Abstract
Rapid detection and accurate identification of methicillin-resistant staphylococci are critical for the effective management of infections caused by these organisms. We describe a multiplex PCR-based assay for the direct detection of methicillin-resistant staphylococci from blood culture bottles (BacT/Alert; Organon-Teknika, Durham, N.C.). A simple lysis method followed by a multiplex PCR assay designed to detect the nuc, mecA, and bacterial 16S rRNA genes was performed. A total of 306 blood culture specimens were collected over a period of 10 months from June 1998 to April 1999, consisting of 236 blood cultures growing staphylococci (including 124 methicillin-resistant Staphylococcus spp.), 50 positive blood cultures which grew organisms other than staphylococci, and 20 blood cultures that were negative for bacterial and fungal pathogens after 5 days of incubation and terminal subculture. DNA extraction, PCR, and detection could be completed in 2.5 h. Of the positive blood cultures with staphylococci, the multiplex PCR assay had a sensitivity and specificity of 99.2% and 100%, respectively. Our results show that rapid, direct detection of methicillin-resistant staphylococci is possible, allowing clinicians to make prompt and effective decisions for the management of patients with staphylococcal bacteremia.
The prevalence of methicillin resistance in nosocomial staphylococci has increased in the past decade (4, 5, 21). There has also been a recent increase in community-acquired methicillin-resistant Staphylococcus aureus (MRSA) infections in patients without apparent recognized risk factors (7). MRSA infections are associated with significant morbidity and mortality, especially in patients with bacteremia (22). The rapid detection of methicillin resistance in staphylococci from bacteremic patients is essential for the prompt institution of effective antimicrobial therapy.
Several investigators have described PCR-based assays for detection of methicillin-resistant staphylococci from clinical samples (8, 12, 14, 20, 23). Some reports have shown that a pure culture of the organism is required (20), while other methods involving direct detection from clinical specimens require long DNA extraction protocols (14). Jaffe et al. reported a rapid method for extracting DNA from blood cultures, but it required several individual PCRs to detect all necessary products for identification of staphylococci (8). While claiming to be more rapid than conventional culture and susceptibility testing, some methods may still include many steps for DNA extraction and long PCR protocols requiring several hours (12, 23). We describe a rapid and simple multiplex PCR-based method to detect methicillin-resistant staphylococci directly from positive blood culture bottles, dramatically reducing the time for identification from 24 to 48 h to approximately 3 h.
MATERIALS AND METHODS
Clinical specimens and conventional identification.
A total of 306 blood culture (BacT/Alert; Organon-Teknika, Durham, N.C.) specimens were collected over a period of 10 months from June 1998 to April 1999 from the microbiology departments of the Sunnybrook and Women's College Health Sciences Centre and the St. Vincent Catholic Medical Centers, consisting of 236 positive blood cultures with staphylococci, 50 positive blood cultures which grew organisms other than staphylococci, and 20 blood cultures that were negative for bacterial and fungal pathogens after 5 days of incubation and terminal subculture. All positive specimens were aliquoted into 1.5-ml microcentrifuge tubes as soon as a positive signal and Gram stain showed the presence of bacteria. The negative blood cultures were aliquoted after 5 days of incubation showing no positive signal and no organisms by Gram stain. The microcentrifuge tubes were stored at −70°C until testing.
Staphylococcal isolates were identified by standard methods, including Gram stain, catalase, latex agglutination (Pastorex Plus; Bio-Rad, Mississauga, Canada) and tube coagulase (10). Susceptibility testing was performed with the Vitek GPS-SV card as well as the oxacillin agar screen plate with 6 μg of oxacillin per ml (18). Confirmation of oxacillin resistance was made by PCR for detection of the mecA gene (13). Organisms from the positive blood cultures yielding pathogens other than staphylococci were identified with conventional laboratory tests.
DNA extraction directly from positive blood culture bottles.
