Abstract
In an ongoing molecular epidemiology study, human immunodeficiency virus-negative patients with first-time pulmonary tuberculosis from a high-incidence community were enrolled. Mycobacterium tuberculosis strains were identified by restriction fragment length polymorphism analysis with two fingerprinting probes. Of 131 patients, 3 (2.3%) were shown to have a mixture of strains in one or two of their serial cultures. This study further investigated these cases with disease caused by multiple M. tuberculosis strains in the context of the molecular epidemiology of the study setting.
Most studies, using a variety of typing techniques, attribute an episode of active tuberculosis to a single bacterial clone. Multiple disease episodes caused by reinfection have been shown (4, 12), but there is limited reporting of mixed infection during a single episode of tuberculosis. Epidemiology laboratories may note mixed Mycobacterium tuberculosis cultures, but these are likely to be excluded from further analysis as possible laboratory contamination if not confirmed by serial culture, as is the general practice in our laboratory.
Phage typing and restriction fragment length polymorphic (RFLP) analysis with IS6110 have illustrated the presence of more than one M. tuberculosis strain cultured from different anatomical sites in single patients (1, 6). Initial mixed infection by a drug-resistant strain and a drug-sensitive strain was suggested, where the drug-resistant strain became the main infecting strain (10). Similarly, a drug-sensitive strain and later a drug-resistant strain with a related, but not identical, IS6110 RFLP pattern were cultured from a patient's sputum samples (3), while another patient produced a culture containing two unrelated strains, which could be differentiated on single-colony cultures (3). Further cases have been described where patients were simultaneously infected by more than one strain (9, 14), with variance in the band intensities of the different strains (14). In high-prevalence settings such as the Madrid, Spain, prison system and South African mines, mixed-strain infections have been identified, often corresponding to human immunodeficiency virus (HIV) positivity (4, 7). In those reports, the patients were chronically exposed to infectious sources of M. tuberculosis in their places of work (3, 7, 9) or by their imprisonment with high levels of exposure while HIV positive (4). In some reports, the treatment regimens were not adhered to (10, 14).
In this study, we report on three cases where multiple M. tuberculosis strains were cultured during one disease episode from HIV-negative patients who resided in a community with a very high tuberculosis incidence. The infecting strains were drug sensitive, and serial cultures confirmed the dual nature of the cultures. We discuss these cases in the context of the molecular epidemiology of the study region and their impact on our understanding of disease presentation.
MATERIALS AND METHODS
In an ongoing study since May 1999, patients with first-time pulmonary tuberculosis from five neighboring primary health care clinics in metropolitan Cape Town, South Africa, were enrolled. The disease dynamics and very high tuberculosis incidence (251 cases per 100,000 population annually) of part the community were known (12, 13), and unpublished data suggest that the incidence would be similar and equally high for the larger region.
Patients were enrolled in the study only if they were HIV negative with first-time pulmonary tuberculosis (smear and culture positive). Patients were managed according to the DOTS (directly observed treatment, short course) strategy and received a regimen consisting of isoniazid, rifampin, ethambutol, and pyrazinamide during a 2-month intensive phase and isoniazid and rifampin during a 4-month consolidation phase. Treatment of patients was initiated at diagnosis. Sputum was collected at diagnosis and at follow-up visits during treatment. Sputum was decontaminated by a standard NaOH-Na-citrate method. In short, equal volumes of 4% NaOH and 2.9% sodium citrate were added to sputum treated with Sputagest (Mast Diagnostics, Merseyside, United Kingdom) to a final volume of twice the volume of the treated sputum. The samples were incubated at 37°C with shaking (330 rpm) for 20 min. Two volumes of phosphate buffer (0.05 M, pH 6.8) were added, and the samples were centrifuged at 3,000 rpm (1,900 × g) for 20 min. The pellet was resuspended in phosphate buffer to a volume of 2.5 ml. The concentrated sediment was inoculated into BACTEC12B medium (Becton Dickinson, Sparks, Md.), and drug resistance profiles were determined as described by the manufacturer (S. H. Siddiqi, BACTEC 460TB system product and procedure manual, 1995). Bacterial DNA for DNA fingerprinting was extracted from Lowenstein-Jensen slant cultures subcultured from the BACTEC cultures (6). The IS6110-3′ fingerprinting probe for RFLP analysis was used according to standardized methodology (11). Hybridization patterns were visualized using an enhanced chemiluminescence kit (Amersham Pharmacia, Buckinghamshire, England). Analysis of the RFLP data with GelCompar (version 4.0; Applied Maths, BVBA, Kortrijk, Belgium) enabled the comparison of strain genotype data with data generated in a molecular epidemiology study conducted at two of the five neighboring clinics since 1992 (13). Strains were grouped into strain families according to an IS6110-3′ similarity index of ≥65%, and each strain was further identified by an IS-3′ cluster number (13). Additional fingerprinting with the direct repeat (DRr) probe (8) allowed the identification of more than one different strain genotype in a culture, as it usually generates only one or two hybridizing bands per strain. Inclusion of patient serial cultures allowed the confirmation of strain genotype. Patients were all followed up prospectively, and chest radiographs taken at diagnosis, at the end of intensive phase, and at the end of treatment were read in a systematic way without knowledge of results. Possible laboratory contamination was assessed by correlating the strain identity of serial cultures and of cultures processed on the same day.
