Abstract
In October 2001, a letter containing a large number of anthrax spores was sent through the Brentwood post office in Washington, D.C., to a United States Senate office on Capitol Hill, resulting in contamination in both places. Several thousand people who worked at these sites were screened for spore exposure by collecting nasal swab samples. We describe here a screening protocol which we, as a level A laboratory, used on very short notice to process a large number of specimens (3,936 swabs) in order to report preliminary results as quickly as possible. Six isolates from our screening met preliminary criteria for Bacillus anthracis identification and were referred for definitive testing. Although none of the isolates was later confirmed to be B. anthracis, we studied these isolates further to define their biochemical characteristics and 16S rRNA sequences. Four of the six isolates were identified as Bacillus megaterium, one was identified as Bacillus cereus, and one was an unidentifiable Bacillus sp. Our results suggest that large-scale nasal-swab screening for potential exposure to anthrax spores, particularly if not done immediately postexposure, may not be very effective for detecting B. anthracis but may detect a number of Bacillus spp. that are phenotypically very similar to B. anthracis.
The threat of biological warfare became a reality in October 2001 when dissemination of spores of Bacillus anthracis, the causative agent of anthrax, occurred through letters sent through the United States postal system, resulting in several cases of anthrax infection. Since dispatch of the contaminated letters followed soon after the terrorist plane crashes in New York City and Washington, D.C., high anxiety prevailed as the nation tried to assess the magnitude of this new bioterrorist attack. The anthrax infections and associated deaths were followed closely by the press and the American public, as the governmental, medical, and scientific communities attempted to deal with real rather than hypothetical anthrax exposures. Although inhalation anthrax had not been reported in the United States since 1978 (2, 15), sporadic cases were reported in other countries, and an outbreak occurred in Sverdlovsk (in the former Soviet Union) in 1979 (12). Guidelines have been established to facilitate prompt and effective response to a bioterrorism event by the public health care system (4, 6, 9). Criteria for laboratory diagnosis of anthrax have also been established (3), and the role of the clinical microbiology laboratory has been addressed (10, 13). Anthrax spores can be aerosolized relatively easily and can resist environmental stresses for a long period. Spores are in the ideal size range (2 to 6 μm) for reaching the human lower respiratory tract (7). Consequently, B. anthracis is considered to be one of the organisms that have the highest risk for public health and that could cause mass casualties if used in biowarfare (14).
In Washington, D.C., the opening of a highly contaminated letter in a Capitol Hill (CH) office, followed soon after by evidence of postal worker exposures at the Brentwood post office (PO) facility, led to the decision to screen all persons who had some degree of risk due to time spent in these locations. In total, we received 3,936 nasal swabs over a 5-day period; 689 of these specimens were from CH, and 3,247 specimens were from the PO. The CH samples were sent to us from the National Naval Medical Center (NNMC) and were only a small fraction of the total number of specimens obtained from the CH episode. The PO specimens were all sent directly to our laboratory. In responding to the demands of this continuously evolving situation, our concerns were (i) that the microbiological diagnostic needs of the National Institutes of Health Clinical Center patients not be compromised while we were addressing a large influx of critical specimens from CH and PO personnel, (ii) that we have sufficient material resources to cover the expected needs as well as the capacity to handle any possible additional specimens, (iii) that we be able to plant and process thousands of specimens as quickly as possible, (iv) that we be confident in the accuracy of our results, and (v) that we attend to the safety concerns of our own laboratory staff.
In this report, we describe our level A laboratory protocol for rapidly accommodating this large number of specimens on relatively short notice. We also describe the screening procedure we used and the detection of six B. anthracis-like isolates that needed to be forwarded for definitive identification. While none of these isolates proved to be B. anthracis, they all had some features suggestive of that organism. We therefore thought it would be useful to provide some data regarding these organisms, as they might be representative of B. anthracis-like isolates that could be found during large-scale nasal-culture screening.
MATERIALS AND METHODS
Logistical arrangements.
We decided that the most efficient and reliable way to handle specimen labeling, planting, and work-up would be to use an “assembly line” system that was separate from the routine work being done on the National Institutes of Health Clinical Center specimens. Two teams were set up, each consisting of one person for computer entry of the nasal-swab specimens, one person to check specimen identifiers with the submitted handwritten list of persons cultured, one person to assure that the swab and plate labels were identical, two persons to plant the specimens, and one person to provide assistance as needed. Team personnel were medical technologists and staff members at the Ph.D. and M.D. levels.
