Abstract
As part of a longitudinal study of antimicrobial resistance among salmonellae isolated from swine, we studied 484 Salmonella enterica subsp. enterica serovar Typhimurium (including serovar Typhimurium var. Copenhagen) isolates. We found two common pentaresistant phenotypes. The first was resistance to ampicillin, chloramphenicol, streptomycin, sulfamethoxazole, and tetracycline (the AmCmStSuTe phenotype; 36.2% of all isolates), mainly of the definitive type 104 (DT104) phage type (180 of 187 isolates). The second was resistance to ampicillin, kanamycin, streptomycin, sulfamethoxazole, and tetracycline (the AmKmStSuTe phenotype; 44.6% of all isolates), most commonly of the DT193 phage type (77 of 165 isolates), which represents an unusual resistance pattern for DT193 isolates. We analyzed 64 representative isolates by amplified fragment length polymorphism (AFLP) analysis, which revealed DNA fingerprint similarities that correlated with both resistance patterns and phage types. To investigate the genetic basis for resistance among DT193 isolates, we characterized three AmKmStSuTe pentaresistant strains and one hexaresistant strain, which also expressed resistance to gentamicin (Gm phenotype), all of which had similar DNA fingerprints and all of which were collected during the same sampling. We found that the genes encoding the pentaresistance pattern were different from those from isolates of the DT104 phage type. We also found that all strains encoded all of their resistance genes on plasmids, unlike the chromosomally encoded genes of DT104 isolates, which could be transferred to Escherichia coli via conjugation, but that the plasmid compositions varied among the isolates. Two strains (strains UT08 and UT12) had a single, identical plasmid carrying blaTEM (which encodes ampicillin resistance), aphA1-Iab (which encodes kanamycin resistance), strA and strB (which encode streptomycin resistance), class B tetA (which encodes tetracycline resistance), and an unidentified sulfamethoxazole resistance allele. The third pentaresistant strain (strain UT20) was capable of transferring by conjugation two distinct resistance patterns, AmKmStSuTe and KmStSuTe, but the genes were carried on plasmids with slightly different restriction patterns (differing by a single band of 15 kb). The hexaresistant strain (strain UT30) had the same plasmid as strains UT08 and UT12, but it also carried a second plasmid that conferred the AmKmStSuGm phenotype. The second plasmid harbored the gentamicin resistance methylase (grm), which has not previously been reported in food-borne pathogenic bacteria. It also carried the sul1 gene for sulfamethoxazole resistance and a 1-kb class I integron bearing aadA for streptomycin resistance. We also characterized isolates of the DT104 phage type. We found a number of isolates that expressed resistance only to streptomycin and sulfamethoxazole (the StSu phenotype; 8.3% of serovar Typhimurium var. Copenhagen strains) but that had AFLP DNA fingerprints similar or identical to those of strains with genes encoding the typical AmCmStSuTe pentaresistance phenotype of DT104. These atypical StSu DT104 isolates were predominantly cultured from environmental samples and were found to carry only one class I integron of 1.0 kb, in contrast to the typical two integrons (InC and InD) of 1.0 and 1.2 kb, respectively, of the pentaresistant DT104 isolates. Our findings show the widespread existence of multidrug-resistant Salmonella strains and the diversity of multidrug resistance among epidemiologically related strains. The presence of resistance genes on conjugative plasmids and duplicate genes on multiple plasmids could have implications for the spread of resistance factors and for the stability of multidrug resistance among Salmonella serovar Typhimurium isolates.
The frequency of resistance among food-borne pathogens has increased dramatically, presumably due to the extensive use of antimicrobial agents in human and veterinary medicine (8, 26). Furthermore, resistance to combinations of several classes of antimicrobials has led to the emergence of multidrug-resistant (MDR) strains that may pass from food animals to humans (30, 48). One important pathogen known to harbor multiple resistance factors is Salmonella, one of the leading causes of food-borne bacterial diseases. It is estimated that the annual economic costs due to food-borne Salmonella infections in the United States are $2.4 billion (http://www.ers.usda.gov).
MDR Salmonella isolates have been reported since the 1960s (1), with the resistance patterns of Salmonella serovars of public health importance often associated with specific phage types. One notable MDR strain is Salmonella enterica subsp. enterica serovar Typhimurium definitive type 104 (DT104). It was first recognized in the United Kingdom (39) and since has been reported in many parts of the world (6, 16, 18, 19, 27) and from various host species including food animals and pets (12, 22, 43, 46) as well as from processed ready-to-eat meat products (47). DT104 strains are commonly known to be pentaresistant, exhibiting resistance to ampicillin, chloramphenicol, streptomycin, sulfamethoxazole, and tetracycline (the AmCmStSuTe resistance phenotype). The drug resistance spectra of MDR strains of Salmonella serovars have also been expanding in recent years (13, 23, 28, 40, 49).
Another important phage type of serovar Typhimurium often exhibiting multidrug resistance is DT193, an MDR strain responsible for outbreaks in humans in the late 1980s and early 1990s, mainly in Europe. Two of the major outbreaks due to DT193 (in Italy and the United Kingdom) were traced to contaminated pork products (25, 32). Preliminary analysis of the resistance determinants has shown that the isolates collected from the outbreak in the United Kingdom carried their resistance factors on conjugative plasmids (17). This phage type has been among the most common MDR strains that we identified among swine isolates (15). Since at least two DT193 outbreaks affecting humans have been traced to contaminated pork products within the last decade (25, 32) and since serovar Typhimurium isolates with similar resistance patterns have been reported to be increasingly prevalent in recent years (1998 report of the National Antimicrobial Resistance Monitoring System [http://www.cdc.gov/ncidod/dbmd/narms/annual/1998_anu.htm]), we thought that it would be important to characterize the genetic basis of multidrug resistance in these strains commonly isolated from swine in our study.
The genetic characterization of antimicrobial resistance genes as well as their location and diversity is important in identifying factors involved in resistance, understanding the diversity of MDR strains, identifying genetic linkages among markers, understanding potential transfer mechanisms, and developing efficient detection methods. In this study, we identified two predominant pentaresistant MDR phenotypes among serovar Typhimurium (and serovar Typhimurium var. Copenhagen) isolates, determined the genetic diversities of these isolates using amplified fragment length polymorphism (AFLP) DNA fingerprinting techniques, identified the resistance genes involved, and determined the locations and diversities of these resistance genes.
MATERIALS AND METHODS
Collection of Salmonella isolates.
We collected fecal samples from two swine production companies using all-in and all-out management systems. Samples from a total of 6 nursery farms and 18 finishing farms were included in the study. At each visit, 96 fecal samples and 30 feed samples were collected. In addition, environmental samples were collected from each barn. Ten drag swab samples were collected from cleaned and apparently disinfected empty barns at the finishing stage before pigs were moved in from nursery barns. Sample collection was performed for three replicates (cohorts of pigs) between 1997 and 2000. A total of 7,452 samples (6,912 fecal samples and 540 drag swab environmental samples) were collected for this study. With an average prevalence of 21% per group, a total of 1,565 Salmonella isolates of 30 different serovars were detected. The most common serovar, serovar Typhimurium (including serovar Typhimurium var. Copenhagen), constitutes more than 75% of the total. In this study, a subset of the total, 484 Salmonella serovar Typhimurium and Salmonella serovar Typhimurium var. Copenhagen isolates, was characterized.