One milliliter of sterile, distilled, deionized water was added to a 100-μl aliquot of each sample, mixed by inversion, and incubated at room temperature for 5 min. After centrifugation at 16,000 × g for 1 min, the supernatant was discarded, and the pellet was resuspended in 100 μl of Triton X-100 lysis buffer (100 mM NaCl [Sigma Chemicals, St. Louis, Mo.], 10 mM Tris-HCl [Sigma, pH 8], 1 mM EDTA [Sigma, pH 9], and 1% Triton X-100 [Sigma]). Five microliters of lysostaphin (1 mg of lysostaphin per ml [Sigma]) was added, mixed, and incubated at 37°C for 10 min. This suspension was then boiled for 10 min. After cooling to room temperature for 5 min, the sample was centrifuged at 16,000 × g for 1 min, and 1 μl of the supernatant was used for PCR.
DNA extraction from bacterial colonies.
An aliquot of each frozen blood culture sample was inoculated onto solid medium. A 1-μl loopful of organism grown either on a Columbia agar plate with 5% sheep blood or on chocolate agar was used. This was inoculated into a 100-μl aliquot of Triton X-100 lysis buffer containing 2 μl of a 1-mg/ml solution of lysostaphin. The suspension was then incubated in a 37°C water bath for 10 min and boiled for an additional 10 min. The suspension was cooled at room temperature for 5 min and centrifuged at 16,000 × g for 1 min. A 1-μl aliquot of the supernatant was used for PCR testing.
PCR amplification.
Primers used for multiplex PCR are listed in Table 1. Multiplex PCR was performed in a 25-μl volume with 1× PCR buffer (Roche Molecular Systems), 3 mM MgCl2, a 200 μM concentration of each deoxynucleoside triphosphate, 2.5 U of Taq polymerase (Roche), 0.2 μM each of the 16S rRNA and nuc primers, and 0.5 μM mecA primer with 1 μl of template DNA (as prepared above). Thermocycling conditions in a GeneAmp 9600 thermocycler (Applied Biosystems) were as follows: 94°C for 2 min, followed by 25 cycles of 94°C for 15 s, 55°C for 30 s, and 72°C for 30 s, with a final extension at 72°C for 10 min. Thermocycling required 1.5 h to complete.
TABLE 1.
Primers used for multiplex PCR detection of methicillin-resistant staphylococci
Primers for detection of the bacterial 16S rRNA (16S) gene (11), mecA (17), and nuc (3) were used, giving products of 798 bp, 533 bp, and 270 bp, respectively. Electrophoresis at 100 V for 30 min was performed to separate the PCR amplicons on a 1% 1× TBE (Tris-borate-EDTA)-agarose gel. Gels were then stained with ethidium bromide and photographed under UV illumination. Multiplex PCR was also performed from each positive blood culture with three to five bacterial colonies (approximately 1-μl loopful) after overnight incubation on Columbia agar with 5% sheep blood or chocolate agar in an atmosphere of 5% CO2. Thermocycling conditions in a GeneAmp 9600 thermocycler were modified as follows: 94°C for 2 min, followed by 30 cycles of 94°C for 1 s and 55°C for 15 s, with a final 10-min extension at 72°C. Thermocycling required 1 h to complete (13). PCR control organisms were used with each batch of samples and included Staphylococcus aureus ATCC 43300 (MRSA) and methicillin-sensitive S. aureus (MSSA) ATCC 29213. PCR products were visualized as above.
Seeding experiments.
A suspension of S. aureus ATCC 43300 (MRSA) (>108 CFU/ml) was made in 1 ml of phosphate-buffered saline. This suspension was serially diluted, and each dilution was plated for colony counts. Each dilution was then centrifuged and resuspended in 100 μl of a 5-day negative blood culture. The sample was treated and processed as above. One microliter from each dilution was then used for PCR. Original concentrations of organisms were calculated from standard plating on Columbia agar supplemented with 5% sheep blood. The seeding experiments were performed in duplicate to ensure accuracy and reproducibility.
