Abstract
Two outbreaks of respiratory tract illness associated with prolonged cough occurring in 1998 and 1999 in New York State were investigated. A PCR test for Bordetella pertussis was primarily used by a private laboratory to confirm 680 pertussis cases. Several clinical specimens had positive culture results for B. pertussis during both outbreaks, which confirmed that B. pertussis was circulating during the outbreaks. However, testing by the New York State Department of Health reference laboratory suggested that some of the PCR results may have been falsely positive. In addition, features of the outbreak that suggested that B. pertussis may not have been the primary agent of infection included a low attack rate among incompletely vaccinated children and a significant amount of illness among patients testing PCR negative for B. pertussis. These investigations highlight the importance of appropriate clinical laboratory quality assurance programs, of the limitations of the PCR test, and of interpreting laboratory results in context of clinical disease.
Pertussis, also known as whooping cough, is a highly contagious disease caused by the bacterium Bordetella pertussis. Following the introduction of diphtheria and tetanus toxoids and whole-cell pertussis vaccine (DTP) in 1946, the incidence of pertussis in the United States declined, reaching a nadir in 1976 (1, 5). However, periodic epidemics of pertussis continue to occur at 3- to 5-year intervals, and pertussis remains endemic to the United States (1, 3, 12). Since 1976 the number of pertussis cases reported annually has increased fivefold to >7,000 in 1998 to 2000, with an increasing number of cases reported among adolescents and adults (12, 38). Evidence also suggests that pertussis cases may be grossly underreported (33). Because pertussis is thought to be a somewhat uncommon disease in the United States and the clinical presentation resembles that for other illnesses associated with prolonged cough (8, 32, 37), it is often not considered in the differential diagnosis by health care providers (7, 31).
Confirming the diagnosis of pertussis in the laboratory is challenging. Isolation of B. pertussis in culture, the traditional diagnostic standard for pertussis, has nearly 100% specificity and is widely used (9, 19, 25). However, because B. pertussis is fastidious, the sensitivity of culture can vary greatly and is dependent on the stage of illness at the time of specimen collection, the technique used for specimen collection, specimen adequacy and transport, and culture conditions. Under ideal conditions, the typical culture positivity rate can be greater than 50%; however, the rate is usually lower because of the reasons given above, as well as prior pertussis vaccinations, concurrent antibiotic therapy, and long elapsed time (e.g., more than 3 weeks) since cough onset (13, 19, 24, 30). Seven to ten days may be required to isolate and confirm B. pertussis, precluding rapid culture confirmation (17).
Sensitive and specific PCR assays have been developed by several investigators to amplify and detect B. pertussis DNA (10, 29, 35, 36). A primary advantage of this diagnostic method is the rapid turnaround time, which typically amounts to a few hours (2). However, PCR assays require a validated protocol, sophisticated technology, training, and rigorous quality assurance (QA). If handled improperly, reagents and assays can become contaminated with PCR amplicons or cellular DNA, leading to false-positive test results (34). Furthermore, PCR assays cannot differentiate between dead and viable organisms (11, 16). Accordingly, PCR-positive results for B. pertussis DNA may not indicate infection (14), and the predictive value of PCR assays for cases of pertussis has not been well established (23). No commercial, FDA-approved B. pertussis PCR assays are available, nor have any assays been standardized or validated among laboratories (26). Nevertheless, the exquisite sensitivity of PCR-based assays over culture for fastidious organisms often makes them the technique of choice for many laboratories. In New York State (NYS), laboratories are permitted to perform these assays if documented validation studies and protocols have been approved by the NYS Department of Health/Wadsworth Center (NYSDOH/WC).
In 1997, the Council of State and Territorial Epidemiologists (CSTE) and the Centers for Disease Control and Prevention (CDC) began accepting positive PCR results for public health surveillance and as part of the criteria for confirmed pertussis cases (6). In 1999, 13% of the 7,298 reported pertussis cases in the United States were confirmed using PCR (38).