Isolation, identification, and antimicrobial resistance.
We isolated salmonellae using conventional methods, as described previously (2). Antimicrobial susceptibility was tested with the Vitek Jr. (VITEK system manual, 1996; Biomerieux Vitek, Hazelwood, Mo.) semiautomated system for 11 antimicrobials. The antimicrobials (and the respective breakpoints for resistance, given in parentheses) were amikacin (32 μg/ml), amoxicillin-clavulanic acid (16 μg/ml), ampicillin (16 μg/ml), cefotaxime (16 μg/ml), cephalothin (16 μg/ml), chloramphenicol (16 μg/ml), ciprofloxacin (2 μg/ml), gentamicin (8 μg/ml), piperacillin (32 to 64 μg/ml), tetracycline (8 μg/ml), and trimethoprim-sulfamethoxazole (40 to 80 μg/ml). The breakpoints of the Vitek Jr. system indicated above were matched with the NCCLS standard breakpoints for gram-negative enteric organisms, and the following quality control strains were routinely used: Escherichia coli ATCC 25922, Pseudomonas aeruginosa ATCC 27853, Staphylococcus aureus ATCC 29213, and Enterococcus faecalis ATCC 29212 (29). The Kirby-Bauer disk diffusion method for three more antimicrobials, kanamycin, streptomycin, and sulfamethoxazole, was performed as described previously (15). Serotyping and phage typing were performed at the National Veterinary Services Laboratories, Ames, Iowa.
Identification of resistance gene alleles.
We identified resistance genes either by PCR or, when necessary, by cloning of PCR products and sequencing of the cloned genes. The primers used for PCR are listed in Table 1. We used the following PCR conditions: denaturation at 95°C for 5 min and then 40 cycles of denaturation at 95°C for 1 min, annealing at 53°C for 30 s, and extension at 72°C for 30 s.
TABLE 1.
PCR primers used in identification of resistance genes
| Gene | Oligonucleotide sequencesa | Product size (bp) | Reference or source |
|---|---|---|---|
| blaPSE1 | F, TTT GGT TCC GCG CTA TCT G; R, TAC TCC GAG CAC CAA ATC CG | 150 | 9 |
| blaTEM | F, GCA CGA GTG GGT TAC ATC GA; R, GGT CCT CCG ATC GTT GTC AG | 310 | 9 |
| aphA1-lab | F, AAA CGT CTT GCT CGA GGC; R, CAA ACC GTT ATT CAT TCG TGA | 500 | 14 |
| aadA | F, GTG GAT GGC GGC CTG AAG CC; R, AAT GCC CAG TCG GCA GCG | 528 | 24 |
| strA | F, CTT GGT GAT AAC GGC AAT TC; R, CCA ATC GCA GAT AGA AGG C | 548 | AF273682 |
| strB | F, ATC GTC AAG GGA TTG AAA CC; R, GGA TCG TAG AAC ATA TTG GC | 509 | AF024602 |
| sulA | F, CAC TGC CAC AAG CCG TAA; R, GTC CGC CTC AGC AAT ATC | 360 | AF071555 |
| tetA | F, GCT ACA TCC TGC TTG CCT TC; R, CAT AGA TCG CCG TGA AGA GG | 210 | X61367 |
| tetB | F, TTG GTT AGG GGC AAG TTT TG; R, GTA ATG GGC CAA TAA CAC CG | 659 | 19 |
| tetG | F, CAG CTT TCG GAT TCT TAC GG; R, GAT TGG TGA GGC TCG TTA GC | 844 | 19 |
| grm | F, AAG CGC ACG AAG CGC GGG CTG; R, AAG GCG GGC CTC AAG GAG GTC | 414 | 14 |
| aadB | F, GAG CGA AAT CTG CCG CTC TTG; R, CTG TTA CAA CGG ACT GGC CGC | 310 | 14 |
| aac(6)-I | F, TGA GCA TGA CCT TGC GAT; R, GAA CAG CAA CTC AAC CAG | 337 | AF282595 |
| int | F, GGC ATC CAA GCA GCA AG; R, AAG CAG ACT TGA CCT GA | Variable | 19 |
F, forward; R, reverse.
In order to identify the resistance gene cassette(s) carried on integrons among non-DT104 strains with pentaresistant phenotypes, we cloned and sequenced the integron amplicon. Briefly, class I integron primers with ends compatible with XbaI were designed, and DNA fragments were amplified with primers common to class I integrons by using the PCR conditions listed above. The DNA product was purified and digested with XbaI and cloned into a plasmid vector (pPCR-Script Cam; Stratagene, La Jolla, Calif.). E. coli DH5α was transformed by electroporation, and candidate β-galactosidase-negative white colonies were selected on 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside. We verified the presence of inserts by isolation of plasmid DNA, and inserts were sequenced with an Applied Biosystems automated sequencer.
DNA fingerprinting.
We used a previously described AFLP fingerprinting method (41), with modifications. This method has been shown to have superior discriminatory power and reproducibility for the subtyping of Salmonella serovars (21, 31), and, in addition, we found this method to be both economical and efficient. Briefly, cells were grown overnight on Luria-Bertani broth (Difco, Atlanta, Ga.), and the genomic DNA was purified with a Qiagen DNeasy tissue kit (Qiagen, Valencia, Calif.), as described in the DNAeasy Tissue Kit Handbook (p. 16-18; Qiagen GmbH, Hilden, Germany, 1999) and adjusted to a concentration of 50 ng/μl in a volume of 10 μl of water. The DNA was digested with EcoRI and MseI at 37°C for 1 h. Adapters were then ligated to each end of the restriction fragments with T4 DNA ligase (New England Biolabs) at 16°C overnight. The fragments were then amplified with primers specific for the EcoRI adapter (forward primer, 5′-GACTGCGTACCAAATC) and the MseI adapter (reverse primer, 5′-GATGAGTCCTGAGTAA), ensuring that only those fragments with one EcoRI end and one MseI end were amplified. The conditions for amplification were 94°C for 15 s, 60°C for 30 s with increases of 1 s per cycle for 28 cycles, and then incubation at 72°C for 2 min. The amplified fragments then underwent a second round of amplification, this time with an infrared-labeled EcoRI primer (Licor, Lincoln, Nebr.). An additional adenine was added to this primer at its 3′ end (5′-GACTGCGTACCAAATCA) in order to obtain the fewer bands optimum for band scoring and fingerprint analysis. This final selective amplification consisted of 13 cycles at 94°C for 10 s, 65°C for 30 s, and 72°C for 1 min and then 25 cycles of 94°C for 10 s, 56°C for 30 s, and 72°C for 1 min, with ramping at 1 s/cycle for the final step. The reaction mixture was finally incubated for 2 min at 72°C. The fragments were then separated on a Licor 4200 DNA sequencer. A detailed AFLP protocol can be found at http://www4.ncsu.edu/∼wagebrey/AFLP-Salm.pdf. The bands were scored and analyzed with Quantity One software (version 4.1.1; Bio-Rad, Richmond, Calif.). The fingerprints were analyzed by using the Dice coefficient algorithm, and dendrograms were constructed by the unweighted pair group method with arithmetic averages clustering method.
Conjugation and restriction patterns of plasmids.