RESULTS
Of the 236 blood cultures growing gram-positive cocci in clusters, there were 107 methicillin-resistant coagulase-negative staphylococci, 73 methicillin-susceptible coagulase-negative staphylococci, 36 methicillin-susceptible S. aureus, 17 methicillin-resistant S. aureus, and 3 Micrococcus spp. An additional 50 positive blood cultures were also used in this evaluation, yielding Streptococcus pyogenes from 2 blood cultures, 5 Streptococcus agalactiae, 10 Enterococcus faecalis, 5 Enterococcus faecium, 1 Propionibacterium acnes, 12 Escherichia coli, 4 Serratia marcescens, 3 Enterobacter cloacae, 3 Pseudomonas aeruginosa, 2 Haemophilus influenzae, 2 Neisseria meningitidis, and 1 Bacteroides fragilis.
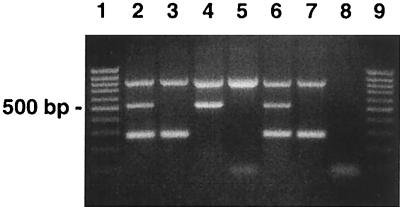
The detection limit of the multiplex PCR assay for the simultaneous detection of the bacterial 16S rRNA gene, mecA, and nuc was calculated to be 105 CFU/ml, based on the seeding experiments. The detection limits of individual PCR assays for the mecA gene, nuc gene, and 16S rRNA gene were 105, 103, and 103, respectively (data not shown). Figure 1 shows the results of the multiplex PCR assay with the 16S rRNA, mecA, and nuc gene products after PCR amplification with different organisms.
FIG. 1.
Agarose gel showing PCR-amplified products of the 16S rRNA, mecA, and nuc genes. Lanes 1 and 9, 100-bp DNA ladder. Lane 2, MRSA, showing all three amplification products (16S at 798 bp, mecA at 533 bp, and nuc at 270 bp). Lane 3, MSSA, with only the 16S and nuc products. Lane 4, coagulase-negative, methicillin-resistant staphylococci with 16S and mecA products only. Lane 5, coagulase-negative, methicillin-sensitive staphylococci, showing only the 16S product. Lane 6, MRSA S. aureus ATCC 43300 control. Lane 7, MSSA S. aureus ATCC 29213 control. Lane 8, reagent control.
The results of multiplex PCR direct testing from blood cultures and from bacterial isolates are summarized in Table 2. Direct multiplex PCR testing with DNA extracted from positive blood cultures did not misidentify any isolate compared to the PCR test results with actual bacterial growth. The total time required to perform the multiplex PCR assay directly from positive blood cultures was 2.5 h. The sensitivity of the PCR assay was 100%, with the 223 positive blood cultures with staphylococci that yielded a positive 16S rRNA amplification product. However, in 13 samples that grew a Staphylococcus species, the 16S rRNA product failed to amplify. Two of these positive blood cultures (both coagulase negative) yielded no product whatsoever, although in the 11 other samples, PCR gave the correct mecA and nuc results. These 13 samples were diluted 1:10, and the multiplex PCR assay was repeated with 1 μl of this diluted template. PCR was now able to amplify not only the mecA and/or nuc products but also the 16S rRNA product in all 13 samples. The discrepant results, compared with organism PCR, are summarized in Table 3.
TABLE 2.
Multiplex PCR identification of 286 positive blood cultures by comparison of organism PCR with direct PCR testing from clinical specimens
| Organism type | No. of strains | Expected PCR resultsa (16S/mecA/nuc) | Direct PCR results, 16S/mecA/nuc (no. of isolates) |
|---|---|---|---|
| CoNS,b methicillin resistant | 107 | +/+/− | +/+/− (100); −/+/− (7) |
| CoNS, methicillin susceptible | 73 | +/−/− | +/−/− (71); −/−/− (2) |
| MSSA | 36 | +/−/+ | +/−/+ (34); −/−/+ (2) |
| MRSA | 17 | +/+/+ | +/+/+ (15); −/+/+ (2) |
| Micrococcus spp. | 3 | +/−/− | +/−/− (3) |
| Escherichia coli | 12 | +/−/− | +/−/− (10); −/−/− (2) |
| Enterobacter cloacae | 3 | +/−/− | +/−/− |
| Serratia marcescens | 4 | +/−/− | +/−/− |
| Pseudomonas aeruginosa | 3 | +/−/− | +/−/− |
| Haemophilus influenzae | 2 | +/−/− | +/−/− |
| Neisseria meningitidis | 2 | +/−/− | +/−/− |
| Bacteroides fragilis | 1 | +/−/− | +/−/− |
| Streptococcus pyogenes | 2 | +/−/− | +/−/− |
| Streptococcus agalactiae | 5 | +/−/− | +/−/− |
| Enterococcus faecalis | 10 | +/−/− | +/−/− |
| Enterococcus faecium | 5 | +/−/− | +/−/− |
| Propionibacterium acnes | 1 | +/−/− | +/−/− |
+, presence of PCR product; −, absence of PCR product.