This report describes investigations of two outbreaks of cough illness that occurred in NYS during 1998 and 1999; both were thought to be primarily due to B. pertussis based on PCR test results. Hundreds of PCR-positive individuals were treated with antibiotic or placed on antibiotic prophylaxis, and some PCR-positive health care workers were furloughed to protect patients from exposure to pertussis. Our investigation of these outbreaks suggests that there was likely overdiagnosis of pertussis, illustrates the danger in overreliance on PCR, and emphasizes the need for better methods and standards for pertussis diagnosis.
MATERIALS AND METHODS
Outbreak investigation.
The outbreaks of pertussis occurred in three small, largely rural contiguous NYS counties. The first outbreak occurred between September 1998 and April 1999 and the second between July and November 1999.
For clinical case ascertainment, we used the CSTE and CDC clinical case definition for pertussis, which requires at least 14 days of cough with either paroxysms, whoop, or post-tussive vomiting, without other apparent cause (4, 6). During outbreaks, 14 days of cough alone are sufficient to meet the clinical case definition. Confirmed cases are defined as either (i) culture-positive with any cough duration, (ii) a clinical case with a positive PCR result for B. pertussis DNA, or (iii) a clinical case epidemiologically linked to a confirmed case. Laboratory confirmation of pertussis during both outbreaks was performed using a PCR assay which targets a repetitive DNA element from the genome of B. pertussis, developed in-house by a private laboratory. Public health officials contacted individuals who tested positive by the PCR to collect clinical and epidemiologic data.
Sample processing, culture, and PCR amplification at NYSDOH/WC laboratories.
Specimens were received as a swab stabbed into Regan-Lowe transport medium. The swab was removed, streaked onto Regan-Lowe plates with and without cephalexin for culture, and then placed in 1 ml of phosphate-buffered saline (pH 7.6), briefly mixed in a vortex apparatus, and removed. This 1-ml suspension was subsequently processed for PCR by centrifugation, followed by lysing of the bacterial pellet with 200 μl of lysis buffer (10 mM Tris [pH 8.3], 2.5 mM MgCl2, 1% Tween 20, 1% NP-40) at 100°C for 10 min. All sample processing was performed using designated pipettors and equipment in a working area used specifically for processing to avoid potential amplicon contamination. Ten microliters of lysed sample was tested in the NYSDOH PCR.
The primer, chosen from a repetitive sequence IS481, on the forward strand was BP-F (5′-ATC-TGC-TGC-ACA-TCG-ACA-TC-3′), and the primer on the reverse strand was BP-R (5′-CGA-TGG-CCA-CGA-AGA-CGA-AGT-C-3′). The PCR was performed in a total sample volume of 100 μl with a final concentration of 1 μM of each primer, 200 μM of each deoxynucleoside triphosphate, 10 mM Tris-HCl (pH 8.3), 50 mM KCl, 2 mM MgCl2, and 2.5 U of AmpliTaq Gold (Perkin-Elmer, Foster City, Calif.), which was used for PCR in a Perkin-Elmer 9600 thermocycler. Thermocycler conditions consisted of 95°C for 11 min followed by 43 cycles of 94°C for 30 s, 60°C for 30 s, and 72°C for 30 s. Amplified products were visualized on an agarose gel.
Parallel testing studies and proficiency testing.
The following laboratory studies were performed to assess the reliability and validity of the PCR test at the private laboratory.
(i) During outbreak 1, two nasopharyngeal swabs were obtained concurrently from 33 symptomatic persons. To determine the validity of B. pertussis PCR results at the private laboratory, one specimen from each person was transported to both the private laboratory and at ambient temperature in Reagan-Lowe medium to the NYSDOH/WC pertussis laboratory for PCR testing for B. pertussis. NYSDOH/WC utilized a gene target within the same repeat of IS481, yet located just downstream of the private laboratory's target, to obtain a similar level of sensitivity. This choice would also avoid amplification of potential amplicon contamination present in samples prepared at the private laboratory. Samples were partially lysed, and an aliquot of crude DNA was amplified by PCR.