We performed conjugation experiments to determine whether antimicrobial resistance markers were located on conjugative plasmids. Candidate MDR donor strains were mated with a spontaneous rifampin-resistant derivative of E. coli K-12 strain MG1655. Single colonies of each of the donor and the recipient were mixed on Luria-Bertani agar, and the mixture was incubated for 6 h at 37°C. The mixture was then transferred to a selective plate containing rifampin and one of the antimicrobials to be tested and incubated at 37°C overnight. Transconjugants were further purified and confirmed to be E. coli rather than spontaneous rifampin-resistant salmonellae by growth on MacConkey agar. Selective plating and PCR amplification were used to test for the presence of other resistance markers. The antibiotics used in the selective plates (and their respective concentrations, given in parentheses) were as follows: ampicillin (100 μg/ml), kanamycin (50 μg/ml), tetracycline (25 μg/ml), and gentamicin (16 μg/ml).
To determine the size and restriction fragment patterns of plasmids conjugated into E. coli, plasmid DNA was purified from each transconjugant and digested with HindIII. The fragments were run on 1% agarose in 1× Tris-acetate-EDTA buffer. The size of each plasmid was determined by adding all fragment sizes.
RESULTS
Phenotypic characterization of MDR serovar Typhimurium and serovar Typhimurium var. Copenhagen.
In a longitudinal study of swine salmonellosis in commercial swine production systems at multiple sites in North Carolina, we analyzed a total of 484 isolates (156 Salmonella serovar Typhimurium isolates and 328 Salmonella serovar Typhimurium var. Copenhagen isolates) collected in three replicate samplings of 18 groups of pigs. We found two common pentaresistant MDR phenotypes: resistance to ampicillin, chloramphenicol, streptomycin, sulfamethoxazole, and tetracycline (the AmCmStSuTe phenotype; 36.2%) or resistance to ampicillin, kanamycin, streptomycin, sulfamethoxazole, and tetracycline (the AmKmStSuTe phenotype; 44.6%). Phage typing resulted in the classification of the serovar Typhimurium isolates (including serovar Typhimurium var. Copenhagen isolates) into eight different phage types: DT104, DT193, DT21, DT208, DT12, U302, DT169, and DT120. Analysis of isolates with the aforementioned two pentaresistance patterns showed that DT104 predominated among isolates with the AmCmStSuTe resistance pattern (106 of 121 isolates for which the phage type was obtained), while phage type U302 was less commonly represented among isolates with this pentaresistant phenotype (11 of 121 isolates). The isolates with the AmKmStSuTe pentaresistance pattern were predominantly of the DT193 phage type (53 of 59 isolates) among serovar Typhimurium var. Copenhagen isolates and the DT21 phage type among the serovar Typhimurium isolates (82 of 106 isolates).
Genetic diversity of MDR strains.
In order to determine the extent of genetic diversity among these MDR strains of Salmonella serovar Typhimurium (including Salmonella serovar Typhimurium var. Copenhagen), we performed DNA fingerprinting using AFLP analysis of 64 isolates originating from eight farms. The dendrogram in Fig. 1 shows the similarities of these strains on the basis of the fragment sizes of the amplified bands. Isolates with the same letter code (e.g., UT) were collected from the same farm on the same visit. The results show that two major clusters of Typhimurium and serovar Typhimurium var. Copenhagen were identified. The first cluster (Fig. 1, cluster 1, A to C) included 19 of the 64 isolates fingerprinted. The predominant resistance pattern was pentaresistance of the AmKmStSuTe phenotype (15 of 19 isolates), but isolates with the AmKmStSuTeGm (1 of 19 isolates), StTe (1 of 19 isolates), and AmCmStSuTe (2 of 19 isolates) phenotypes were also seen. In this cluster, only one isolate was found from an environmental sample (a drag swab of the barn floor), and this isolate exhibited the AmCmStSuTe resistance phenotype. The predominant phage type in this cluster was DT193 (11 of 19 isolates), followed by DT12 (4 of 19 isolates), DT104 (1 of 19 isolates), and U302 (1 of 19 isolates), along with two untypeable isolates. Isolates in this cluster were derived from only four of the eight farms.
FIG. 1.
Genetic diversity between the two common pentaresistant MDR Salmonella serovar Typhimurium phenotypes based on AFLP fingerprinting. Two major clusters are shown: cluster 1, the AmKmStSuTe resistance phenotype cluster (with three subclusters, subclusters 1A to 1C); and cluster 2, the DT104 cluster (with five subclusters, subclusters 2A to 2E). ∗, isolates collected from environmental samples (drag swabs); §, isolates in cluster 1 that encode β-lactam resistance using blaPSE-1; the remainder of the isolates with ampicillin resistance in cluster 1 carry blaTEM; ¶, isolates in cluster 2 carrying blaTEM; the remainder of the isolates with ampicillin resistance in cluster 2 carry blaPSE-1.
The second cluster (Fig. 1, cluster 2, A to E) included 45 of the 64 isolates fingerprinted. The predominant resistance phenotypes in this cluster were AmCmStSuTe (31 of 45 isolates) and StSu (9 of 45 isolates). Rarely, isolates with the hexaresistance patterns were also found: UF19 with the AmCmStSuTeCe phenotype (2 of 45 isolates) and UH03 with the AmCmStSuTeGm phenotype (1 of 45 isolates). The remaining two isolates, UO02 and UM06, showed a pentaresistance pattern (the AmKmStSuTe phenotype), which was the predominant resistance phenotype of cluster 1. The predominant phage type in the second cluster was DT104 (41 of 45 isolates), with isolates of this phage type commonly exhibiting one of two resistance phenotypes: AmCmStSuTe or StSu. In addition to DT104, one isolate of each of phage types DT193, DT21, DT169, and U302 was found within this cluster. MDR strains in this cluster were isolated from all eight farms: 15 (34%) from environmental samples (drag swabs) and 30 (66%) from fecal samples.
Characterization of non-DT104 pentaresistant serovar Typhimurium and serovar Typhimurium var. Copenhagen isolates.
We have identified a group of MDR strains with the AmKmStSuTe resistance pattern. Strains of phage types DT193, DT12, DT21, and DT208 exhibited this resistance phenotype, the most common one being DT193. This phage type has been implicated in disease outbreaks, but the resistance profile that we have most often seen (AmKmStSuTe) is different from those that have commonly been observed in previous studies: either AmStSuTe (3, 11, 17) or Te alone (17). We chose four isolates for genetic characterization: isolates UT08, UT12, and UT20 (all with the AmKmStSuTe resistance phenotype) and isolate UT30 (with the AmKmStSuTeGm resistance phenotype). We chose these isolates for two reasons. First, all four isolates were collected from the same farm at the same time. Thus, characterization of these isolates enabled us to study the similarity (or diversity) of phenotypically similar isolates obtained from the same environment. Second, we found that UT08 and UT12 had identical fingerprints, consistent with their identical resistance phenotypes, but that UT20 had a different fingerprint, although it shared the same pentaresistance phenotype. In addition, we noticed that the fingerprint of the hexaresistant UT30 strain was similar to those of UT08 and UT12, even though its resistance pattern included resistance to gentamicin (the Gm phenotype). Thus, we were interested in studying further this disparity between fingerprint similarity and resistance phenotype.
Conjugal transfer of resistance genes and plasmid characterization.