CoNS, coagulase negative staphylococci.
TABLE 3.
Discrepant results of direct PCR testing for 15 positive blood cultures compared with organism PCRa
| Organism type (no. of isolates) | Organism PCR (16S/mecA/nuc) | Direct PCR
|
|
|---|---|---|---|
| Neat (16S/mecA/nuc) | 1:10 diluted template (16S/mecA/nuc) | ||
| MRSA (2) | +/+/+ | −/+/+ | +/+/+ |
| MSSA (2) | +/−/+ | −/−/+ | +/−/+ |
| MRCoNS (7) | +/+/− | −/+/− | +/+/− |
| MSCoNS (2) | +/−/− | −/−/− | +/−/− |
| E. coli (2) | +/−/− | −/−/− | +/−/− |
+, presence of PCR product; −, absence of PCR product. MRCoNS and MSCoNS, methicillin-resistant and susceptible, coagulase-negative staphylococci, respectively.
Fifty positive blood cultures with nonstaphylococcal organisms and 20 negative blood cultures were used to assess the specificity of the assay. None of the positive blood cultures that grew organisms other than staphylococci showed amplification products corresponding to either the mecA or nuc product (Table 2). The assay was successful in amplifying the 16S rRNA gene product from other bacteria when present. Only two such specimens initially failed to show a product after PCR amplification. In these samples, the multiplex PCR assay was able to detect the 16S rRNA gene product after a 1:10 dilution of the template. Multiplex PCR did not amplify any product from the 20 negative blood cultures.
DISCUSSION
Rapid identification of S. aureus and detection of methicillin resistance are essential for early initiation of appropriate antimicrobial therapy and to limit the inappropriate use of glycopeptide agents. We chose a multiplex PCR assay to detect the 16S rRNA gene (used as an internal control), the mecA gene (responsible for true methicillin resistance in staphylococci), and the nuc gene (found only in S. aureus and not in coagulase-negative staphylococci). Current standard laboratory methods for detecting oxacillin resistance require at least 48 to 72 h for isolation, identification, and susceptibility testing. We have described a rapid method for detection of methicillin-resistant staphylococci directly from positive blood culture specimens in less than 3 h. The assay also accurately distinguished S. aureus from coagulase-negative staphylococci.
The sensitivity of this multiplex PCR assay when used directly on extracted DNA from positive blood cultures was 105 CFU/ml with seeded 5-day negative blood cultures with a control MRSA strain (S. aureus ATCC 43300). This should be a sufficient detection limit, since the presence of microorganisms is easily seen on Gram stain prior to PCR testing, and none of the positive blood cultures containing staphylococci were misidentified or missed. A random subset of 60 positive blood cultures with staphylococci were subjected to colony counts, with counts ranging from 105 to 109 CFU/ml (data not shown).
Brakstad et al. noted a decreased PCR sensitivity when testing the nuc PCR assay with S. aureus in the presence of blood and urine, achieving a sensitivity of 103 CFU, while the detection limit with S. aureus cells suspended in saline was <10 CFU (3). We were unable to match this sensitivity, perhaps because of the multiplex format, the nature of the specimens, and our shortened thermocycling conditions with a GeneAmp 9600 thermocycler. Murakami et al. noted that in bacterial suspensions used for PCR for detection of the mecA gene, a detection limit of 4 × 105 CFU/ml could be achieved (17). This somewhat lower sensitivity may be due to the fact that mecA is a single-copy gene.