RESULTS
From May 1999 to September 2001 (28 months), RFLP data were available for 210 patients, of whom 136 had serial cultures. Strain identity was confirmed in 131 of the cases, while in 5 cases (3.7%) a serial culture differed from the initial strain and was excluded. Patients were all HIV negative at inclusion in the study and were reported to have first-time pulmonary tuberculosis.
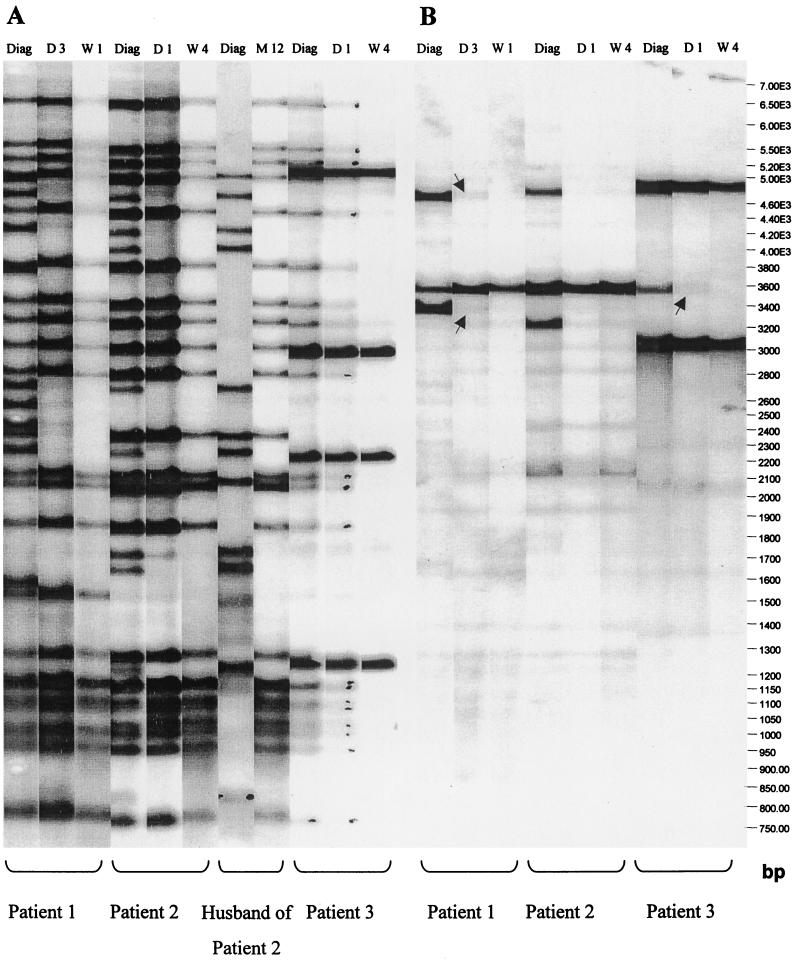
Among the 131 patients, RFLP analysis identified seven cultures with a possible mixture of strains. After repeat RFLP analysis and exclusion of possible laboratory contamination due to coprocessing of identical stains on the same date, three cases (2.3%) were confirmed. Mixed strains were grown from subsequent cultures from patients 1 and 3, and one culture from patient 2. Table 1 lists the RFLP patterns (strain family and IS-3′ cluster number) and the DRr fragment sizes for patients 1 to 3. Figure 1 shows the IS-3′ and DRr RFLP patterns.
TABLE 1.
