Abstract
A method for reliable quantification of Pneumocystis carinii in research models of P. carinii pneumonia (PCP) that is more convenient and reproducible than microscopic enumeration of organisms would greatly facilitate investigations of this organism. We developed a rapid quantitative touchdown (QTD) PCR assay for detecting P. carinii f. sp. carinii, the subspecies of P. carinii commonly used in research models of PCP. The assay was based on the single-copy dihydrofolate reductase gene and was able to detect <5 copies of a plasmid standard per tube. It was reproducibly quantitative (r = 0.99) over 6 log values for standards containing ≥5 copies/tube. Application of the assay to a series of 10-fold dilutions of P. carinii organisms isolated from rat lung demonstrated that it was reproducibly quantitative over 5 log values (r = 0.99). The assay was applied to a recently reported in vitro axenic cultivation system for P. carinii and confirmed our microscopy findings that no organism multiplication had occurred during culture. For all cultures analyzed, QTD PCR assays showed a decrease in P. carinii DNA that exceeded the expected decrease due to dilution of the inoculum upon transfer. In conclusion, a rapid, sensitive, and reproducible quantitative PCR assay for P. carinii f. sp. carinii has been developed and is applicable to in vivo as well as in vitro systems. The assay should prove useful for conducting studies in which quantification of organism burden or growth assessment is critical, such as in vitro antimicrobic susceptibility testing or in vivo immunopathological experiments.
Accurate quantification of Pneumocystis carinii is essential for the correct interpretation of many research experiments of this organism. Since the organism cannot be readily grown in vitro, the traditional quantification method for P. carinii is enumeration of organisms by microscopic examination, which is difficult and cumbersome due to the organism's growth in clusters and the presence of different morphological forms (14, 19). Different stains vary in their sensitivities of detection as well as in their abilities to detect the different forms of P. carinii. Definitive diagnosis of P. carinii pneumonia (PCP) often relies on identification of cysts, which frequently occur within clusters of trophozoites. However, quantification of cysts is not an optimal method for measurement of organism burden, since the cyst-to-trophozoite ratio may change under different conditions and stages of PCP (21, 22).
Quantification by a molecular method would eliminate difficulties in visual enumeration of organisms and would also be independent of the observer. Assessment of organism burden is important in animal models of immunopathogenesis, and accurate quantification is the cornerstone of a number of in vivo and in vitro studies, for example, those pertaining to antimicrobial susceptibilities. Various molecular approaches involving total DNA measurement or semiquantitative PCR methods using end point detection have previously been reported but are currently not broadly utilized (9, 14, 16).
We undertook the development of a real-time PCR assay to quantify P. carinii f. sp. carinii, the common subspecies infecting the immunosuppressed laboratory rat, which is the most commonly used in vivo model of PCP (2, 19). We then assessed this new assay during studies attempting to establish the continuous axenic culture system recently described by Merali et al. (14).
MATERIALS AND METHODS
Cloned template.
The P. carinii f. sp. carinii dihydrofolate reductase (DHFR) gene (665 bp; GenBank accession no. AF322061) (3, 12) was cloned into the pCRII vector (Invitrogen) and used as a standard. After propagation and purification of the plasmid, the concentration of DHFR gene copies (number of copies per microliter) was derived from the A260 optical density measurement and the plasmid molecular weight. Tenfold serial dilutions (10−2 to 105 DHFR gene copies/μl) of the linearized plasmid in water were prepared with glycogen (33.3 μg/ml) as the DNA carrier.
P. carinii.
P. carinii organisms from the lungs of corticosteroid-treated Sprague-Dawley rats (obtained by contract from Indiana University) were partially purified by Ficoll-Hypaque density gradient centrifugation as previously described (7). P. carinii pellets were inoculated immediately into the culture medium for in vitro cultivation purposes or stored at −70°C for subsequent quantitative touchdown (QTD) PCR analysis. For analysis by QTD PCR and microscopy, the pellet (0.7 ml) was thawed and resuspended 20-fold in phosphate-buffered saline (PBS), and 10-fold serial dilutions in PBS were prepared. The number of DHFR gene copies per tube (QTD PCR results) was converted to the number of whole organisms per milliliter of original sample by multiplying by a factor of 30 (because of the concentration of the 200-μl sample used during DNA extraction and the utilization of 5 μl of DNA extract per PCR tube).
