated.
Relationships between PFGE pattern, mucA mutation, and phenotype.
Twelve of the 19 (63%) isolates belonging to pulsotype I or its subtypes (the dominant strain) shared the same mucA mutation profile (only the silent mutation at nt 342, which was common to all 59 isolates tested); in contrast, only 12 of the 31(39%) isolates of other pulsotypes exhibited this mucA mutation pattern (P = 0.09 by the χ2 test). Of the 19 isolates comprising the dominant strain, only 5 had potentially functional mucA mutations, and only 1 of these had a change that led to a stop codon. There was no evidence of an association between the RFLP and PFGE pattern or specific mucA mutations and the phenotype among the 30 isolates for which detailed colony characteristics were available (data not shown).
DISCUSSION
This study provides the first published data on the genetic relatedness of P. aeruginosa isolates from adult CF patients in Australia. In agreement with other studies, there was substantial diversity among the strains. Nonetheless, all belonged to the same lineage on the basis of the 75% similarity cutoff designated by Grothues and Tummler (8).
The widely held concept that most CF patients harbor the same environmentally acquired strain of P. aeruginosa over many years has been challenged by several recent reports of cross infection [9, 10, 13; K. Armstrong, G. Nixon, and J. Carlin, Pediatr. Pulmonol. 20(Suppl. 20):A393, 2000]. Nonetheless, the transmissibility of P. aeruginosa isolates between CF patients remains a contentious issue. The finding that more than one-third of the patients in the study were infected with the same strain and that another patient harbored a closely related strain supports the circulation of a transmissible strain in this center. Interestingly, two patients in this group also harbored an unrelated strain(s), raising the possibility that these two patients had been superinfected by the dominant strain. There had been no coincident outpatient or inpatient visits for the patients infected with the same strain, and there had been no known social contacts between the patients over the previous 3 years. However, as all patients evaluated in the study were chronically infected with P. aeruginosa, the possibility of transmission through interactions prior to this epoch cannot be excluded. Investigations are under way to determine the overall prevalence of this dominant strain both in patients from this CF center and in the environment as a first step in differentiating among person-to-person transmission, acquisition from a common source (including a nosocomial source), and independent acquisition of common environmental strains.
As expected, almost all of the patients evaluated in the study were colonized with multiple strains of P. aeruginosa, determined on the basis of phenotypic characteristics, and just over one-half of the patients were infected with the same clone or minor variants of it. The results of recent investigations have suggested that this phenotypic variability reflects the ability of this organism to adapt and survive in the face of host immune mechanisms, the extensive use of antibiotic therapy, and the heterogeneity of the deteriorating lungs of CF patients (22, 24). Evidence has emerged indicating that these adaptations promote the selection of hypermutable or mutator strains that may be characterized by alterations in the DNA repair and error avoidance genes (14, 15). The randomly amplified polymorphic DNA patterns of the mutators were shown to be patient specific. This indicates that it is unlikely that our dominant strain is a mutator, although further investigations will be needed for confirmation. This study has confirmed a lack of a relationship between phenotype and genotype.
This study has extended the investigation of Boucher and colleagues (2), which provided the first data on the prevalence and nature of mucA mutations in clinical isolates of P. aeruginosa. Whereas the study of Boucher et al. (2) mainly documented mucA mutations in single isolates from individual patients, the present survey has focused on multiple isolates. As in the previous study, most mutations were located within the third quarter of the mucA gene, and many involved frameshifts arising from single base deletions or C-to-T mutations resulting in premature stop codons. Interestingly, the specific changes in our isolates were markedly different; the deletion of a G residue at nt 430, which affected the homopolymeric stretch of five G residues between positions 425 and 430, the most common mutation in the previous study, was present in only four isolates from three of our patients. Conversely, the A-to-G transition at nt 342, present in all of our isolates, was not reported previously. Our findings support the theory of Boucher et al. (2) that mucA mutations deregulating alginate production provide one pathway to mucoidy. Nonetheless, it was noteworthy that the overall percentage of potentially biologically significant mucA mutations in this study (44%) was markedly lower than that (84%) reported previously (2), indicating that pathways to mucoidy vary geographically. Further studies will be needed to investigate alternative pathways to mucoidy, including mutations in other genes within the mucABCD complex (2), posttranslational modification, or an unrelated mechanism such as exposure to oxygen radicals (12). The mechanisms underpinning the considerable variation in the amount of alginate produced by individual strains of P. aeruginosa remain obscure. This study provided no evidence of any association between specific mucA mutations and the degree of mucoidy, but much larger studies will be needed for confirmation of this result. This is the first study relating RFLP and PFGE patterns to mucA mutations. It seems that mucA serves as a marker of genetic instability that is not reflected by the RFLP and PFGE patterns. The most interesting finding was a trend toward an association between the dominant strain and a lack of functional mucA mutations that may well become significant in studies with larger numbers of patients. It seems that the dominant strain predominantly uses strategies other than mucA mutations for conversion to mucoidy. It may also have properties for enhanced survival or transmission.