(ii) During outbreak 2, the NYSDOH/WC Bacteriology Proficiency Testing Laboratory prepared 10 specimens consisting of culture suspensions or sterile media, four with viable B. pertussis cells, and six with either viable E. coli or normal saline, which were tested for B. pertussis in a blind fashion by PCR at both the private and NYSDOH/WC pertussis laboratories. During outbreak 2, the private laboratory changed the PCR primers for their B. pertussis testing on October 17, 1999, and retested the 10 blinded specimens with the new primers.
Day care study.
To assess whether transmission of cough illness was consistent with pertussis vaccination status and exposure during outbreak 1, we investigated all licensed day care centers known to have two or more children with confirmed pertussis (positive PCR and a cough duration for ≥14 days). Data were collected for all children attending these centers using a standard questionnaire completed by parents and center employees. Vaccination records available at the day care center determined vaccination status.
RESULTS
Summary of outbreaks.
Between 15 September 1998 and 14 November 1999, 3,769 individuals from outbreaks 1 and 2 had a PCR assay for B. pertussis DNA (Table 1). Of these, 1,297 (34%) had a positive PCR result; only 607 (47%) persons with a positive PCR result met the CSTE/CDC pertussis case definition.
TABLE 1.
Summary of Pertussis testing and case ascertainment by outbreak, New York, 15 September 1998 to 14 November 1999
| Parameter | Outbreak 1, no. (%) | Outbreak 2, no. (%) |
|---|---|---|
| PCR tested | 2,362 | 1,407 |
| PCR tested positive | 746 (32) | 551 (39) |
| Met Clinical Case Definition | 400 (54) | 207 (38) |
| Culture tested | 38 | 377 |
| Culture tested positive | 2 (5) | 3 (1) |
| Epidemiologically linked | 29 | 39 |
| Total confirmed casesa | 431 (18) | 249 (18) |
Based on Council of State and Territorial Epidemiologists and the Centers for Disease Control and Prevention pertussis case definition.
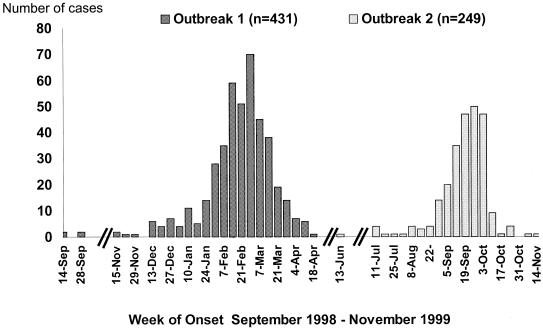
During this 13-month time period, there were 431 (outbreak 1) and 249 (outbreak 2) persons with an acute cough illness that met the confirmed pertussis case definition (Fig. 1). A similar proportion of patients tested positive by PCR for B. pertussis DNA in both outbreaks (Table 1). Only 5 (0.8%) of the 680 cases were confirmed by positive B. pertussis culture results.
FIG. 1.
Cases of cough illness by week of onset (pertussis outbreaks in NYS, September 1998 to November 1999).
Besides cough, additional clinical symptoms reported by case patients were similar between the two outbreaks with the exception of whoop. In outbreaks 1 and 2, respectively, paroxysms of cough were reported in 294 (68%) and 167 (67%) confirmed cases, post-tussive vomiting was reported in 173 (40%) and 67 (27%) cases, and whoop was reported in 210 (49%) and 57 (23%) cases; all three symptoms were experienced by 53 (13%) and 30 (12%) case patients in the two outbreaks. Thirty-eight case patients (9%) in outbreak 1 and 5 case patients (2%) in outbreak 2 were hospitalized. No deaths occurred.
Among the 680 confirmed cases, the highest incidence was among infants (age, <12 months). However, the incidence among infants was higher in outbreak 2 (953 per 100,000) than in outbreak 1 (558 per 100,000). In outbreak 1, almost one-half of the 431 cases (n = 181 [42%]) occurred among children aged <5 years; 113 cases occurred among children aged 7 months to 4 years. Of those 113 children, 98 (87%) had received three or more doses of a pertussis-containing vaccine, 5 (4%) had received one or two doses, and 3 (3%) were unvaccinated. The vaccination status for seven children was unknown. For outbreak 2, 20% of the 249 cases occurred among children aged <5 years and there were 30 cases among children aged 7 months to 4 years. For outbreak 2, parent report of vaccination status was incomplete and could not be analyzed.