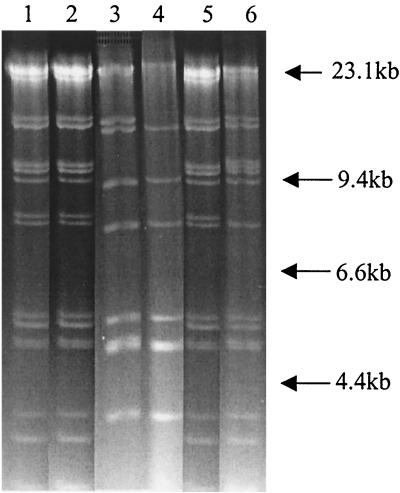
To determine whether any of the resistance genes of these MDR strains were located on conjugative plasmids, we conjugated each of the four donors (isolates UT08, UT12, UT20, and UT30) with a rifampin-resistant derivative of E. coli K-12 strain MG1655. We found that all of the resistance markers in all four isolates could be transferred by conjugation and thus were encoded on conjugative plasmids. However, the numbers and restriction patterns of the conjugative plasmids carrying resistance markers varied within and between strains. For UT08 and UT12, conjugation with selection for resistance to ampicillin, kanamycin, or tetracycline always yielded transconjugants resistant to all five antimicrobials to which the donor strains were resistant. Restriction enzyme analysis revealed that UT08 and UT12 each transfer approximately 140 kb of plasmid DNA harboring all five of the resistance genes (Fig. 2, lanes 1 and 2). These plasmids also had identical restriction fragment patterns when they were cut with HindIII (Fig. 2) and other enzymes (data not shown). Thus, it is likely that these two strains, which have identical resistance patterns and fingerprints and which carry the same plasmid, are in fact clonal. UT30, however, was able to transfer by conjugation plasmids with two distinct resistance patterns and restriction maps. Despite the similarity of its fingerprint to those of UT08 and UT12, it could transfer by conjugation a plasmid encoding resistance to amikacin, kanamycin, streptomycin, sulfamethoxazole, and tetracycline as well as one producing resistance to amikacin, kanamycin, streptomycin, sulfamethoxazole, and gentamicin. The first of these, which encoded the AmKmStSuTe resistance phenotype, was identical to the plasmids isolated from UT08 and UT12; it was 140 kb and had the same HindIII restriction pattern as the plasmids isolated from UT08 and UT12 (Fig. 2, lane 5). The second plasmid, which encoded the AmKmStSuGm resistance phenotype, was approximately 145 kb and had a restriction pattern different from those of the other plasmids (Fig. 2, lane 6). Thus, it is possible that UT30 is a derivative of UT08 (and/or UT12) that has acquired a second conjugative plasmid that encodes resistance to gentamicin and duplicates the resistance to the other four antimicrobials already present in these strains. UT20 also carried two conjugative plasmids with restriction patterns different from those of the plasmids found in the other three strains: an 85-kb plasmid (pUT20A) carried genes encoding resistance to all five antimicrobials found in UT20 (Fig. 2, lane 3), and a second plasmid (pUT20B) of about 70 kb harbored genes encoding resistance to kanamycin, streptomycin, sulfamethoxazole, and tetracycline (Fig. 2, lane 4). Apart from the presence of a 15-kb band in pUT20A, the two plasmids from UT20 exhibited similar restriction patterns. The similarities of the restriction patterns of these two plasmids suggest that they may in fact be related plasmids.
FIG. 2.
HindIII restriction fragment patterns of the four plasmids isolated from isolates of phage type DT193 isolated from the same location (farm) at the same time with similar pentaresistance phenotypes (AmKmStSuTe and AmKmStSuTeGm with the class I integron). The lanes and antimicrobial resistance phenotypes of these plasmids are as follows: 1, pUT08, AmKmStSuTe; 2, pUT12, AmKmStSuTe; 3, pUT20A, AmKmStSuTe; 4, pUT20B KmStSuTe; 5, pUT30A, AmKmStSuTe; 6, pUT30B, AmKmStSuGm (with the class I integron).
Identification of antimicrobial resistance genes.
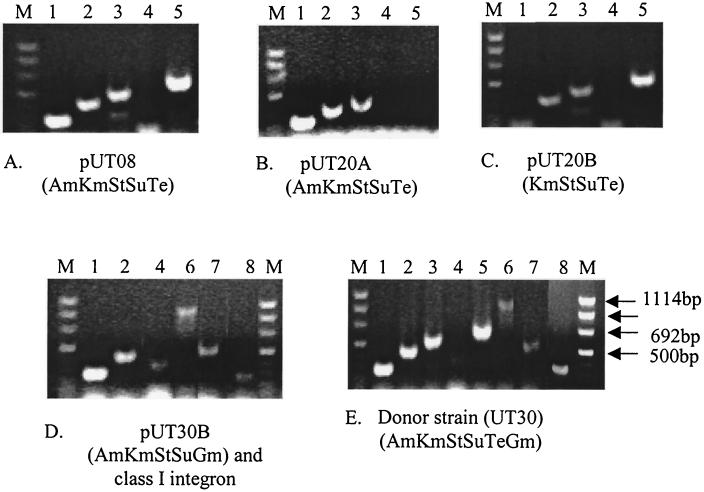
To characterize the genetic basis of resistance in the four plasmids isolated and described above, we identified resistance genes using PCR and cloning. Since strains UT08, UT12, and UT20 appear to carry the same plasmid, we used the plasmid originally obtained from UT08 (pUT08) to represent the other two in these studies. Unlike DT104, which encodes β-lactam resistance using the blaPSE-1 β-lactamase gene, all three plasmids with a phenotype of resistance to β-lactams (pUT08, pUT20A, and pUT30) encoded ampicillin resistance by means of the blaTEM gene (Fig. 3A, B, D, and E). Further PCR testing of an additional 59 MDR isolates with the AmKmStSuTe resistance pattern revealed that they all carried the blaTEM gene (data not shown). We found that all plasmids encoding kanamycin resistance did so by means of the aminoglycoside phosphotransferase gene aphA1-1ab and that all plasmids encoding tetracycline resistance had tetB (Fig. 3A to E). However, we identified two different streptomycin resistance alleles: streptomycin-6-phosphotransferase (strA and strB) was the sole streptomycin resistance determinant in three of the plasmids (pUT08, pUT20A, and pUT20B). On plasmid pUT30B, however, we also found the aminoglycoside adenyltransferase gene (aadA), in addition to strAB (Fig. 3D and E). Using a primer pair homologous to regions of class I integrons that flank the antibiotic resistance cassettes (Table 1), we amplified by PCR an approximately 1-kb product from plasmid pUT30B. Cloning and sequencing of this product revealed that aadA is encoded on this class I integron. We found sulfamethoxazole resistance encoded by the dihydropteroate synthase gene (sul1) in pUT30, but we were not able to detect this gene on the other three plasmids (Table 2). The gene responsible for sulfonamide resistance on these plasmids remains to be identified. To identify the gene responsible for gentamicin resistance, we designed primers specific for three distinct bacterial genes known to encode resistance to gentamicin [aac(6)-I, aadB, and grm] and attempted to amplify each from plasmid pUT30. We found that we could detect only the gentamicin resistance methylase (grm) of Micromonospora purpurae, an allele not previously identified in pathogenic bacteria (Fig. 3E). The presence of both a class I integron and the gentamicin resistance gene on the same plasmid derived from UT30 could have important implications in the expansion of the multidrug resistance spectrum, with the possibility of additional resistance genes being captured by the integron and the potential for the horizontal transfer of resistance genes within or between bacterial species.