Organisms cultivated and frozen in dimethyl sulfoxide as previously described (14) were kindly provided by A. B. Clarkson, Jr., and S. Merali for inoculation of in vitro cultures.
In vitro cultivation.
Either freshly isolated organisms from two rats or two frozen aliquots were used for initiation of primary cultures. Axenic in vitro cultures were performed as previously described (14). Briefly, the cultures were maintained in collagen-coated Transwell inserts held in appropriate multiwell plates (Costar) and incubated at 31°C in ambient air. The volumes of the media in the insert and below the insert were as previously described (14). The basic medium was minimal essential medium with Earle's salts (Gibco) supplemented as previously described (14). S-adenosyl-l-methionine (SAM; Sigma or Research Biochemical International) was added twice daily. In initial experiments, stock solutions of medium containing SAM (500 μg/ml) were freshly prepared, and the medium below the inserts was exchanged twice daily as initially reported (14). In later experiments, freshly prepared SAM stock solution (10 mM) was added separately twice daily to the culture wells to yield a final concentration of 500 μM as subsequently reported (15), and the medium below the inserts was exchanged daily.
When the wells were harvested, the organisms were suspended by agitation and the volume in the insert was removed. Subcultures were immediately performed, and slides were prepared for microscopy. The remaining volume was stored at −70°C for later analysis.
Microscopy.
For assessing in vitro growth by direct microscopy, sets of two replicate slides were prepared. One set was prepared with 2 μl of the harvested material, and additional sets were prepared with diluted material corresponding to the inoculated density when passaged into new subcultures. Each set of replicate slides was stained with Diff-Quik (Dade Behring) and by an indirect immunofluorescent-antibody assay using a previously described monoclonal antibody, 7D7, known to stain rat-derived Pneumocystis (10).
DNA extraction and amplification.
DNA was extracted by using the NucliSens kit (Organon Teknika) according to the manufacturer's recommendations. A 200-μl volume of each specimen was used per extraction. Primers RDHFR11 (5′-GTT GCA CTT ACA ACT TCT TAT GG-3′) and RDHFR12 (5′-TAG ATC CAG AGA TTC ATT TCG AG-3′) were designed to amplify a 234-bp segment of the single-copy DHFR gene (3, 12). The primers were commercially synthesized (Operon Technology). For PCR, all reactions were performed in glass capillaries (Roche) with a final reaction volume of 20 μl of 1× LightCycler-FastStart DNA Master Hybridization Probes reaction mixture (Roche) containing FastStart Taq, reaction buffer, deoxynucleoside triphosphate, a 0.5 μM concentration of each primer, a 0.2 μM concentration of each fluorescence resonance energy transfer (FRET) probe, 5.0 mM MgCl2, and 1 U of heat-labile uracil-DNA glycosylase (UNG) (from BMTU 3346; Roche). The FastStart Taq ensures hot start. In the reaction mixture, dUTP was substituted for dTTP. Five microliters of extracted DNA or cloned template per tube was analyzed. All samples were kept on ice during preparation. Tubes were incubated at room temperature (20.0 to 25.0°C) for 10 min to allow UNG activity to take place. Thermocycling and detection were performed on the LightCycler (Roche). An initial preheating step of 10 min at 95°C was used to activate the DNA polymerase, inactivate UNG, and melt double-stranded DNA. Then a touchdown procedure was utilized, consisting of 11 cycles as follows: 5 s at 95°C; annealing for 10 s at temperatures decreasing from 65 to 55°C, with a decrease of 1°C per cycle; and an extension step of 20 s at 72°C. An additional 34 cycles with annealing at 55°C were performed. At least two negative controls were included in each PCR experiment.