Effective infection control programs are based on knowledge of the organisms and how they are spread. The molecular techniques used in this study facilitate tracking of strains within communities of CF patients and can help monitor the emergence of virulent strains within individuals. The role of bacterial molecular typing as an integral part of CF infection control programs warrants further investigation.
Acknowledgments
This study was supported by a National Health and Medical Research Council of Australia project grant.
We acknowledge the assistance provided by Carmel Moriarty and Paul Torzillo.
REFERENCES
- 1.Barth, A. L., and T. L. Pitt. 1995. Auxotrophy of Burkholderia (Pseudomonas) cepacia from cystic fibrosis patients. J. Clin. Microbiol. 33:2192-2194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Boucher, J. C., H. Mudd, and V. Deretic. 1997. Mucoid Pseudomonas aeruginosa in cystic fibrosis: characterization of muc mutations in clinical isolates and analysis of clearance in a mouse model of respiratory infection. Infect. Immun. 65:3838-3846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Boukadida, J., M. De Montalembert, G. Lenoir, P. Scheinmann, M. Veron, and P. Berche. 1993. Molecular epidemiology of chronic pulmonary colonisation by Pseudomonas aeruginosa in cystic fibrosis. J. Med. Microbiol. 38:29-33. [DOI] [PubMed] [Google Scholar]
- 4.Breitenstein, S., S. Walter, J. Bosshammer, U. Romling, and B. Tummler. 1997. Direct sputum analysis of Pseudomonas aeruginosa macrorestriction fragment genotypes in patients with cystic fibrosis. Med. Microbiol. Immunol. 186:93-99. [DOI] [PubMed] [Google Scholar]
- 5.Cheng, K., R. L. Smyth, J. R. Govan, C. Doherty, C. Winstanley, N. Denning, D. P. Heaf, H. van Saene, and C. A. Hart. 1996. Spread of beta-lactam-resistant Pseudomonas aeruginosa in a cystic fibrosis clinic. Lancet 348:639-642. [DOI] [PubMed] [Google Scholar]
- 6.Govan, J. R., and V. Deretic. 1996. Microbial pathogenesis in cystic fibrosis: mucoid Pseudomonas aeruginosa and Burkholderia cepacia. Microbiol. Rev. 60:539-574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Grothues, D., U. Koopmann, H. von der Hardt, and B. Tummler. 1988. Genome fingerprinting of Pseudomonas aeruginosa indicates colonization of cystic fibrosis siblings with closely related strains. J. Clin. Microbiol. 26:1973-1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Grothues, D., and B. Tummler. 1991. New approaches in genome analysis by pulsed-field gel electrophoresis: application to the analysis of Pseudomonas species. Mol. Microbiol. 5:2763-2776. [DOI] [PubMed] [Google Scholar]
- 9.Hunfeld, K. P., C. Schmidt, B. Krackhardt, H. G. Posselt, J. Bargon, Y. Yahaf, V. Schafer, V. Brade, and T. A. Wichelhaus. 2000. Risk of Pseudomonas aeruginosa cross-colonisation in patients with cystic fibrosis within a holiday camp—a molecular-epidemiological study. Wien. Klin. Wochenschr. 112:329-333. [PubMed] [Google Scholar]
- 10.Jones, A. M., J. R. Govan, C. J. Doherty, M. E. Dodd, B. J. Isalska, T. N. Stanbridge, and A. K. Webb. 2001. Spread of a multiresistant strain of Pseudomonas aeruginosa in an adult cystic fibrosis clinic. Lancet 358:557-558. [DOI] [PubMed] [Google Scholar]
- 11.Mahenthiralingam, E., M. E. Campbell, J. Foster, J. S. Lam, and D. P. Speert. 1996. Random amplified polymorphic DNA typing of Pseudomonas aeruginosa isolates recovered from patients with cystic fibrosis. J. Clin. Microbiol. 34:1129-1135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mathee, K., O. Ciofu, C. Sternberg, P. W. Lindum, J. I. Campbell, P. Jensen, A. H. Johnsen, M. Givskov, D. E. Ohman, S. Molin, N. Hoiby, and A. Kharazmi. 1999. Mucoid conversion of Pseudomonas aeruginosa by hydrogen peroxide: a mechanism for virulence activation in the cystic fibrosis lung. Microbiology 145:1349-1357. [DOI] [PubMed] [Google Scholar]