Laboratory results.
The PCR assays used by NYSDOH/WC in this outbreak investigation had an analytical sensitivity of approximately 30 organisms of B. pertussis (data not shown [NYSDOH/WC, 2001]). Moreover, the assay was specific for B. pertussis and Bordetella holmesii and did not react with other Bordetella spp. or other common respiratory pathogens (data not shown [NYSDOH/WC, 2001]). At the NYSDOH/WC laboratory, this PCR assay typically produces threefold more positives than culture methods.
(i) Of the 33 primary respiratory specimens that were tested by PCR for B. pertussis DNA by both the private laboratory and NYSDOH/WC during outbreak 1, 20 specimens tested positive at the private laboratory compared with 8 at NYSDOH/WC. Six tested positive at both laboratories (Table 2).
TABLE 2.
Additional laboratory studies, pertussis outbreaks, New York, September 15, 1998 to November 14, 1999
| Laboratory study | nc | Results of PCR for B. pertussis DNA
|
|
|---|---|---|---|
| Private laboratory | NYSDOH/WC | ||
| PCRa | 33 | 20 PCR positive | 8 PCR positive |
| 13 PCR negative | 25 PCR negative | ||
| QA testb | 10 | 4 PCR positive for B. pertussis DNA | 4 PCR positive for B. pertussis DNA |
| 2 nonpertussis specimens PCR positive | 6 nonpertussis specimens PCR negative | ||
| 4 nonpertussis PCR negative | |||
PCR for B. pertussis DNA was performed by the private laboratory and NYSDOH/WC pertussis laboratory on dual samples from the same patients.
Specimens were prepared by NYSHD/WC Proficiency Testing Laboratory (four with B. pertussis) and blind PCR tested by the private laboratory and NYSDH/WC pertussis laboratory.
n, no. of samples studied.
(ii) During outbreak 2, PCR testing of the 10 blinded proficiency test samples resulted in the private laboratory correctly classifying 8 samples: all four specimens containing viable B. pertussis and four of the six samples that did not. The remaining two B. pertussis-negative samples were falsely identified as positive for B. pertussis. NYSDOH/WC's PCR test correctly identified the presence or absence of B. pertussis DNA for all 10 specimens in a blinded evaluation.
Following the change on 17 October 1999 of the PCR primers (to facilitate the amplification of an entirely new region of the repetitive element) used at the private laboratory, there was a decline in the proportion of PCR tests that were positive. In the two weeks prior to the change of primers, 44% of the 598 specimens PCR tested by the private laboratory were positive, compared with 10% of the 251 specimens tested in the two weeks following the change of primers. Moreover, retesting of the blinded proficiency test samples using the new primers resulted in correct identification of all 10 samples.
Day care study.
Seven licensed day care centers, with a total of 278 children in attendance, which each had two or more PCR-positive confirmed cases of pertussis among children during outbreak 1 were studied. All 278 children were younger than 5 years. A total of 24 children were PCR-confirmed cases, for an overall attack rate of 9%. Of these 24 children, 16 (67%) were fully vaccinated (three doses of diphtheria and tetanus toxoids and whole-cell pertussis vaccine plus two booster doses) against pertussis. Notably, 28 undervaccinated children (6 unvaccinated and 22 partially vaccinated) attending these centers were exposed to PCR-confirmed cases via shared classrooms but did not develop any symptoms. None of the undervaccinated children were receiving any chemoprophylaxis at the time of exposure.