IS6110-3′ RFLP identities of the strains isolated from cultures of three patients with mixed-strain cultures and sizes of the fragments generated with the DRr probe
| Patient and stage | Date | No. of strains | IS-3′ identity | Sizes (kb) of DRr fragments |
|---|---|---|---|---|
| 1 | ||||
| Diagnosis | 07/12/00 | 2 | F13-71, F29-211 | 4.71, 3.38; 3.57 |
| Day 3 | 07/14/00 | 2 | F13-71, F29-211 | 4.71, 3.38; 3.57 |
| Wk 1 | 07/19/00 | 1 | F29-211 | 3.57 |
| Wk 2 | 07/27/00 | 1 | F29-211 | 3.57 |
| 2 | ||||
| Diagnosis | 03/08/01 | 2 | F29-212, F11-492 | 3.57; 4.79, 3.23 |
| Day 1 | 03/09/01 | 1 | F29-212 | 3.57 |
| Wk 4 | 04/05/01 | 1 | F29-212 | 3.57 |
| 3 | ||||
| Diagnosis | 03/13/01 | 2 | F140-330, F29-1 | 4.84, 3.04; 3.57 |
| Day 1 | 03/14/01 | 2 | F140-330, F29-1 | 4.84, 3.04; 3.57 |
| Wk 4 | 04/10/01 | 1 | F140-330 | 4.84, 3.04 |
FIG. 1.
(A) IS6110-3′ RFLP patterns for the mixed and follow-up cultures of three patients as well as the initial infecting and relapse strains isolated (and confirmed by serial culture) from the husband of patient 2. (B) DRr RFLP patterns generated by cultures from three patients with mixed-strain cultures, where the mixture of strains is indicated by the DR patterns of two strains. The arrows indicate low-intensity DR bands. Diag, diagnosis; D, day; W, week.
Patient 1.
Patient 1 is a 44-year-old unemployed female smoker who was ill for 3 weeks before diagnosis with smear-positive pulmonary tuberculosis in July 2000. There was no close contact with other tuberculosis patients. Chest radiography at diagnosis showed unilateral consolidation in the right upper lobe and a small right pleural effusion but no cavities. No fibrosis, indicating previous pulmonary tuberculosis, was detected. The patient was compliant and completed treatment. The clinical response, serial sputum smears, and chest radiography confirmed a good response to therapy. All of her cultures were drug sensitive and remained smear positive to week 4. Cultures at diagnosis, day 3, week 1, and week 2 were fingerprinted. The culture at diagnosis was a mixture of two strains (F13-71 and F29-211). In the day 3 culture, the strain which was faint at diagnosis (F29-211) became more prominent and persisted in follow-up cultures. The DRr probe confirmed the presence of two strains in the diagnosis culture and, with additional exposure time, also in the day 3 culture.
Patient 2.
Patient 2 is a 36-year-old female smoker who was ill for 2 weeks before diagnosis (March 2001) with smear-positive pulmonary tuberculosis. A strong family history of pulmonary tuberculosis exists (father previously treated, mother treated in 1999 [with no RFLP data], and husband [see below]). A chest radiograph at diagnosis revealed consolidation in all lung lobes with cavities in the right upper lobe. There was fibrosis in the right upper lobe, indicating possible previous pulmonary tuberculosis. The patient was compliant and completed treatment. Serial sputum smears and chest radiography confirmed a good response to therapy. All cultures were drug sensitive. Sputum samples remained smear and culture positive up to week 4 and culture positive at week 8. DNA from diagnosis, day 1, and week 4 cultures was available for RFLP analysis. Two strains, F29-212 and F11-492, were identified in the diagnosis culture. The DRr probe confirmed the presence of two strains in the diagnosis culture. The husband of this patient was also enrolled in this study and was diagnosed with tuberculosis (F11-492) in December 1999 and subsequently with reinfection disease in December 2000 (month 12) with F29-212. Neither of the strains was identified in any other patients in the study.
Patient 3.
Patient 3 is a 30-year-old unemployed male smoker who was ill for 5 months before diagnosis with smear-positive pulmonary tuberculosis in March 2001. No history of close contact with other tuberculosis patients was recorded. The diagnosis chest radiograph revealed bilateral consolidation with cavities in the right and left upper lobes. No radiological signs of previous pulmonary tuberculosis were detected. The patient was compliant during the intensive phase but thereafter defaulted and moved out of the area. Sputum smear examination was negative at the end of the intensive phase (month 2), and chest radiography showed improvement. All cultures were drug sensitive. Sputum samples were smear and culture positive to week 4 and only culture positive at week 8. RFLP analysis was done for the diagnosis, day 1, and week 4 cultures. Two strains (F140-330 and F29-1) were identified in the diagnosis and day 1 cultures. The F140-330 strain remained prominent. The DRr probe confirmed the presence of two strains in the diagnosis and day 1 cultures.