- 13.McCallum, S. J., J. Corkill, M. Gallagher, M. J. Ledson, C. A. Hart, and M. J. Walshaw. 2001. Superinfection with a transmissible strain of Pseudomonas aeruginosa in adults with cystic fibrosis chronically colonised by P aeruginosa. Lancet 358:558-560. [DOI] [PubMed] [Google Scholar]
- 14.Oliver, A., R. Canton, P. Campo, F. Baquero, and J. Blazquez. 2000. High frequency of hypermutable Pseudomonas aeruginosa in cystic fibrosis lung infection. Science 288:1251-1253. [DOI] [PubMed] [Google Scholar]
- 15.Oliver, A., F. Bacquero, and J. Blazquez. 2002. The mismatch repair system (mutS, mutL and uvrD genes) in Pseudomonas aeruginosa: molecular characterization of naturally occurring mutants. Mol. Microbiol. 43:1641-1650. [DOI] [PubMed] [Google Scholar]
- 16.Pujana, I., L. Gallego, G. Martin, F. Lopez, J. Canduela, and R. Cisterna. 1999. Epidemiological analysis of sequential Pseudomonas aeruginosa isolates from chronic bronchiectasis patients without cystic fibrosis. J. Clin. Microbiol. 37:2071-2073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Romling, U., D. Grothues, U. Koopmann, B. Jahnke, J. Greipel, and B. Tummler. 1992. Pulsed-field gel electrophoresis analysis of a Pseudomonas aeruginosa pathovar. Electrophoresis 13:646-648. [DOI] [PubMed] [Google Scholar]
- 18.Rose, B. R., C. H. Thompson, L. A. Chantrill, M. H. Tattersall, and Y. E. Cossart. 1992. Prevalence and distribution of human papillomavirus type-16 DNA in pelvic lymph nodes of patients with cervical cancer and in women with no history of cervical abnormality. Int. J. Cancer 52:225-228. [DOI] [PubMed] [Google Scholar]
- 19.Rose, B. R., C.H. Thompson, J. Zhang, M. Stoeter, A. Stephen, H. Pfister, M. H. N. Tattersall, and Y. E. Cossart. 1997. Sequence variation in the upstream regulatory region of HPV 18 isolates from cervical cancers. Gynecol. Oncol. 66:282-289. [DOI] [PubMed] [Google Scholar]
- 20.Sader, H. S., A. C. Pignatari, I. L. Leme, M. N. Burattini, R. Tancresi, R. J. Hollis, and R. N. Jones. 1993. Epidemiologic typing of multiply drug-resistant Pseudomonas aeruginosa isolated from an outbreak in an intensive care unit. Diagn. Microbiol. Infect. Dis. 17:13-18. [DOI] [PubMed] [Google Scholar]
- 21.Silbert, S., A. L. Barth, and H. S. Sader. 2001. Heterogeneity of Pseudomonas aeruginosa in Brazilian cystic fibrosis patients. J. Clin. Microbiol. 39:3976-3981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sniegowski, P. D., P.J. Gerrish, and R. E. Lenski. 1997. Evolution of high mutation rates in experimental populations of E. coli. Nature 387:703-705. [DOI] [PubMed] [Google Scholar]
- 23.Struelens, M. J., V. Schwam, A. Deplano, and D. Baran. 1993. Genome macrorestriction analysis of diversity and variability of Pseudomonas aeruginosa strains infecting cystic fibrosis patients. J. Clin. Microbiol. 31:2320-2326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Taddei, F., M. Radman, J. Maynard-Smith, B. Toupance, P. H. Gouyon, and B. Godelle. 1997. Role of mutator alleles in adaptive evolution. Nature 387:700-702. [DOI] [PubMed] [Google Scholar]
- 25.Tenover, F. C., R. D. Arbeit, R. V. Goering, P. A. Mickelsen, B. E. Murray, D. H. Persing, and B. Swaminathan. 1995. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J. Clin. Microbiol. 33:2233-2239. [DOI] [PMC free article] [PubMed] [Google Scholar]