DISCUSSION
The results of epidemiological and laboratory studies of these two outbreaks of cough illness illustrate the potential problems and consequences of relying exclusively on a PCR test to confirm pertussis. We confirmed by culture that pertussis was circulating in the community during these outbreaks. However, a significant epidemiological feature of one of these outbreaks was that unvaccinated children exposed to PCR-confirmed cases in day care centers did not become ill, whereas fully vaccinated children did. Thus, B. pertussis may not have been the primary or only cause of these outbreaks. Moreover, a positive PCR test due to transient colonization with the agent (15, 18, 20, 21, 22, 27, 28) or a falsely positive test could lead to misdiagnosis of pertussis in persons with cough illness due to other etiologies. Indeed, our results suggested that diagnostic reliance on a PCR assay for B. pertussis DNA together with probable false-positive test results likely overestimated the role of pertussis in these outbreaks.
The PCR for B. pertussis DNA was used by a private laboratory to confirm the majority of cases in these two outbreaks (93% in outbreak 1 and 83% in outbreak 2). When compared with testing at the state reference laboratory, the majority of the B. pertussis PCR results from the private laboratory were not substantiated (Table 2). Moreover, during proficiency testing of 10 blinded specimens, the private laboratory reported false-positive results for one-third of the non-B. pertussis specimens. After changing primers towards the end of the second outbreak, the private laboratory retested the 10 samples and correctly identified all 10 specimens. This suggested that DNA contamination may have caused an uncertain number of false-positive results during the two outbreaks. PCR false-positive results can be due to DNA contamination, which may have been exacerbated by the larger-than-normal volume of samples processed during the outbreaks.
The overdiagnosis of pertussis during these outbreaks had important public health consequences. These included the unnecessary treatment of asymptomatic individuals with a positive PCR result for B. pertussis DNA who may not have harbored B. pertussis, as well as inappropriate chemoprophylaxis for their close contacts. In outbreak 2, more than 1,100 persons in close contact with an individual with a positive PCR result for B. pertussis DNA were advised to take a 2-week course of chemoprophylaxis with a macrolide antibiotic. Staff at doctor offices, hospitals, and county health departments were overwhelmed with testing demands and follow-up of individuals who tested PCR positive for B. pertussis DNA. During outbreak 2, 10% of the 500-member staff of a community hospital, many of whom were asymptomatic, were furloughed for 5 days as a result of positive PCR tests.
A QA program for pertussis PCR assays is paramount and needs to include all aspects of the assay from specimen extraction to amplicon detection. Some QA program components (e.g., reagent controls for identity, stability, and potency and functional controls for equipment) are commonly applied to pertussis PCR assays. Functional controls for the procedure are needed as there is no acknowledged reference standard and no universal results acceptance criteria to monitor for test failures. In addition, assays should be revalidated when changing significant assay parameters or reagents, such as thermocyclers, specimen extraction procedures, and primers. Although proficiency testing is an important part of QA, sanctioned and certified testing is not readily available and in-house performance is often assessed with samples that fail to resemble clinical specimens, are limited in number, and include too few samples per panel. In addition, in-house testing often does not challenge reproducibility, sensitivity, or turn-around time. For these reasons, there is a pressing need for standardization of methods and reference reagents as well as for complete QA programs including proficiency testing. Laboratories currently using PCR for B. pertussis DNA may benefit from evaluating their QA programs to ensure the best use of this test, particularly if it is used exclusively for confirming the diagnosis. Based on the results of these investigations, the following standards should be followed by all laboratories performing PCR for B. pertussis DNA: (i) maintaining culture capabilities for B. pertussis, especially during times of suspected higher pertussis incidence so that the diagnosis can be confirmed for at least some cases, (ii) running one negative control through the entire processing step for every 7 samples, (iii) running small batches of samples (10 to 12 per run), (iv) performing proficiency testing regularly in a blind fashion to ensure proper test and personnel performance, and (v) incorporating hierarchical acceptance criteria monitoring for test failures, including positive and negative functional controls, reevaluating multiple consecutive positive specimens, and confirming that positive specimens were collected from persons meeting the pertussis clinical case definition.