Amplicon detection.
FRET detection probes were designed to anneal with a 1-base gap between the probes. The following probes were commercially synthesized (Idaho Technology) and labeled with Red 640 as the reporter: RDHFR13 (5′-CCT CTT TTG TAC CAA CTT TTG ATT CAT T-fluorescein-3′) and RDHFR14 (5′-Red 640-GAA TCG ATG AAT GTT GTA TTG ATG GG-phosphate-3′).
Data analysis.
All acquired fluorescence data were analyzed by using LightCycler software. In theory, true quantification is achieved with real-time PCR since the cycle number in which amplicons become detectable is proportional to the logarithm of the initial number of templates. In each experiment, at least three standards of cloned template (105, 103, and 10 copies/μl of specimen) were included to generate a standard curve for quantification of positive specimens.
RESULTS
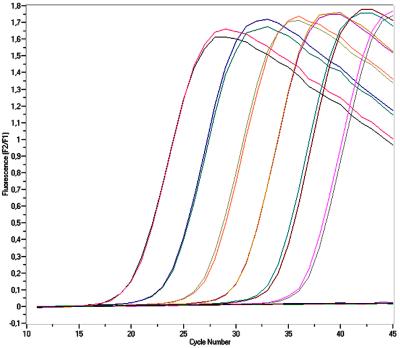
We utilized a real-time PCR format that is quantitative, rapid, and highly reproducible (5) (Fig. 1). We chose the single-copy DHFR gene as the PCR target since this would allow direct quantitation of the number of organisms from the number of DHFR gene copies detected by the assay. The goal was to develop an assay that would be applicable to in vivo as well as in vitro studies of PCP.
FIG. 1.
Real-time detection of PCR products: fluorescence data acquired during an experiment testing cloned standards in duplicate. Standard concentrations of 5 × 100 to 5 × 105 copies/tube were positive. The standards became positive approximately 3.3 cycles apart, corresponding to the 10-fold increase in concentration.
Cloned template.
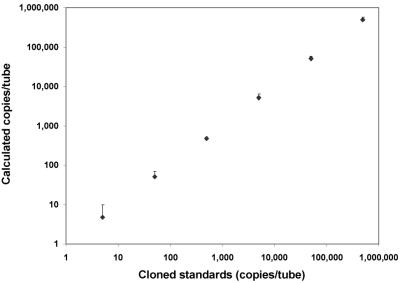
The performance of the QTD PCR assay was evaluated by analyzing 10-fold serial dilutions of the cloned DHFR gene template in 20 separate experiments. Standard curves (cycle number versus log concentration) were constructed by linear regression, and the calculated values for standards were recorded. All of the 9 standards with 0.05 copies/tube were negative, 4 of the 11 standards with 0.5 copies/tube were positive (mean, 1.9), and 12 of the 13 standards with 5 copies/tube were positive, indicating a sensitivity of <5 copies per tube. Plotting all standards with ≥5 copies/tube together (n = 94), the correlation coefficient (r) was found to be 0.99 (Fig. 2). Thus, the assay was quantitative over 6 log values down to 5 copies per tube and had the potential to be useful for quantitation of P. carinii organisms.
FIG. 2.
QTD PCR applied to cloned template: correlation between measured and calculated concentrations of P. carinii DHFR gene copies using serial dilutions of plasmid DNA. Values represent the mean (+1.96 standard deviations [SD]) calculated concentrations (⧫) of standards (5 to 500,000 copies/tube) assayed by QTD PCR (r = 0.99; n = 94).