FIG. 3.
PCR amplification of antimicrobial resistance genes among four plasmids isolated from Salmonella serovar Typhimurium phage type DT193 isolates. The identification number and resistance phenotype of each plasmid are indicated under each panel. The amplified genes shown in the five panels (A to E) are as follows: β-lactamase TEM (blaTEM) (lanes 1), aminoglycoside phosphotransferase (aphA1-Iab) (lanes 2), streptomycin-6-phosphotransferase (strA) (lanes 3), dihydropteroate synthase (sul1) (lanes 4), class B tetracycline resistance (tetB) (lanes 5), class I integron (int1) (lanes 6), aminoglycoside adenylyltransferase (aadA) (lanes 7), and gentamicin resistance methylase (grm) (lanes 8). Lanes M, molecular size markers.
TABLE 2.
Summary of antimicrobial resistance genes and class I integrons identified among Salmonella isolates of phage types DT193, DT104, and U302
| Phage type | Plasmid or pattern | Gene(s) for resistance to antimicrobials
|
Class I integron size/location | |||||
|---|---|---|---|---|---|---|---|---|
| Amikacin | Kanamycin | Streptomycin | Sulfamethoxazole | Tetracycline | Gentamicin | |||
| DT193 | pUT08 | blaTEM | aphA1-Iab | strAB | NDa | tetB | ||
| DT193 | pUT12 | blaTEM | aphA1-Iab | strAB | ND | tetB | ||
| DT193 | pUT20A | blaTEM | aphA1-Iab | strAB | ND | ND | ||
| DT193 | pUT20B | aphA1-Iab | strAB | ND | tetB | |||
| DT193 | pUT30A | blaTEM | aphA1-Iab | strAB | ND | tetB | ||
| DT193 | pUT30B | blaTEM | aphA1-Iab | strAB | sul1 | grm | 1.0 kb/plasmid | |
| DT193 | pUT30B | blaTEM | aphA1-Iab | aadA | sul1 | grm | 1.0 kb/plasmid | |
| DT104 | AmCmStSuTe | blaPSEl | aadA | sul1 | tetG | 1.0 and 1.2 kb/chromosome | ||
| DT104 | StSu | aadA | sul1 | 1.0 kb/chromosome | ||||
| U302 | AmKmStSuTe | blaTEM | aphA1-Iab | strAB | tetA | 1.0 kb/ND | ||
| U302 | AmCmStSuTe | blaPSEl | aadA | sul1 | tetG | 1.0 and 1.2 kb/ND | ||
ND, not determined.
Class I integron polymorphism among DT104 isolates.
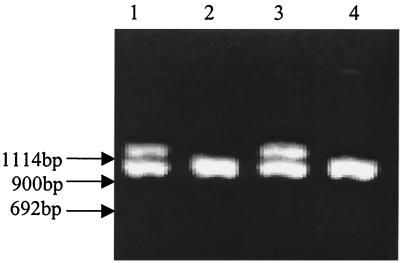
Among Salmonella serovar Typhimurium isolates, strains of phage type DT104 are known to be some of the MDR strains most important to public health. Strains of this phage type also most commonly have a pentaresistant AmCmStSuTe resistance phenotype. It has recently been shown that pentaresistant DT104 strains harbor two class I integrons, encoding resistance to ampicillin, streptomycin, and sulfamethoxazole, and an intervening region encoding resistance to chloramphenicol and tetracycline, all within a span of 13 kb (7). However, in this study, we have identified DT104 isolates with the StSu resistance pattern (8.3% of serovar Typhimurium var. Copenhagen isolates). These isolates had epidemiological significance in that they were more commonly isolated from an environmental source, i.e., drag swabs of cleaned barns (seven of the nine isolates). To characterize these StSu isolates, we first performed DNA fingerprinting to determine the genetic similarity between DT104 isolates with pentaresistance and those with the StSu resistance phenotype. As shown in the dendrogram in Fig. 1, in most cases StSu isolates exhibited a fingerprint identical to that of at least one pentaresistant DT104 isolate. Based upon this observation, we examined the genetic basis of resistance of the StSu DT104 isolates from our study. Although pentaresistant DT104 is known to harbor two integrons of 1.0 and 1.2 kb (InC and InD, respectively), the StSu strains evaluated in this study carried only one class I integron of 1.0 kb (Fig. 4), implying the absence of part of the pentaresistant architecture of DT104. Pentaresistant DT104 harbors two class I integrons on a 13-kb chromosomal region, and these integrons encode the complete pentaresistance phenotype. In strains of the StSu resistance phenotype, the integron encoding the β-lactam resistance and the intervening segment encoding chloramphenicol and tetracycline resistance may have been excised. In addition to the lack of one integron, we verified the absence of ampicillin, chloramphenicol, and tetracycline resistance genes by PCR testing of the known resistance genes (Table 2).
FIG. 4.
PCR amplicons of the class I integron(s) from DT104 strains with two different antimicrobial resistance phenotypes: AmCmStSuTe strains isolated from fecal samples (lanes 1 and 3) and StSu strains isolated from environmental samples (drag swabs) (lanes 2 and 4). Product sizes of approximately 1.0 and 1.2 kb are shown.
DISCUSSION
An increased incidence of MDR Salmonella serovar Typhimurium has been widely reported in the last decade, presumably due to the extensive use of antimicrobial agents in human and veterinary medicine. Here we have described the common resistance patterns and genetic characterization of MDR Salmonella serovar Typhimurium strains cultured from commercial swine operations. We found two common pentaresistance patterns: AmCmStSuTe and AmKmStSuTe. The former resistance pattern was most commonly found in strains of the DT104 phage type, not an unexpected finding, since numerous studies have shown the association of pentaresistance of the AmCmStSuTe type with DT104. We further found, however, that the majority of pentaresistant DT104 isolates (92 of 93 tested) were of the Copenhagen variant of serovar Typhimurium. It is difficult to assess whether DT104 is universally associated with the Copenhagen variant since serotyping schemes in past studies have often not differentiated between serovar Typhimurium and serovar Typhimurium var. Copenhagen. It is clear, however, that there is an association between host species and some Salmonella serovars and that serovar Typhimurium var. Copenhagen is adapted to the swine host (36). It is also known that swine-associated phage types have been reported to be common causes of human salmonellosis (4, 35, 45). Thus, our observations that serovar Typhimurium var. Copenhagen was common among pigs and was associated with phage type DT104 suggest that contaminated pork products present a substantial risk for the acquisition of strains with this type of multiresistance by humans. In addition to DT104, 11 of 14 phage type U302 isolates also exhibited pentaresistance of the AmCmStSuTe type. Genetic characterization also revealed that these U302 isolates carried the same resistance genes that DT104 carries (Table 2), a finding consistent with recent reports (33, 42). Although this phage type is infrequently isolated in most present studies, its close relationship to DT104 may signify its potential public health importance. Its genetic architecture also presents a challenge for the identification of DT104 by using amplification of resistance alleles (9), since the two phage types encode identical resistance genes.