Many of the confounding PCR diagnostic test results we experienced throughout these outbreaks reflect the current state of PCR technology in general and of pertussis diagnosis in particular. PCR for B. pertussis DNA assays evolved rapidly, and numerous protocols have been reported; because of the sensitive and timely results that they provide compared with the diagnostic standard (B. pertussis culture), these assays have moved from reference laboratories and academic institutions to smaller treatment facilities. Consequently, PCR testing may soon ascend from presumptive to diagnostic status and become the standard of care. However, no commercial product is available; no standardized protocol, reagents, or reporting format are widely accepted; and no strategy for unifying assays currently exists. Other challenges to the wide acceptance of PCR for B. pertussis DNA are recognized: clinicians must be properly educated in interpreting PCR test results, and the clinical utility of results is poorly defined because predictive values for these tests are unknown.
We support the contention that B. pertussis PCR assays can be a valuable tool in the diagnosis of this infectious agent. At the NYSDOH/WC laboratory, an ongoing study of culture versus PCR on hundreds of samples has revealed that PCR typically is threefold more likely to detect a positive B. pertussis sample (D. Bopp, unpublished data [NYSDOH/WC, 2001]). The clinical significance of this difference is unknown, although at least half of the PCR-positive and culture-negative samples were from patients who had an acute cough illness that met the pertussis case definition.
Widespread availability of sensitive and specific clinical laboratory tests for diagnosis of pertussis is essential if the burden of pertussis disease is to be fully documented. PCR, when well performed, can serve as a valuable adjunct to culture and a presumptive test to detect B. pertussis. PCR may become more reliable with the addition of a traditional hybridization step using a specific probe or by use of probe-based assays. Because of the implications for public health programs, it is essential that validated diagnostic tests performed with appropriate QA be available to support case investigation and outbreak control activities.
Acknowledgments
We thank the following people for their assistance in case investigations, data collection, and public health response to the outbreaks: Robert Berke and Marcia Clark, Chautauqua County Health Department; Jack Schwartz, Erie County Health Department; Elaine Roman, Niagara County Health Department; and Robert Bentkowski, Debra Hilfstein, Colleen O'Connor-Walker, Patricia O'Hanlon, and Margaret Oxtoby, New York State Department of Health. We also thank Mary McCauley, Susan Chu, and Jane Seward at the National Immunization Program for their editorial and critical review of the manuscript.
REFERENCES
- 1.Bass, J. W., and R. R. Wittler. 1994. Return of epidemic pertussis in the United States. Pediatr. Infect. Dis. J. 13:343-345. [DOI] [PubMed] [Google Scholar]
- 2.Birkebaek, N. H., I. Heron, and K. Skjodt. 1994. Bordetella pertussis diagnosed by polymerase chain reaction. APMIS 102:291-294. [PubMed] [Google Scholar]
- 3.Centers for Disease Control and Prevention. 1993. Resurgence of pertussis—United States, 1993. Morb. Mortal. Wkly. Rep. 42:952-953, 959-960. [PubMed] [Google Scholar]
- 4.Centers for Disease Control and Prevention. 1999. Pertussis, p. 67-83. In Epidemiology and prevention of vaccine-preventable diseases, 5th ed. Centers for Disease Control and Prevention, Atlanta, Ga.
- 5.Centers for Disease Control and Prevention. 1997. Pertussis vaccination: use of acellular pertussis vaccines among infants and young children. Recommendations of the Advisory Committee on Immunization Practices. Morb. Mortal. Wkly. Rep. 46(RR-7):1-25. [PubMed] [Google Scholar]
- 6.Council of State and Territorial Epidemiologists. 1997. Council of State and Territorial Epidemiologists National Meeting, Position Statement 9. Council of State and Territorial Epidemiologists, Saratoga Springs, N.Y.
- 7.Deeks, S., G. De Serres, N. Boulianne, B. Duval, L. Rochette, C. Dery, and S. Halperin. 1999. Failure of physicians to consider the diagnosis of pertussis in children. Clin. Infect. Dis. 20:840-846. [DOI] [PubMed] [Google Scholar]
- 8.Edwards, K. M., M. D. Decker, and E. A. Mortimer, Jr. 1999. Pertussis vaccine, p. 293-344. In S. A. Plotkin, W. A. Orenstein (ed.), Vaccines, 3rd ed. WB Saunders, Philadelphia, Pa.