The PO specimens were transferred to our laboratory immediately after they were collected (within approximately 1 to 3 h). Fifteen specimens from the PO were misplaced at the collection site and arrived 3 days after collection. The arrival of CH specimens was slightly delayed, since they were first sent to the NNMC. Planting of specimens was done as expeditiously as possible, and all specimens were planted the day of receipt, except for the few specimens that were received after midnight, which were kept in a 4°C refrigerator and planted at approximately 7 a.m. After the specimens were planted, the plates were placed in boxes holding approximately 100 plates each and incubated in two large incubators designated exclusively for the screening cultures. The cultures were examined within 24 h of planting, and those negative for suspicious colonies were reexamined after approximately 48 h of incubation. Specimen planting and plate examination were done in a biological safety cabinet. Personnel were gowned and gloved and used sleeve protectors. To eliminate variability in the evaluation of cultures, only three experienced individuals (one microbiologist and two senior medical technologists) did the assessment of all the culture plates. Any plates with colonies suggestive of B. anthracis (nonhemolytic, large gray-white colonies) were transferred to our biosafety level 3 laboratory for Gram staining and motility testing. The approximate turnaround time to report the presumptive results was 48 to 72 h after the specimens were collected.
Laboratory supplies.
Once the number of anticipated specimens was determined, a request for urgent delivery of 4,000 tryptic soy agar plates with 5% (vol/vol) sheep blood (SBA plates; BBL, Cockeysville, Md.) was placed; these were received within 24 h. An additional set of 1,500 SBA plates was provided by the NNMC, and 2,000 SBA plates were subsequently ordered as a backup supply. An adequate supply of plastic disposable loops (Fisher Scientific, Pittsburgh, Pa.) and motility test medium (Remel Laboratories, Lenexa, Kans.) was also ensured.
Nasal swabs.
The CH swabs were received in a variety of commercially available swab transport containers, all of which contained a moist transport medium. The PO swabs were collected using dry Dacron or rayon swabs, which were placed in individual plastic sandwich bags.
Culture protocol.
With some modifications, we followed the established guidelines for the presumptive identification of B. anthracis for level A laboratories (3). The specimens from each swab were planted on one SBA plate by rotating the swab over one-third of the plate. The inoculated area of the plate was then streaked to obtain isolated colonies by first using one edge of a disposable loop for the second third of the plate, and then the other edge of the loop for the final third of the plate. This method obviated the need for multiple loops for each specimen or for taking the time to flame wire loops between streak areas or between specimens. The disinfectant solution, a 1:10 dilution of 6.15% sodium hypochlorite (Clorox), was used for loop disposal and cleaning of workbenches, as recommended (5). All plates were incubated at 35°C in ambient air and examined first after overnight incubation, with a final examination after 48 h of incubation. Gram stains were performed on nonhemolytic or weakly hemolytic colonies that were morphologically consistent with Bacillus spp. or on colonies of uncertain morphology. Before being stained, the slides were fixed with 100% methanol for 1 min to avoid aerosolization that might have occurred with heat fixing in a flame. Organisms that were large gram-positive rods resembling Bacillus spp. were isolated to obtain pure cultures and tested for motility in semisolid motility test medium. Nonhemolytic, nonmotile isolates were sent for definitive testing in order to quickly confirm the presence or absence of B. anthracis. Beta-lactamase testing was not included as part of the screening process, since although B. anthracis is usually beta-lactamase negative, a strain selected for bioterrorism might be beta-lactamase positive. B. anthracis-like isolates were sent through the NNMC to the Division of Microbiology of the Armed Forces Institute of Pathology (AFIP), Department of Infectious and Parasitic Diseases Pathology, Department of Defense, Washington, D.C., for definitive testing.
Identification of Bacillus spp. referred for definitive testing.
We proceeded with further characterization of the isolates sent to the AFIP and found not to be B. anthracis to determine which species of nonmotile, nonhemolytic Bacillus had been isolated that were suggestive of B. anthracis. These isolates were tested for the following morphological and biochemical properties: colony morphology; cell width; motility; spore shape and position; swelling of the cell by the spore; anaerobic growth on SBA; growth at 50°C on SBA; lecithinase activity in egg yolk agar; hydrolysis of casein, starch, and gelatin; production of arginine dihydrolase and indole; nitrate reduction; and acid production from inulin, mannitol, and salicin. Biochemical test results were obtained from API 20E strips (bioMérieux Vitek, Inc., Hazelwood, Mo.) and from standard microbiological test media (Remel Laboratories) as needed. Production of beta-lactamase was tested by using BBL DrySlide Nitrocefin (Becton Dickinson and Company, Sparks, Md.). In addition to biochemical testing, 16S rRNA sequencing (Microseq Full Gene; Applied Biosystems, Foster City, Calif.) was also performed on these strains, and the sequences were compared with the GenBank database.