P. carinii organisms isolated from in vivo rat model.
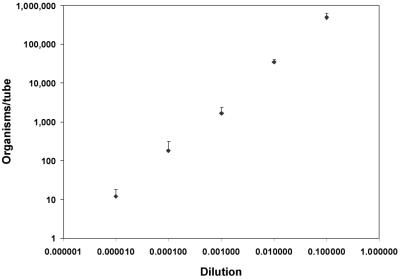
To evaluate the assay's ability to quantitate whole organisms, DNA was extracted from 10-fold serial dilutions of rat-derived P. carinii organisms and assayed. In two separate experiments, five replicates were tested at dilutions of 10−1 to 10−8, and two replicates were tested at dilutions of 10−9 to 10−11. All PCR tubes with dilutions of 10−7 or more (n = 16) were negative, and one of the five tubes with dilutions of 10−6 (0.7 copies/tube) was positive. All tubes diluted 10−1 to 10−5 were positive (n = 25) and, as shown in Fig. 3, yielded an r of 0.99. Thus, the assay was quantitative over at least 5 log values, down to 12 copies/tube, when applied to whole organisms. By extrapolation, the original pellet of organisms had a concentration of 2.9 × 109 organisms per ml. Uninfected healthy rat DNA was negative by this assay.
FIG. 3.
QTD PCR applied to whole organisms: correlation between the calculated number of DHFR gene copies and the dilution factor of whole organisms. Values represent the mean (+1.96 SD) calculated concentrations (⧫) of 10-fold dilutions of resuspended pelleted P. carinii organisms isolated from rat lung (r = 0.99; n = 25).
In vitro cultures of P. carinii.
We attempted to reproduce a culture system recently reported to be successful in axenically culturing P. carinii (14), with assessment of organism growth by light microscopy. Culture growth was further assessed by QTD PCR. To determine the optimal inoculation density for primary cultures, the density of the inoculum for initiation of cultures was varied (with differences in density of as much as 104-fold). The intervals between subculturing ranged from 3 to 51 days. Dilutions of 1:10 and/or 1:100 were performed at each passage with up to six passages per culture. In one experiment, replicate subcultures were initiated simultaneously and then harvested at different time points for evaluation of the growth rate over time. A total of 95 cultures (primary cultures or subcultures) were performed.
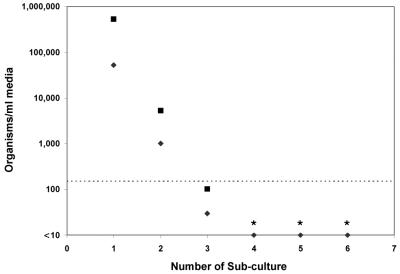
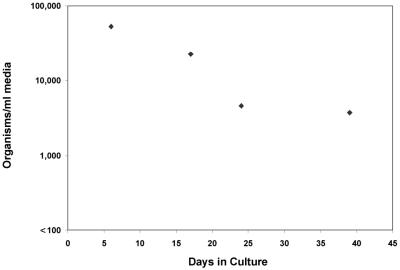
Only semiquantitative counting of P. carinii organisms was found to be possible by microscopy. With this method, growth was not observed at any point in any of the cultures, whether they were derived from frozen samples or from freshly isolated organisms from rat lung (data not shown). The QTD PCR findings were consistent with this observation; in three experiments, harvested material from 39 cultures showed a median decrease in the number of P. carinii DHFR gene copies of 84% (range, 19 to 100%), with no increase over time in any culture. Data from one experiment are shown in Fig. 4. In this experiment, the primary culture was inoculated with 5.4 × 106 organisms/ml, as quantitated by QTD PCR, and the first subculture was passaged (1:10) 10 days later. Passages 4, 5, and 6 were negative by QTD PCR, indicating <150 organisms/ml of medium. In another experiment, replicate subcultures initiated simultaneously at day 0 and harvested at different time points also showed a decrease in P. carinii DNA at all time points (Fig. 5). This sampling method circumvented any potentially negative effects caused by repetitive sampling and disruption of organisms in a single well (14).
FIG. 4.