In our study, when the nuc PCR assay was performed in a nonmultiplexed format, a sensitivity of 103 CFU/ml could be achieved. However, with the mecA PCR assay in a nonmultiplexed format, a sensitivity of only 105 CFU/ml was possible. There are many factors that may affect the kinetics of a single PCR with just one set of primers, and many more factors may play a role in a multiplex PCR. Even the size of the amplification product may influence how much product is visible after gel electrophoresis detection.
Numerous reports in the literature have noted problems of inhibition of PCR assays when attempting to use clinical specimens that contain blood or blood by-products. These inhibitory substances can include sodium polyanetholesulfonic acid, heme and its metabolic products, and certain acidic polysaccharides, which can negatively affect PCR-based methods (6, 16). In addition, Al-Soud et al. have reported that components such as human plasma immunoglobulin G (1) and lactoferrin (2) can also be major inhibitors of PCR. In a recent report describing a multiplex PCR-based method for detection of methicillin-resistant staphylococci, Mason et al. noted 4 of 77 samples which failed to produce any amplifiable PCR product (14). Although not addressed, inhibition of the PCR assay may have occurred. Another report by Lem et al. noted that inhibition occurred in 2 of 100 samples, although no attempt was made to perform additional testing to resolve the inhibited samples (12).
The 16S rRNA gene primers included in our multiplex PCR assay served as an internal control to ensure the integrity of each PCR. The majority of positive blood cultures were correctly identified with our direct multiplex PCR assay. However, in a small number of specimens, PCR did not initially amplify the 16S rRNA gene product. Subsequent dilution of the template preparations appeared to have diluted out these interfering substances or inhibitors and allowed amplification of the 16S rRNA gene product as well as the mecA and/or nuc product. Millar et al. reported that extraction of DNA from BacT/Alert blood culture bottles containing sodium polyanetholesulfonic acid may be problematic for PCR. Serial dilutions of the prepared DNA template resulted in removal of the inhibitor, allowing amplification of the bacterial DNA (15).
We describe here a simple yet rapid method for detection of methicillin-resistant staphylococci from positive blood cultures. Although there have been several reports in the literature describing PCR-based methods for direct detection of resistant staphylococci (8, 12, 14, 20, 23), none have been optimized to allow both rapid DNA extraction and rapid PCR and detection. A potential limitation of this method is that the assay would not be able to distinguish the presence of MRSA in a blood culture from a culture that yielded a mixed growth of coagulase-negative and methicillin-susceptible staphylococci. However, such a mixed culture is unlikely to occur frequently; of the 236 positive blood cultures that grew staphylococci, none yielded more than one species.
Although PCR testing has been limited to reference centers and highly specialized laboratories, PCR testing in a routine diagnostic microbiology laboratory is a possibility. Reports in the literature have described the routine use of PCR methods and have shown that successful integration of molecular detection with conventional microbiology techniques can be achieved (9, 19). The costs associated with PCR-based methods may not be as prohibitive as they once were. Techniques for batch preparation and aliquoting of PCR reagents, DNA controls, and extraction buffers have made routine PCR testing much easier to perform. For our multiplex PCR assay for the detection of methicillin-resistant staphylococci, the reagent and supplies cost for reporting the PCR test result of one specimen is $7.65 (Canadian dollars). This includes three controls run with each test. The total “hands-on” technologist time is approximately 30 min (includes extraction, PCR setup, agarose gel electrophoresis setup, and documentation of results). At $30.00 (Canadian dollars)/h, the total cost of a PCR assay for one specimen is $22.65 (Canadian dollars). However, the cost per specimen is considerably less if samples are batched. For the present study, a batch of 38 samples (including controls) was tested at a time, and the cost of the PCR assay, including reagents, disposables, and labor, was $3.29 (Canadian dollars) per specimen.