DISCUSSION
In this study, we have illustrated the presence of multiple M. tuberculosis strains in cultures from patients residing in an area with a high tuberculosis incidence. The availability of serial cultures taken from diagnosis and follow-up visits resulted in the identification of five cases where serial isolates did not confirm strain identity. Rigorous auditing of laboratory records for coprocessing led to the removal of two potential dual-infection cases to eliminate possible laboratory error. It was established that cultures from the three patients in this study were not coprocessed with any cultures which could be the source of one of the two strains observed in the mixed-strain cultures of the patients. The dual nature of the infecting strains was confirmed by serial culture for two of the three cases. Both of the RFLP probes used could identify more than one strain in the mixed-strain cultures.
This study is the first to report on mixed-strain disease in a high-incidence community setting. Mixed strains were not observed in serial cultures from a comprehensive study in The Netherlands (5), where there is a low tuberculosis incidence. Despite the high degree of reinfection reported for this community (12), the observation of mixed-strain cultures was limited. It is therefore unlikely that, using the present methodology, the identification of multiple-strain cultures will have an impact on the interpretation of molecular epidemiological studies. The sensitivity of bacterial culture and DNA fingerprinting for the identification of more than one M. tuberculosis strain may, however, be limited and allow a mixture of strains to be identified only when the strains are present in the sputum within optimal limits. A minimum ratio of 1:9 was suggested for the detection of both strains in a mixture of strains (5).
Mixed strains cultured from sputum during a first episode of tuberculosis alter our understanding of the host response during numerous transmission events and of subsequent disease development. The simultaneous presence of more than one active M. tuberculosis strain and the high percentage of reinfection disease in high-incidence communities (12) may imply that infection with one strain does not protect against infection with another strain. This significantly affects the development of vaccines, as a single vaccine strain may not confer protection against other strains.
The different intensities of the two infecting strains observed in the mixed-strain cultures indicate some interaction between the different infecting strains. The prominence of one strain in a mixed-strain culture was previously observed (6, 14), as well as the persistence of one strain in follow-up cultures (14). The more prominent strain in the diagnosis culture of patient 1, however, almost disappeared at day 3, and the less prominent strain alone persisted to week 2. It appears, therefore, that the onset of treatment may affect the survival of strains differently and the observation of mixed-strain infections may be limited to serial cultures taken prior to or shortly after the onset of treatment.
All three patients presented with different variants of F29 strains, probably reflecting the high occurrence of this strain family in the area. Strain family F29 is part of the globally relevant Beijing strain family (2), with a historic as well as a recent component in the study region (unpublished data). The F29 strains identified in the cultures of these patients were not clonal and did not appear to be evolutionarily related. The F29-211 strain identified in patient 1 was cultured from four other patients enrolled at the same clinic. This strain type was not observed elsewhere and may be part of a recent outbreak. The F29-212 strain cocultured from patient 2 was additionally cultured only from a confirmed reinfection disease episode in her husband. The other strain in the diagnosis culture of patient 2 was initially (December 1999) cultured from her husband. Neither of these strains was cultured from other patients enrolled in the study. Patient 2 was most likely infected with the F11-492 strain during her husband's initial disease episode but only reactivated more recently while she was also exposed to the strain responsible for reinfection of her husband. Her chest radiograph showed fibrosis, possibly indicating undetected previous tuberculosis. The F29-1 strain cocultured from patient 3 was shown to form a large endemic F29 IS-3′ cluster (unpublished data) and was also cultured from six more patients in the present study. These patients were enrolled at different clinics, and it is proposed that the endemic nature of F29-1 strains enhances opportunity for infection. Similarly the F140-330 strain (the other infecting strain for patient 3) has a high frequency in the region (data not shown). For both patients 1 and 3, no contact with known tuberculosis patients was reported, indicating that casual transmission may contribute considerably to disease dynamics.
We conclude that immunocompetent individuals in a community setting may be infected with and express more than one drug-sensitive M. tuberculosis strain. Using the present methodology, the limited observation of mixed strains, even in a setting with a high level of secondary infections, will not greatly affect the interpretation of molecular epidemiology data. The observation of mixed strains may be limited to early serial cultures, after which the interaction of the strains or antituberculosis treatment may result in the persistence of a single strain. Both casual (nonidentified) and household contacts were indicated to have contributed to the coinfection of patients.
Acknowledgments
We thank GlaxoSmithKline for financial support of this project.
We thank M. de Kock for technical assistance. We thank the nursing staff and social workers involved with the study and the patients for complying with all of the demands made on them.