Quantitation of P. carinii organisms passaged serially (⧫). For each subculture, a 1:10 dilution was performed. For reference, the calculated inoculated amount of DNA for each subculture is graphed to show the expected value if there were no replication and no loss of DNA during culture (▪). The dashed line indicates the level of sensitivity and lower limit of quantitation (5 copies/tube equal to 150 organisms/ml). The first subculture was derived from a primary culture inoculated to a density of 5.4 × 106 copies/ml by passage after 10 days of incubation. ∗, subcultures 4, 5, and 6 were negative by QTD PCR.
FIG. 5.
P. carinii culture over time: quantitative results from four replicate subcultures (♦). A primary culture was inoculated to a density of 5.4 × 106 copies/ml and subsequently passaged (1:10) 10 days later (day 0) into four replicate subcultures, which were then assayed at the indicated time intervals.
DISCUSSION
This QTD PCR assay is able to detect fewer than 5 copies of the rat-derived P. carinii DHFR gene per tube, which correlates to fewer than 150 organisms/ml of specimen. Using cloned templates (Fig. 2) and organisms isolated from infected rat lung (Fig. 3), the assay yielded reproducible results over several log values. The slightly higher correlation for cloned templates compared to that for whole organisms (Fig. 2 and 3) was possibly due to heavy organism clumping, which may have compromised the accuracy of the serial dilutions. The unusual heavy clumping was possibly due to the use of pelleted frozen organisms for the dilution series. We compared quantitation by light microscopy with that by QTD PCR but found that clumping and the difficulty in identifying organisms permitted only a semiquantitative estimation by Diff-Quik staining. Investigators should find the QTD PCR assay to be an easier yet more reliable and accurate method for quantification of the number of organisms in rats, in culture, or in other research studies.
We undertook axenic cultivation of P. carinii by using a previously reported method (14) but were unsuccessful. A drop in organism density was estimated by microscopy for all cultures. With QTD PCR, a steady decrease in P. carinii DNA during culture was observed and no increase in DNA was demonstrated at any time in any of the cultures. The decrease in DNA was likely due to degradation of DNA from dead organisms. There are several potential explanations for our inability to reproduce the continuous axenic cultivation previously reported (14, 15). First, although the fresh rat lung-derived organisms used for inoculation of primary cultures were morphologically intact by microscopy, they were not assessed for viability. We also used frozen samples reported to retain viability (14), but it is possible that these organisms were also not viable. A second explanation involves the stability of SAM, which has been reported to be a key factor in the successful cultivation of P. carinii (14). P. carinii is reported to be a SAM auxotroph (15). SAM is a key intermediate metabolite (e.g., as a methyl-group donor and precursor of polyamines), and to our knowledge, no other organism has been reported to be a SAM auxotroph (other than SAM-requiring mutants [1, 15]). SAM is a very unstable compound, so methodological differences in preparation, storage, or administration may have affected our culture attempts.
Our QTD PCR method has a number of advantages over standard PCR assays. A touchdown protocol can increase the specificity and sensitivity of assays (4). Initial high annealing temperatures reduce formation of spurious PCR products that might interfere with amplification of the specific target in low-copy-number samples. The annealing temperature is then gradually lowered to a level that increases yield. We have recently reported that a touchdown procedure can be incorporated into a real-time PCR assay without compromising detection by FRET probes, which ensures efficient and specific amplification in low-copy-number samples, thereby increasing the sensitivity (8). No manipulation of amplicons is required at any step during real-time PCR, lessening the possibility of contamination problems. To further reduce contamination risks, UNG and dUTP were used for prevention of amplicon carryover (20). The entire assay has a turnaround time of <4 h, including DNA extraction, which further increases the potential utility of this procedure.
A recent study utilized real-time PCR with the LightCycler to quantitate rat- and human-derived organisms but used SYBR green rather than specific probes for detection of the amplified product (6). We feel that a gene-specific confirmatory probe is essential for maximization of the specificity of this method. The authors of the latter study (6) noted that rat-derived DHFR primers (18) amplified human-derived P. carinii RNA. However, we and others have recently shown that those primers amplify rat-derived but not human-derived P. carinii DHFR genes (11, 17). Molecular viability assays targeting P. carinii f. sp. carinii mRNA have also been described recently (6, 13). However, such assays are not suitable for quantifying the numbers of organisms, since the number of mRNA molecules per organism is unknown and since one cannot assume that the amount of mRNA per organism is the same under all conditions.