In addition to the large number of DT104 isolates cultured in this study, we also identified phage types with pentaresistance of the AmKmStSuTe type. This pattern was found predominantly in two phage types, DT193 (77 of 165 isolates) and DT21 (82 of 165 isolates). As was the case for DT104, there was a clear association of phage type with serovar; the great majority of DT193 isolates with pentaresistance of the AmKmStSuTe type were serovar Typhimurium var. Copenhagen, while most DT21 isolates were serovar Typhimurium. The spread of MDR DT193 was reported as early as 1978 in studies by Threlfall et al. (38) in bovine and human isolates, which showed that phage type conversion of MDR DT204 by the acquisition of resistance factors of specific compatibility group I1 was important in the emergence and expansion of phage type DT193. Further characterization of DT193 in several studies showed that this phage type usually exhibits the AmStSuTe or Te resistance pattern (3, 11, 17, 18). However, we consistently found DT193 isolates with the AmKmStSuTe pentaresistance pattern, which was uncommon in previous studies. Rarely, isolates from our study also carried resistance to gentamicin. These findings thus imply that DT193 is extending its spectrum of multiresistance.
We performed DNA fingerprinting using AFLP analysis in order to discern genetic similarities among a representative sample of the serovar Typhimurium var. Copenhagen isolates obtained in this study. This analysis divided the isolates into two groups (clusters 1 and 2; Fig. 1) composed primarily of phage types DT193 and DT104 with the AmKmStSuTe and AmCmStSuTe resistance patterns, respectively, with 90 to 100% similarity within each group. The similarity between and within groups was expected, since most isolates of serovar Typhimurium have been shown to be genetically very similar (5). Our finding is also consistent with those of earlier studies that showed that diversity among DT193 strains was based upon differences in plasmid compositions (17). DT104 has also been proposed to have originated as a single clone which spread epidemically (10, 20), predicting similar fingerprints among isolates. The heterogeneity that we found in each cluster is presumably due to recent genetic events such as deletions, insertions, or mutations of one or a few loci or by the acquisition of plasmids. The identical fingerprints between pentaresistant and StSu DT104 isolates suggest that these isolates are genotypically closely related and that the loss of resistance genes is likely to be a recent genetic event (discussed further below). We did find, however, that a few isolates were genetically more similar to members of the group dominated by the opposite serovar and the opposite resistance pattern (for example, strains UU01 and FV06 have the AmCmStSuTe pattern and are phage types DT104 and U302, respectively, but are similar to strains with the AmKmStSuTe pattern, while UO02 and UF02 are phage types DT193 and DT21, respectively, but map to the cluster with the AmCmStSuTe pattern). There is evidence that subtle changes in the bacterial genome insufficient to change the fingerprint pattern may cause a change in phage type (44), thus suggesting the need for phenotypic methods complementary to genotypic fingerprinting for the ultimate discrimination of strains.
We characterized the molecular basis of resistance of the DT193 isolates in this study both because this phage type represents a significant cause of food-borne disease and because we had identified the AmKmStSuTe resistance pattern in many of these isolates, a pattern similar to that for pentaresistant DT104 isolates and previously not commonly reported among DT193 isolates. We chose for study four isolates that were both phenotypically and genotypically similar, as well as epidemiologically related, in order to investigate the means by which differences in antibiotic resistance can be encoded in similar strains. We found that all resistance genes were encoded on conjugative plasmids that could be efficiently transferred to E. coli. It has long been recognized that antimicrobial resistance genes among Salmonella serovars can be carried on conjugative plasmids; however, we isolated plasmids of different sizes and numbers and with different resistance gene contents from strains collected from the same location and time, strengthening the evidence of genetic heterogeneity of epidemiologically related Salmonella serovar Typhimurium isolates. We found, for example, that the addition of gentamicin resistance in one strain was due to the acquisition of a second conjugative plasmid that also encoded duplicative resistance to ampicillin, kanamycin, streptomycin, and sulfamethoxazole. Gentamicin resistance was encoded by a homologue of grm of M. purpurae, a gene that, to our knowledge, has not previously been found in pathogenic bacteria. We also found this plasmid to have a class I integron harboring aadA, which encodes streptomycin resistance, and sul1, which encodes sulfamethoxazole resistance and which is commonly found on class I integrons. It is unclear whether the duplication of resistance in this strain provided any selective advantage (for example, aadA also encodes resistance to spectinomycin and so might broaden the spectrum of resistance). The presence of an integron on this plasmid is of particular concern. Integrons are capable of trapping one or more resistance gene cassettes and furnishing a promoter for the efficient expression of antimicrobial resistance genes (37), leading to the rapid evolution of multiresistance. Salmonella strains carrying this plasmid might thus acquire new resistance factors and might easily transfer those resistance genes within and between species by means of conjugation.
In addition to the many AmCmStSuTe pentaresistant DT104 isolates cultured in this study, we also found atypical DT104 strains with only the StSu resistance pattern, a pattern that was observed in 8.3% of DT104 isolates. Similar atypical DT104 strains have also been reported previously (11, 34); however, none of those reports examined the epidemiological significance of these strains or performed detailed genetic characterizations. We detected these StSu isolates predominantly from environmental samples consisting of drag swabs collected from cleaned and apparently disinfected barns before they were filled with new groups of pigs. As expected from their phenotype, PCR analysis of isolates with the StSu resistance phenotype showed that they did not carry genes encoding resistance to ampicillin, chloramphenicol, and tetracycline. We also found that, in contrast to the 1.0- and 1.2-kb integrons of pentaresistant DT104, isolates with the StSu phenotype had only one integron of 1.0 kb. We hypothesized that pentaresistant DT104 isolates excise a chromosomal fragment encoding ampicillin, chloramphenicol, and tetracycline resistance, including the 1.2-kb integron, by homologous recombination in the presence or absence of some unidentified selective pressure. Since most of the StSu DT104 isolates were isolated from cleaned barns, it is possible that the loss of this chromosomal piece provides a selective advantage for survival during the cleaning and disinfecting procedures. Alternatively, expression of all five resistance factors may reduce the competitive fitness of Salmonella strains when growing outside of the animal host, in the absence of antimicrobial agents, thus allowing the selective proliferation of strains that have reduced their levels of expression of resistance genes by deleting them.
The results of this study have important implications for public health. We found MDR Salmonella serovar Typhimurium isolates to be common on commercial swine farms. A frequent isolate was phage type DT193, which had the AmKmStSuTe resistance phenotype and which is classified as one of the phage types of public health importance. We have shown here that DT193 isolates encode the genes for resistance to all five antimicrobials on plasmids that can easily be transferred by conjugation. This poses a significant threat not only through the food-borne salmonellosis attributed to pork consumption but also through the horizontal transfer of resistance determinants to commensal organisms of the human digestive tract or other pathogens of importance to human health. Studies on the epidemiological significance of this pentaresistant drug resistance phenotype on human health are under way. The high prevalence of phage type DT104, which has been known to cause outbreaks globally for the last decade, poses an additional threat through the food-borne acquisition of this pentaresistant strain.
Acknowledgments
This work was supported by funding from the FDA (FD-U-001880-01) and the North Carolina Pork Producers Council (to C.A.).
We thank Paul Orndorff for critical review of the manuscript.