For laboratories that wish to use commercially available test kits for detecting methicillin-resistant staphylococci, two commercially available systems for detection of methicillin resistance mediated by the mecA gene are Food and Drug Administration cleared for use in diagnostic laboratories. These are the Velogene kit (Alexon-Trend, Ramsey, Minn.) and the MRSA-Screen (Denka-Seiken, Tokyo, Japan). Although these tests are excellent for detection of either the mecA gene (Velogene) or the product of the mecA gene, PBP2a (MRSA-Screen), both require overnight growth of an organism for testing.
The results presented in this study have shown that rapid DNA extraction with a multiplex PCR-based assay is possible, with results available in less than 3 h. Further optimization of multiplex PCR-based assays, coupled with improved PCR technology, such as real-time PCR systems, may lead to improved assay sensitivity, overcoming inhibition problems, and even more rapid results, allowing clinicians to make prompt decisions for the effective management of patients with staphylococcal bacteremia.
Acknowledgments
We are grateful for support from the OAML Research Trust Small Grants Program (no. SGP-98-009).
REFERENCES
- 1.Al-Soud, W. A., and P. Rådström. 2001. Purification and characterization of PCR-inhibitory components in blood cells. J. Clin. Microbiol. 39:485-493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Al-Soud, W. A., L. J. Jönsson, and P. Rådström. 2000. Identification and characterization of immunoglobulin G in blood as a major inhibitor of diagnostic PCR. J. Clin. Microbiol. 38:345-350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brakstad, O. G., K. Aasbakk, and J. Maeland. 1992. Detection of Staphylococcus aureus by polymerase chain reaction amplification of the nuc gene. J. Clin. Microbiol. 30:1654-1660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Centers for Disease Control and Prevention NNIS System. 2001. National nosocomial infections surveillance (NNIS) system report, data summary from January 1992-June 2001, issued August 2001. Am. J. Infect. Control 29:404-421. [DOI] [PubMed] [Google Scholar]
- 5.Diekema, D. J., M. A. Pfaller, F. J. Schmitz, J. Smayevsky, J. Bell, R. N. Jones, M. Beach, and the SENTRY Participants Group. 2001. Survey of infections due to Staphylococcus species: frequency of occurrence and antimicrobial susceptibility of isolates collected in the United States, Canada, Latin America, Europe, and the Western Pacific region for the SENTRY antimicrobial surveillance program, 1997-1999. Clin. Infect. Dis. 32(Suppl. 2):S114-S132. [DOI] [PubMed] [Google Scholar]
- 6.Fredricks, D. N., and D. A. Relman. 1998. Improved amplification of microbial DNA from blood cultures by removal of the PCR inhibitor sodium polyanetholesulfonate. J. Clin. Microbiol. 36:2810-2816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Herold, B. C., L. C. Immergluck, M. C. Maranan, D. S. Lauderdale, R. E. Gaskin, S. Boyle-Vavra, C. D. Leitch, and R. S. Daum. 1998. Community-acquired methicillin-resistant Staphylococcus aureus in children with no identified predisposing risk. JAMA 279:593-598. [DOI] [PubMed] [Google Scholar]
- 8.Jaffe, R. I., J. D. Lane, S. V. Albury, and D. M. Niemeyer. 2000. Rapid extraction from and direct identification in clinical samples of methicillin-resistant staphylococci with the PCR. J. Clin. Microbiol. 38:3407-3412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jayaratne, P., and C. Rutherford. 1999. Detection of methicillin-resistant Staphylococcus aureus (MRSA) from growth on mannitol salt oxacillin agar with PCR for nosocomial surveillance. Diagn. Microbiol. Infect. Dis. 35:13-18. [DOI] [PubMed] [Google Scholar]
- 10.Kloos, W. E., and T. L. Bannerman. 1999. Staphylococcus and Micrococcus, p. 264-282. In P. R. Murray, E. J. Baron, M. A. Pfaller, F. C. Tenover, and R. H. Yolken (ed.), Manual of clinical microbiology. ASM Press, Washington, D.C.