In conclusion, we have developed a rapid, sensitive, and reproducible method for quantitating P. carinii f. sp. carinii by PCR that has the potential to replace the tedious and cumbersome quantitative methods currently used in animal studies and that provides a reliable and accurate method for other research efforts, such as in vitro cultivation and antimicrobial drug efficacy testing.
Acknowledgments
Hans Henrik Larsen was supported by the Copenhagen HIV Programme, Danish Medical Research Council grant no. 9900484, Dagmar Marshall's Foundation, and Snedkermester Sophus Jacobsen og hustru Astrid Jacobsens Foundation.
We thank Salim Merali and Allen B. Clarkson, Jr., New York University School of Medicine, for providing frozen cultured organisms for seeding of cultures and Thomas Jonassen, Copenhagen University, for providing material from an uninfected healthy rat.
REFERENCES
- 1.Cherest, H., and Y. Surdin-Kerjan. 1978. S-adenosyl methionine requiring mutants in Saccharomyces cerevisiae: evidences for the existence of two methionine adenosyl transferases. Mol. Gen. Genet. 163:153-167. [DOI] [PubMed] [Google Scholar]
- 2.Dei-Cas, E., M. Brun-Pascaud, V. Bille-Hansen, A. Allaert, and E. M. Aliouat. 1998. Animal models of pneumocystosis. FEMS Immunol. Med. Microbiol. 22:163-168. [DOI] [PubMed] [Google Scholar]
- 3.Edman, J. C., U. Edman, M. Cao, B. Lundgren, J. A. Kovacs, and D. V. Santi. 1989. Isolation and expression of the Pneumocystis carinii dihydrofolate reductase gene. Proc. Natl. Acad. Sci. USA 86:8625-8629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hecker, K. H., and K. H. Roux. 1996. High and low annealing temperatures increase both specificity and yield in touchdown and stepdown PCR. BioTechniques 20:478-485. [DOI] [PubMed] [Google Scholar]
- 5.Heid, C. A., J. Stevens, K. J. Livak, and P. M. Williams. 1996. Real time quantitative PCR. Genome Res. 6:986-994. [DOI] [PubMed] [Google Scholar]
- 6.Kaiser, K., M. Rabodonirina, and S. Picot. 2001. Real time quantitative PCR and RT-PCR for analysis of Pneumocystis carinii hominis. J. Microbiol. Methods 45:113-118. [DOI] [PubMed] [Google Scholar]
- 7.Kovacs, J. A., J. L. Halpern, J. C. Swan, J. Moss, J. E. Parrillo, and H. Masur. 1988. Identification of antigens and antibodies specific for Pneumocystis carinii. J. Immunol. 140:2023-2031. [PubMed] [Google Scholar]
- 8.Larsen, H. H., H. Masur, J. A. Kovacs, V. J. Gill, V. A. Silcott, P. Kogulan, J. Maenza, M. Smith, D. R. Lucey, and S. H. Fischer. 2002. Development and evaluation of a quantitative, touch-down, real-time PCR assay for diagnosing Pneumocystis carinii pneumonia. J. Clin. Microbiol. 40:490-494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lee, C. H., N. L. Bauer, M. M. Shaw, M. M. Durkin, M. S. Bartlett, S. F. Queener, and J. W. Smith. 1993. Proliferation of rat Pneumocystis carinii on cells sheeted on microcarrier beads in spinner flasks. J. Clin. Microbiol. 31:1659-1662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lundgren, B., J. A. Kovacs, N. N. Nelson, F. Stock, A. Martinez, and V. J. Gill. 1992. Pneumocystis carinii and specific fungi have a common epitope, identified by a monoclonal antibody. J. Clin. Microbiol. 30:391-395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ma, L., L. Borio, H. Masur, and J. A. Kovacs. 1999. Pneumocystis carinii dihydropteroate synthase but not dihydrofolate reductase gene mutations correlate with prior trimethoprim-sulfamethoxazole or dapsone use. J. Infect. Dis. 180:1969-1978. [DOI] [PubMed] [Google Scholar]