REFERENCES
- 1.Anderson, E. S., and M. J. Lewis. 1965. Drug resistance and its transfer in Salmonella typhimurium. Nature 206:579-583. [DOI] [PubMed] [Google Scholar]
- 2.Bager, F., and J. Petersen. 1991. Sensitivity and specificity of different methods for the isolation of Salmonella from pigs. Acta Vet. Scand. 32:473-481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baggesen, D. L., and F. M. Aarestrup. 1998. Characterisation of recently emerged multiple antibiotic-resistant Salmonella enterica serovar Typhimurium DT104 and other multiresistant phage types from Danish pig herds. Vet. Rec. 143:95-97. [DOI] [PubMed] [Google Scholar]
- 4.Baggesen, D. L., and H. C. Wegener. 1994. Phage types of Salmonella enterica ssp. enterica serovar typhimurium isolated from production animals and humans in Denmark. Acta Vet. Scand. 35:349-354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Beltran, P., J. M. Musser, R. Helmuth, J. J. Farmer III, W. M. Frerichs, I. K. Wachsmuth, K. Ferris, A. C. McWhorter, J. G. Wells, A. Cravioto, et al. 1988. Toward a population genetic analysis of Salmonella: genetic diversity and relationships among strains of serotypes S. choleraesuis, S. derby, S. dublin, S. enteritidis, S. heidelberg, S. infantis, S. newport, and S. typhimurium. Proc. Natl. Acad. Sci. USA 85:7753-7757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Besser, T. E., C. C. Gay, J. M. Gay, D. D. Hancock, D. Rice, L. C. Pritchett, and E. D. Erickson. 1997. Salmonellosis associated with S. typhimurium DT104 in the USA. Vet. Rec. 140:75.. [PubMed] [Google Scholar]
- 7.Briggs, C. E., and P. M. Fratamico. 1999. Molecular characterization of an antibiotic resistance gene cluster of Salmonella typhimurium DT104. Antimicrob. Agents Chemother. 43:846-849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bronzwaer, S. L., O. Cars, U. Buchholz, S. Molstad, W. Goettsch, I. K. Veldhuijzen, J. L. Kool, M. J. Sprenger, and J. E. Degener. 2002. A European study on the relationship between antimicrobial use and antimicrobial resistance. Emerg. Infect. Dis. 8:278-282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Carlson, S. A., L. F. Bolton, C. E. Briggs, H. S. Hurd, V. K. Sharma, P. J. Fedorka-Cray, and B. D. Jones. 1999. Detection of multiresistant Salmonella typhimurium DT104 using multiplex and fluorogenic PCR. Mol. Cell. Probes 13:213-222. [DOI] [PubMed] [Google Scholar]
- 10.Casin, I., J. Breuil, A. Brisabois, F. Moury, F. Grimont, and E. Collatz. 1999. Multidrug-resistant human and animal Salmonella typhimurium isolates in France belong predominantly to a DT104 clone with the chromosome- and integron-encoded beta-lactamase PSE-1. J. Infect. Dis. 179:1173-1182. [DOI] [PubMed] [Google Scholar]
- 11.Daly, M., and S. Fanning. 2000. Characterization and chromosomal mapping of antimicrobial resistance genes in Salmonella enterica serotype Typhimurium. Appl. Environ. Microbiol. 66:4842-4848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Davies, A., P. O'Neill, L. Towers, and M. Cooke. 1996. An outbreak of Salmonella typhimurium DT104 food poisoning associated with eating beef. Commun. Dis. Rep. CDR Rev. 6:R159-R162. [PubMed] [Google Scholar]
- 13.Fey, P. D., T. J. Safranek, M. E. Rupp, E. F. Dunne, E. Ribot, P. C. Iwen, P. A. Bradford, F. J. Angulo, and S. H. Hinrichs. 2000. Ceftriaxone-resistant Salmonella infection acquired by a child from cattle. N. Engl. J. Med. 342:1242-1249. [DOI] [PubMed] [Google Scholar]
- 14.Frana, T. S., S. A. Carlson, and R. W. Griffith. 2001. Relative distribution and conservation of genes encoding aminoglycoside-modifying enzymes in Salmonella enterica serotype Typhimurium phage type DT104. Appl. Environ. Microbiol. 67:445-448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gebreyes, W. A., P. R. Davies, W. E. Morrow, J. A. Funk, and C. Altier. 2000. Antimicrobial resistance of Salmonella isolates from swine. J. Clin. Microbiol. 38:4633-4636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Glynn, M. K., C. Bopp, W. Dewitt, P. Dabney, M. Mokhtar, and F. J. Angulo. 1998. Emergence of multidrug-resistant Salmonella enterica serotype Typhimurium DT104 infections in the United States. N. Engl. J. Med. 338:1333-1338. [DOI] [PubMed] [Google Scholar]
- 17.Hampton, M. D., E. J. Threlfall, J. A. Frost, L. R. Ward, and B. Rowe. 1995. Salmonella typhimurium DT193: differentiation of an epidemic phage type by antibiogram, plasmid profile, plasmid fingerprint and salmonella plasmid virulence (spv) gene probe. J. Appl. Bacteriol. 78:402-408. [DOI] [PubMed] [Google Scholar]
- 18.Kariuki, S., C. Gilks, J. Kimari, J. Muyodi, P. Waiyaki, and C. A. Hart. 1999. Analysis of Salmonella enterica serotype Typhimurium by phage typing, antimicrobial susceptibility and pulsed field gel electrophoresis. J. Med. Microbiol. 48:1037-1042. [DOI] [PubMed] [Google Scholar]
- 19.Lai-King, N. G., M. R. Mulvey, I. Martin, G. A. Peters, and W. Johnson. 1999. Genetic characterization of antimicrobial resistance in Canadian isolates of Salmonella serovar Typhimurium DT104. Antimicrob. Agents Chemother. 43:3018-3021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Leegaard, T. M., D. A. Caugnat, L. O. Froholm, E. A. Hoiby, and J. Lassen. 2000. Emerging antibiotic resistance in Salmonella typhimurium in Norway. Epidemiol. Infect. 125:473-480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lindstedt, B. A., E. Heir, T. Vardund, and G. Kapperud. 2000. Fluorescent amplified-fragment length polymorphism genotyping of Salmonella enterica subsp. enterica serovars and comparison with pulsed-field gel electrophoresis typing. J. Clin. Microbiol. 38:1623-1627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Low, J. C., B. Tennant, and D. Munro. 1996. Multiple-resistant Salmonella typhimurium DT104 in cats. Lancet 348:1391.. [DOI] [PubMed] [Google Scholar]
- 23.Low, J. C., M. Angus, G. Hopkins, D. Munro, and S. C. Rankin. 1997. Antimicrobial resistance of Salmonella enterica typhimurium DT104 isolates and investigation of strains with transferable apramycin resistance. Epidemiol. Infect. 118:97-103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Madsen, L., F. M. Aarestrup, and J. E. Olsen. 2000. Characterisation of streptomycin resistance determinants in Danish isolates of Salmonella Typhimurium. Vet. Microbiol. 75:73-82. [DOI] [PubMed] [Google Scholar]
- 25.Maguire, H. C., A. A. Codd, V. E. Mackay, B. Rowe, and E. Mitchell. 1993. A large outbreak of human salmonellosis traced to a local pig farm. Epidemiol. Infect. 110:239-246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.McEwen, S. A., and P. J. Fedorka-Cray. 2002. Antimicrobial use and resistance in animals. Clin. Infect. Dis. 34(Suppl. 3):S93-S106. [DOI] [PubMed] [Google Scholar]
- 27.Metzer, E., V. Agmon, N. Andoren, and D. Cohen. 1998. Emergence of multidrug-resistant Salmonella enterica serotype Typhimurium phage-type DT104 among Salmonellae causing enteritis in Israel. Epidemiol. Infect. 121:555-559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Molbak, K., D. L. Baggesen, F. M. Aaerestrup, J. M. Ebbesen, J. Engberg, K. Frydendahl, P. Gerner-Smidt, A. M. Petersen, and H. C. Wegener. 1999. An outbreak of multidrug-resistant Salmonella enterica serotype Typhimurium DT104. N. Engl. J. Med. 341:1420-1425. [DOI] [PubMed] [Google Scholar]
- 29.National Committee for Clinical Laboratory Standards. 1999. Performance standards for antimicrobial disk and dilution susceptibility tests for bacteria isolated from animals; approved standard M31-A. National Committee for Clinical Laboratory Standards, Wayne, Pa.