REFERENCES
- 1.Bates, J. H., W. W. Stead, and T. A. Rado. 1976. Phage type of tubercle bacilli isolated from patients with two or more sites of organ involvement. Am. Rev. Respir. Dis. 114:353-358. [DOI] [PubMed] [Google Scholar]
- 2.Bifani, P. J., B. Mathema, N. E. Kurepina, and B. N. Kreiswirth. 2002. Global dissemination of the Mycobacterium tuberculosis W-Beijing family strains. Trends Microbiol. 10:1:45-52. [DOI] [PubMed] [Google Scholar]
- 3.Braden, C. R., G. P. Morlock, C. L. Woodley, K. R. Johnson, A. C. Colombel, M. D. Cave, Z. Yang, S. E. Valway, I. M. Onorato, and J. T. Crawford. 2001. Simultaneous infection with multiple strains of Mycobacterium tuberculosis. Clin. Infect. Dis. 3:42-47. [DOI] [PubMed] [Google Scholar]
- 4.Chaves, F., F. Dronda, M. Alonso-Sanz, and A. Noriega. 1999. Evidence of exogenous reinfection and mixed infection with more than one strain of Mycobacterium tuberculosis among Spanish HIV-infected inmates. AIDS 13:15-20. [DOI] [PubMed] [Google Scholar]
- 5.De Boer, A. S., K. Kremer, M. W. Borgdorff, P. E. W. de Haas, H. F. Heersma, and D. van Soolingen. 2000. Genetic heterogeneity in Mycobacterium tuberculosis isolates reflected in IS6110 restriction fragment length polymorphism patterns as low-intensity bands. J. Clin. Microbiol. 38:4478-4484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Du Plessis, D. G., R. Warren, M. Richardson, J. J. Joubert, and P. D. van Helden. 2001. Demonstration of reinfection and reactivation in HIV-negative autopsied cases of secondary tuberculosis: multilesional genotyping of Mycobacterium tuberculosis utilizing IS6110 and other repetitive element-based DNA fingerprinting. Tuberculosis 81:211-220. [DOI] [PubMed] [Google Scholar]
- 7.Godfrey-Faussett, P., P. Sonnenberg, S. C. Shearer, M. C. Bruce, C. Mee, L. Morris, and J. Murray. 2000. Tuberculosis control and molecular epidemiology in a South African gold-mining community. Lancet 356:1066-1071. [DOI] [PubMed] [Google Scholar]
- 8.Hermans, W. M., D. van Soolingen, E. M. Bik, P. E. W. de Haas, W. D. Dale, and J. D. A. van Embden. 1991. Insertion element IS987 from Mycobacterium bovis BCG is located in a hot-spot integration region for insertion elements in Mycobacterium tuberculosis complex strains. Infect. Immun. 59:2695-2705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pavlic, M., F. Allerberger, M. P. Dierich, and W. M. Prodinger. 1999. Simultaneous infection with two drug-susceptible Mycobacterium tuberculosis strains in an immunocompetent host. J. Clin. Microbiol. 37:4156-4157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Thiesen, A., C. Reichel, S. Rüsch-Gerdes, W. H. Haas, J. K. Rockstroh, U. Spengler, and T. Sauerbruch. 1995. Mixed-strain infection with a drug-sensitive and multidrug-resistant strain of Mycobacterium tuberculosis. Lancet 345:1512-1513. [DOI] [PubMed] [Google Scholar]
- 11.Van Embden, J. D. A., M. D. Donald, J. T. Crawford, J. W. Dale, K. D. Eisenach, B. Giquel, P. Hermans, C. Martin, R. McAdam, T. M. Shinnick, and P. M. Small. 1993. Strain identification of Mycobacterium tuberculosis by DNA fingerprinting: recommendations for a standardized methodology. J. Clin. Microbiol. 31:406-409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Van Rie, A., R. W. Warren, M. Richardson, T. C. Victor, R. P. Gie, D. A. Enarson, N. Beyers, and P. D. van Helden. 1999. Exogenous reinfection as a cause of recurrent tuberculosis after curative treatment. N. Engl. J. Med. 341:1174-1179. [DOI] [PubMed] [Google Scholar]
- 13.Warren, R., M. Richardson, G. van der Spuy, T. Victor, S. Sampson, N. Beyers, and P. van Helden. 1999. DNA fingerprinting and molecular epidemiology of tuberculosis: use and interpretation in an epidemic setting. Electrophoresis 20:1807-1812. [DOI] [PubMed] [Google Scholar]
- 14.Yeh, R. W., P. C. Hopewell, and C. L. Daley. 1999. Simultaneous infection with two strains of Mycobacterium tuberculosis identified by restriction fragment length polymorphism analysis. Int. J. Tuberc. Lung Dis. 3:537-539. [PubMed] [Google Scholar]