- 9.Gilligan, P. H., and M. C. Fisher. 1984. Importance of culture in laboratory diagnosis of Bordetella pertussis infections. J. Clin. Microbiol. 20:891-893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Glare, E. M., J. C. Paton, R. R. Premier, A. J. Lawrence, and I. T. Nisbet. 1990. Analysis of a repetitive DNA sequence from Bordetella Pertussis and its application to the diagnosis of pertussis using the polymerase chain reaction. J. Clin. Microbiol. 28:1982-1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Grant, I. R., H. J. Ball, and M. T. Rowe. 2002. Incidence of Mycobacterium paratuberculosis in bulk raw and commercially pasteurized cows' milk from approved dairy processing establishments in the United Kingdom. Appl. Environ. Microbiol. 68:2428-2435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Guris, D., P. Strebel, B. Bardenheier, M. Brennan, R. Tachdjian, E. Finch, M. Wharton, and J. R. Livengood. 1999. Changing epidemiology of pertussis in the United States: increasing reported incidence among adolescents and adults, 1990-1996. Clin. Infect. Dis. 28:1230-1237. [DOI] [PubMed] [Google Scholar]
- 13.Hallander, H. O., E. Reizenstein, B. Renemar, G. Rasmuson, L. Mardin, and P. Olin. 1993. Comparison of nasopharyngeal aspirates with swabs for culture of Bordetella pertussis. J. Clin. Microbiol. 31:50-52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.He, Q., J. Mertsola, H. Soini, M. Skurnik, O. Russkanen, and M. K. Viljanen. 1993. Comparison of polymerase chain reaction with culture and enzyme immunoassay for diagnosis of pertussis. J. Clin. Microbiol. 31:642-645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hussain, F. M., S. Boyle-Vavra, and R. S. Daum. 2001. Community-acquired methicillin-resistant Staphylococcus aureus colonization in healthy children attending an outpatient pediatric clinic. Pediatr. Infect. Dis. J. 20:763-767. [DOI] [PubMed] [Google Scholar]
- 16.Josephson, K. L., C. P. Gerba, and I. L. Pepper. 1993. Polymerase chain reaction detection of nonviable bacterial pathogens. Appl. Environ. Microbiol. 59:3513-3515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Katzko, G., M. Hofmeister, and D. Church. 1996. Extended incubation of culture plates improves recovery of Bordetella spp. J. Clin. Microbiol. 34:1563-1564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Krantz, I., K. Alestig, B. Trollfors, and G. Zackeisson. 1986. The carrier state in pertussis. Scand. J. Infect. Dis. 18:121-123. [DOI] [PubMed] [Google Scholar]
- 19.Kwantes, W., H. M. Joynson, and W. O. Williams. 1983. Bordetella pertussis isolation in general practice: 1977-79 whooping cough epidemic in West Glamorgan. J. Hyg. Camb. 90:149-158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lambert, H. P. 1986. The carrier state: Bordetella pertussis. J. Antimicrob. Chemother. 18(Suppl.):13-16. [DOI] [PubMed] [Google Scholar]
- 21.Linnemann, C. C., J. W. Bass, and M. H. Smith. 1968. The carrier state in pertussis. Am. J. Epidemiol. 88:422-427. [DOI] [PubMed] [Google Scholar]
- 22.Long, S. S., C. J. Welkon, and J. L. Clark. 1990. Widespread silent transmission of pertussis in families: antibody correlates of infection and symptomatology. J. Infect. Dis. 161:480-486. [DOI] [PubMed] [Google Scholar]
- 23.Meade, B. D., and A. Bollen. 1994. Recommendations for use of the polymerase chain reaction in the diagnosis of Bordetella pertussis infections. J. Med. Microbiol. 41:51-55. [DOI] [PubMed] [Google Scholar]
- 24.Medical Research Council. 1951. The prevention of whooping-cough by vaccination: a Medical Research Council investigation. Br. Med. J. 4721-4771. [PMC free article] [PubMed]
- 25.Muller, F. M., J. E. Hoppe, and C. H. Wirsing von Konig. 1997. Laboratory diagnosis of pertussis: state of the art in 1997. J. Clin. Microbiol. 35:2435-2443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Murphy, T., K. Bisgard, and G. Sanden. 2000. Diagnosis and laboratory methods, chapter 2. In Guidelines for the control of pertussis outbreaks, National Immunization Program. Centers for Disease Control and Prevention, Atlanta, Ga.