- 30.O'Brien, T. F. 2002. Emergence, spread, and environmental effect of antimicrobial resistance: how use of an antimicrobial anywhere can increase resistance to any antimicrobial anywhere else. Clin. Infect. Dis. 34(Suppl. 3):S78-84. [DOI] [PubMed] [Google Scholar]
- 31.Olive, D. M., and P. Bean. 1999. Principles and applications of methods for DNA-based typing of microbial organisms. J. Clin. Microbiol. 37:1661-1669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pontello, M., L. Sodano, A. Nastasi, C. Mammina, M. Astuti, M. Domenichini, G. Belluzzi, E. Soccini, M. G. Silvestri, M. Gatti, E. Gerosa, and A. Montagna. 1998. A community-based outbreak of Salmonella enterica serotype Typhimurium associated with salami consumption in northern Italy. Epidemiol. Infect. 120:209-214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pritchett, L. C., M. E. Konkel, J. M. Gay, and T. E. Besser. 2000. Identification of DT104 and U302 phage types among Salmonella enterica serotype Typhimurium isolates by PCR. J. Clin. Microbiol. 38:3484-3488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sandvang, D., F. M. Aarestrup, and L. B. Jensen. 1998. Characterization of integrons and antibiotic resistance genes in Danish multiresistant Salmonella enterica Typhimurium DT104. FEMS Microbiol. Lett. 160:37-41. [DOI] [PubMed] [Google Scholar]
- 35.Schiellerup, P., R. J. Abdul-Redha, D. L. Baggesen, S. L. Andersen, and D. Sandvang. 2001. Five cases of gastroenteritis with multiresistant Salmonella enterica serovar Typhimurium DT104 related to farm animals in Denmark. Ugeskr. Laeger. 163:5677-5678. (In Danish.) [PubMed] [Google Scholar]
- 36.Scholtens, R. T., and G. Caroli. 1971. Role of pigeons in the spread of salmonellosis: incidence of different types of Salmonella typhimurium var. Copenhagen in pigeons, man, and other animals. Antonie Leeuwenhoek 37:473-476. [DOI] [PubMed] [Google Scholar]
- 37.Stokes, H. W., and R. M. Hall. 1989. A novel family of potentially mobile DNA elements encoding site-specific gene-integration functions: integrons. Mol. Microbiol. 3:1669-1683. [DOI] [PubMed] [Google Scholar]
- 38.Threlfall, E. J., L. R. Ward, and B. Rowe. 1978. Spread of multiresistant strains of Salmonella typhimurium phage types 204 and 193 in Britain. Br. Med. J. 2:997.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Threlfall, E. J., J. A. Frost, L. R. Ward, and B. Rowe. 1994. Epidemic in cattle and humans of Salmonella typhimurium DT104 with chromosomally integrated multiple drug resistance. Vet. Rec. 134:577.. [DOI] [PubMed] [Google Scholar]
- 40.Threlfall, E. J., L. R. Ward, and B. Rowe. 1997. Increasing incidence of resistance to trimethoprim and ciprofloxacin in epidemic Salmonella typhimurium DT104 in England and Wales. Eurosurveillance 2:81-84. [DOI] [PubMed] [Google Scholar]
- 41.Vos, P., R. Hogers, M. Bleeker, M. Reijans, T. van de Lee, M. Hornes, A. Frijters, J. Pot, J. Peleman, M. Kuiper, et al. 1995. AFLP: a new technique for DNA fingerprinting. Nucleic Acids Res. 23:4407-4414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Walker, R. A., E. Lindsay, M. J. Woodward, L. R. Ward, and E. J. Threlfall. 2001. Variation in clonality and antibiotic-resistance genes among multiresistant Salmonella enterica serotype typhimurium phage-type U302 (MR U302) from humans, animals, and foods. Microb. Drug Resist. 7:13-21. [DOI] [PubMed] [Google Scholar]
- 43.Wall, P. G., E. J. Threllfall, L. R. Ward, and B. Rowe. 1996. Multiresistant Salmonella typhimurium DT104 in cats: a public health risk. Lancet 348:471.. [DOI] [PubMed] [Google Scholar]
- 44.Ward, L., and E. J. Threlfall. 2001. The emergence of new Salmonella Typhimurium phage type associated with pigs, p. 174-175. In P. J. van der Wolf (ed.), Proceedings of the 4th International Symposium on the Epidemiology and Control of Salmonella and other Foodborne Pathogens in Pork. Salinpork, Leipzig, Germany.
- 45.Wegener, H. C., D. L. Baggesen, and K. Gaarslev. 1994. Salmonella typhimurium phage types from human salmonellosis in Denmark 1988-1993. APMIS 102:521-525. [DOI] [PubMed] [Google Scholar]
- 46.Wells, S. J., P. J. Fedorka-Cray, D. A. Dargatz, K. Ferris, and A. Green. 2001. Fecal shedding of Salmonella spp. by dairy cows on farm and at cull cow markets. J. Food Prot. 64:3-11. [DOI] [PubMed] [Google Scholar]
- 47.White, D. G., S. Zhao, R. Sudler, S. Ayers, S. Friedman, S. Chen, P. F. McDermott, D. D. Wagner, and J. Meng. 2001. The isolation of antibiotic-resistant Salmonella from retail ground meats. N. Engl. J. Med. 345:1147-1154. [DOI] [PubMed] [Google Scholar]
- 48.White, D. G., S. Zhao, S. Simjee, D. D. Wagner, and P. F. McDermott. 2002. Antimicrobial resistance of foodborne pathogens. Microbes Infect. 4:405-412. [DOI] [PubMed] [Google Scholar]
- 49.Winokur, P. L., A. Brueggemann, D. L. DeSalvo, L. Hoffmann, M. D. Apley, E. K. Uhlenhopp, M. A. Pfaller, and G. V. Doern. 2000. Animal and human multidrug-resistant, cephalosporin-resistant Salmonella isolates expressing a plasmid-mediated CMY-2 AmpC β-lactamase. Antimicrob. Agents Chemother. 44:2777-2783. [DOI] [PMC free article] [PubMed] [Google Scholar]