- 12.Ma, L., H. Imamichi, A. Sukura, and J. A. Kovacs. 2001. Genetic divergence of the dihydrofolate reductase and dihydropteroate synthase genes in Pneumocystis carinii from 7 different host species. J. Infect. Dis. 184:1358-1362. [DOI] [PubMed] [Google Scholar]
- 13.Maher, N., S. Vermund, M. Lasbury, C. Lee, M. Bartlett, and T. R. Unnasch. 2000. Development and evaluation of a molecular viability assay for Pneumocystis carinii. J. Clin. Microbiol. 38:1947-1952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Merali, S., U. Frevert, J. H. Williams, K. Chin, R. Bryan, and A. B. Clarkson, Jr. 1999. Continuous axenic cultivation of Pneumocystis carinii. Proc. Natl. Acad. Sci. USA 96:2402-2407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Merali, S., D. Vargas, M. Franklin, and A. B. Clarkson, Jr. 2000. S-adenosylmethionine and Pneumocystis carinii. J. Biol. Chem. 275:14958-14963. [DOI] [PubMed] [Google Scholar]
- 16.O'Leary, T. J., M. M. Tsai, C. F. Wright, and M. T. Cushion. 1995. Use of semiquantitative PCR to assess onset and treatment of Pneumocystis carinii infection in rat model. J. Clin. Microbiol. 33:718-724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ortona, E., P. Margutti, A. De Luca, S. E. Peters, A. E. Wakefield, E. Tamburrini, P. Mencarini, E. Visconti, and A. Siracusano. 1996. Nonspecific PCR products using rat-derived Pneumocystis carinii dihydrofolate reductase gene-specific primers in DNA amplification of human respiratory samples. Mol. Cell Probes 10:187-190. [DOI] [PubMed] [Google Scholar]
- 18.Schluger, N., T. Godwin, K. Sepkowitz, D. Armstrong, E. Bernard, M. Rifkin, A. Cerami, and R. Bucala. 1992. Application of DNA amplification to pneumocystosis: presence of serum Pneumocystis carinii DNA during human and experimentally induced Pneumocystis carinii pneumonia. J. Exp. Med. 176:1327-1333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sloand, E., B. Laughon, M. Armstrong, M. S. Bartlett, W. Blumenfeld, M. Cushion, A. Kalica, J. A. Kovacs, W. Martin, E. Pitt, E. L. Pesanti, F. Richards, R. Rose, and P. Walzer. 1993. The challenge of Pneumocystis carinii culture. J. Eukaryot. Microbiol. 40:188-195. [DOI] [PubMed] [Google Scholar]
- 20.Sobek, H., M. Schmidt, B. Frey, and K. Kaluza. 1996. Heat-labile uracil-DNA glycosylase: purification and characterization. FEBS Lett. 388:1-4. [DOI] [PubMed] [Google Scholar]
- 21.Sukura, A. 1995. Trophozoite-to-cyst ratio increases during recovery from Pneumocystis carinii pneumonia in rats. APMIS 103:300-306. [PubMed] [Google Scholar]
- 22.Tamburrini, E., P. Mencarini, E. Visconti, A. De Luca, M. Zolfo, A. Siracusano, E. Ortona, R. Murri, and A. Antinori. 1996. Imbalance between Pneumocystis carinii cysts and trophozoites in bronchoalveolar lavage fluid from patients with pneumocystosis receiving prophylaxis. J. Med. Microbiol. 45:146-148. [DOI] [PubMed] [Google Scholar]