- 27.Peacock, S. J., I. de Silva, and F. D. Lowy. 2001. What determines nasal carriage of Staphylococcus aureus? Trends Microbiol. 9:605-610. [DOI] [PubMed] [Google Scholar]
- 28.Raymond, J., L. Armand-Lefevre, F. Moulin, H. Dabernat, A. Commeau, D. Gendrel, and P. Berche. 2001. Nasopharyngeal colonization by Haemophilus influenzae in children living in an orphanage. Pediatr. Infect. Dis. J. 20:779-784. [DOI] [PubMed] [Google Scholar]
- 29.Reizenstein, E., B. Johansson, L. Mardin, J. Abens, R. Mollby, and H. O. Hallander. 1993. Diagnostic evaluation of polymerase chain reaction discriminative for Bordetella pertussis. B. parapertussis, and B. bronchiseptica. Diagn. Microbiol. Infect. Dis. 17:185-191. [DOI] [PubMed] [Google Scholar]
- 30.Riitta, H. 1982. The effect of early erythromycin treatment on the infectiousness of whooping cough patients. Acta Paediatr. Scand. 298(Suppl.):10-12. [Google Scholar]
- 31.Sotomayor, J., L. B. Weiner, and J. A. McMillan. 1985. Inaccurate diagnosis in infants with pertussis. An eight-year experience. Am. J. Dis. Child. 139:724-727. [DOI] [PubMed] [Google Scholar]
- 32.Strebel, P., D. Guris, and S. G. Wassilak. 1998. Pertussis. In R. B. Wallace, B. N. Doebbeling, and J. M. Last (ed.), Maxcy-Rosenau-Last, Public health & preventive medicine. Appleton & Lange. Stamford, Conn.
- 33.Sutter, R. W., and S. L. Cochi. 1992. Pertussis hospitalizations and mortality in the United States, 1985-1988. JAMA 267:386-391. [PubMed] [Google Scholar]
- 34.Taranger, J., B. Trollfors, L. Lind, G. Zackrisson, and K. Beling-Holmquist. 1994. Environmental contamination leading to false-positive polymerase chain reaction for pertussis. Pediatr. Infect. Dis. J. 13:936-937. [DOI] [PubMed] [Google Scholar]
- 35.Van der Zee, A., C. Agterberg, M. Peeters, J. Schellekens, and F. R. Mooi. 1993. Polymerase chain reaction assay for pertussis: simultaneous detection and discrimination of Bordetella pertussis and Bordetella parapertussis. J. Clin. Microbiol. 34:2134-2164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wadowsky, R. M., H. Michaels, T. Libert, L. A. Kingsley, and G. D. Ehrlich. 1996. Multiplex PCR-based assay for detection of Bordetella pertussis in nasopharyngeal swab specimens. J. Clin. Microbiol. 34:2645-2649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wirsing Von König, C. H., H. Rott, H. Bogaerts, and H. J. Schmitt. 1988. A serologic study of organisms possibly associated with pertussis-like coughing. Ped. Infect. Dis. J. 17:645-649. [DOI] [PubMed] [Google Scholar]
- 38.Zanardi, L., F. B. Pascual, K. Bisgard, T. Murphy, and M. Wharton. 2002. Pertussis—United States 1997-2000. Morb. Mortal. Wkly. Rep. 51:73-76. [PubMed] [Google Scholar]