- 11.Lacroix, J., K. Jarvi, S. Batra, D. Heritz, and M. Mittelman. 1996. PCR-based technique for the detection of bacteria in semen and urine. J. Microbiol. Methods 26:61-71. [Google Scholar]
- 12.Lem, P., J. Spiegelman, B. Toye, and K. Ramotar. 2001. Direct detection of mecA, nuc and 16S rRNA genes in BacT/Alert blood culture bottles. Diagn. Microbiol. Infect. Dis. 41:165-168. [DOI] [PubMed] [Google Scholar]
- 13.Louie, L., S. O. Matsumura, E. Choi, M. Louie, and A. E. Simor. 2000. Evaluation of three rapid methods for detection of methicillin resistance in Staphylococcus aureus. J. Clin. Microbiol. 38:2170-2173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mason, W. J., J. S. Blevins, K. Beenken, N. Wibowo, N. Ojha, and M. S. Smeltzer. 2001. Multiplex PCR protocol for the diagnosis of staphylococcal infection. J. Clin. Microbiol. 39:3332-3338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Millar, B. C., X. Jiru, J. E. Moore, and J. A. P. Earle. 2000. A simple and sensitive method to extract bacterial, yeast and fungal DNA from blood culture material. J. Microbiol. Methods 42:139-147. [DOI] [PubMed] [Google Scholar]
- 16.Monteiro, L., D. Bonnemaison, A. Vekris, K. G. Petry, J. Bonnet, R. Vidal, J. Cabrita, and F. Mégraud. 1997. Complex polysaccharides as PCR inhibitors in feces: Helicobacter pylori model. J. Clin. Microbiol. 35:995-998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Murakami, K., W. Minamide, K. Wada, E. Nakamura, H. Teraoka, and S. Watanabe. 1991. Identification of methicillin-resistant strains of staphylococci by polymerase chain reaction. J. Clin. Microbiol. 29:2240-2244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.National Committee for Clinical Laboratory Standards. 1999. Performance standards for antimicrobial susceptibility testing; ninth informational supplement, M100-S9. National Committee for Clinical Laboratory Standards, Wayne, Pa.
- 19.Petrich, A., K. Luinstra, B. Page, S. Callery, D. Stevens, A. Gafni, D. Groves, M. Chernesky, and J. B. Mahony. 2001. Effect of routine use of a multiplex PCR for detection of vanA- and vanB-mediated enterococcal resistance on accuracy, cost and earlier reporting. Diagn. Microbiol. Infect. Dis. 41:215-220. [DOI] [PubMed] [Google Scholar]
- 20.Schmitz, F. J., C. R. MacKenzie, B. Hofmann, J. Verhoef, M. Finken-Eigen, H. P. Heinz, and K. Kohrer. 1997. Specific information concerning taxonomy, pathogenicity and methicillin resistance of staphylococci obtained by multiplex PCR. J. Med. Microbiol. 46:773-778. [DOI] [PubMed] [Google Scholar]
- 21.Simor, A. E., M. Ofner-Agostini, E. Bryce, K. Green, A. McGeer, M. Mulvey, S. Paton, and the Canadian Nosocomial Infection Surveillance Program, Health Canada. 2001. The evolution of methicillin-resistant Staphylococcus aureus in Canadian hospitals: 5 years of national surveillance. Can. Med. Assoc. J. 165:21-26. [PMC free article] [PubMed] [Google Scholar]
- 22.Whitby, M., M. L. McLaws, and G. Berry. 2001. Risk of death from methicillin-resistant Staphylococcus aureus bacteraemia: a meta-analysis. Med. J. Aust. 175:264-267. [DOI] [PubMed] [Google Scholar]
- 23.Zheng, X., C. P. Kolbert, P. Varga-Delmore, J. Arruda, M. Lewis, J. Kolberg, F. R. Cockerill, and D. H. Persing. 1999. Direct mecA detection from blood culture bottles by branched-DNA signal amplification. J. Clin. Microbiol. 37:4192-4193. [DOI] [PMC free article] [PubMed] [Google Scholar]