RESULTS
CH cultures.
Of the 689 CH cultures, 22 (3.2%) were found to have Bacillus-like colonies that were strongly beta-hemolytic, and they were not pursued further. Seven cultures (1%) contained Bacillus-like colonies that were nonhemolytic, and they were tested for motility; two of the seven were motile and were not identified further. Of the remaining five isolates, two were eliminated due to the narrow cell width of the organism, wet colonies, and/or significant swelling of the vegetative cells by spores. Three isolates from this group were referred for definitive testing.
PO cultures.
Of the 3,247 PO cultures, 96 (3%) had strongly beta-hemolytic Bacillus-like colonies that were not pursued further. An additional 33 isolates (1%), the hemolytic reactions of which were initially unclear, were subcultured to SBA to repeat determination of hemolysis and were simultaneously tested for motility. Of these, 26 were either beta-hemolytic or motile, leaving 7 nonmotile, non-beta-hemolytic strains for further analysis. Four of the seven had wet colonies and/or had spores that significantly swelled the vegetative cells, and they were not pursued further. Three isolates from this group were referred for definitive testing.
Summary of screening analyses.
In two large-scale screening surveys of nasal cultures for B. anthracis, approximately 3% of the cultures contained beta-hemolytic Bacillus-like colonies that were generally easily discerned after overnight incubation and that needed no further evaluation. Most of the specimens showed the presence of normal flora, including staphylococci, streptococci, and corynebacteria, that can be differentiated from the genus Bacillus by their distinct colony morphologies. A low percentage of cultures contained gram-negative rods, either Enterobacteriaceae or Pseudomonas, which were similarly easy to distinguish by colony morphology. The presence of spreading Proteus was also seen in a few specimens and in these instances interfered with recognition of other bacterial colonies. Approximately 1% (40 isolates) of cultures had Bacillus-like colonies that needed further evaluation, requiring subcultures to determine the presence or absence of beta hemolysis and motility. Only 0.4% (three isolates) of the CH cultures and 0.1% (three isolates) of the PO cultures had isolates that were nonhemolytic and nonmotile. To expedite the screening process, we did not use beta-lactamase production as part of the screening criteria nor did we attempt to demonstrate capsule production prior to referral. Six B. anthracis-like isolates were sent to AFIP for definitive testing.
Nonhemolytic, nonmotile isolates referred for additional specific testing for B. anthracis.
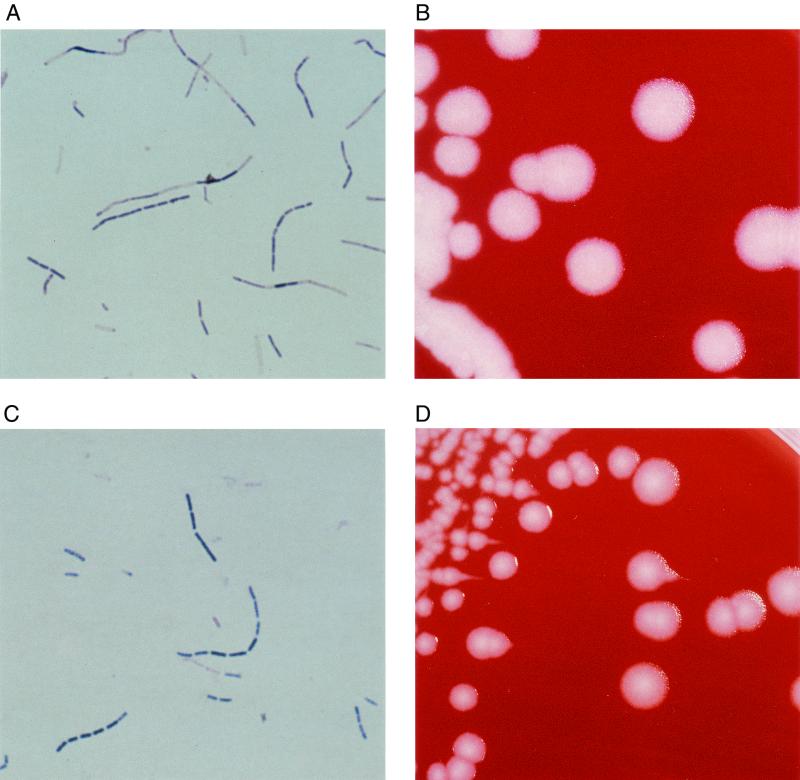
All six isolates (A to F) referred for specific B. anthracis testing were determined by the AFIP not to be B. anthracis. We then further characterized these six B. anthracis-like isolates to obtain more definitive information. The growth and biochemical characteristics of the isolates are shown in Table 1. Identification of the isolates by 16S rRNA sequence analysis, in which the full length of the 16S rRNA gene (∼1,500 bp) was sequenced for each isolate, was also attempted. These sequences were compared for homology with ∼1,500-bp 16S rRNA sequences in the GenBank database. Morphological and biochemical features indicated that four of the isolates (A, C, D, and E) most closely resembled Bacillus megaterium. These four isolates each exhibited the following characteristics typical of B. megaterium: poor anaerobic growth, lack of lecithinase activity, inability to reduce nitrate, and production of acid from mannitol. The four isolates, however, also showed the contradictory findings of good growth at 50°C and lack of acid production from inulin, features not typical of B. megaterium. All four of the isolates produced beta-lactamase. The identification of these four isolates was supported by 16S rRNA sequence analyses; each showed the highest relatedness (>99.5%) to B. megaterium (GenBank accession no. AF142677). Isolate B, by morphological and biochemical criteria, most closely resembled Bacillus cereus, although lecithinase activity was weak. The characteristics of anaerobic growth, failure to grow at 50°C, nitrate reduction, inability to form acid from mannitol, and the production of beta-lactamase, however, were consistent with the B. cereus group. Our sequence analysis of isolate B demonstrated that it most closely aligned with both B. cereus (99.7%) and B. anthracis (99.6%) (GenBank accession no. AF176322 and AF176321, respectively). Isolate F was unidentifiable using biochemical criteria, although its large cell size was consistent with both the B. cereus group and B. megaterium (11). This organism was unusual in that it would not grow on egg yolk agar, so that the lecithinase activity was indeterminate. It was different from other isolates in that it did not produce beta-lactamase. The uniqueness of this organism was confirmed by the 16S rRNA sequence analysis, which showed the closest relationship to several members of the B. cereus group but only at 97% identical bases over the sequence of approximately 1,300 bp. Interestingly, both isolates B and F had colony morphologies consistent with that of B. anthracis (Fig. 1), but all of the definitive tests were determined by the AFIP to be negative. Therefore, isolate B was identified as B. cereus and isolate F was an unidentifiable Bacillus sp.
TABLE 1.
Differential characteristics of Bacillus-like isolates and selected Bacillus species.
| Characteristic | Valuea
|
||||||||
|---|---|---|---|---|---|---|---|---|---|
| Isolate A | Isolate B | Isolate C | Isolate D | Isolate E | Isolate F | B. cereus | B. anthracis | B. megaterium | |
| Motility | − | − | − | − | − | − | + | − | + |
| Hemolysis on SBA | − | W | − | − | − | − | + | − | + |
| Growth at 50°C | + | − | + | + | + | + | − | − | − |
| Anaerobic growthb | T | (+) | T | T | T | (+) | + | + | − |
| Lecithinase activity | − | W | − | − | − | NA | + | + | − |
| Casein hydrolysis | + | + | + | + | + | + | + | + | + |
| Starch hydrolysis | + | + | + | + | + | + | + | + | + |
| Arginine dihydrolase | − | − | − | − | − | − | − | − | − |
| Indole production | − | − | − | − | − | − | − | − | − |
| Gelatin hydrolysis | + | + | + | + | + | + | + | W | + |
| Nitrate reduction | − | + | − | − | − | + | + | + | − |
| β-Lactamase production | + | + | + | + | + | − | + | − | NA |
| Acid from: | |||||||||
| Inulin | − | − | − | − | − | − | − | − | + |
| Mannitol | + | − | + | + | + | + | − | − | + |
| Salicin | + | + | − | − | + | − | + | − | + |
Symbols (except for anaerobic growth): +, positive reaction; −, negative reaction; W, weak reaction; NA, not available.
Symbols for anaerobic growth: T, trace amount of growth; (+), moderate growth; +, good growth; −, no growth.
FIG. 1.
Gram stains and colony morphologies of selected B. anthracis-like isolates. Gram stains of isolates B (A) and F (C) show large gram-positive bacilli. The colonies of isolates B (B) and F (D) are large, flat, gray-white, and nonhemolytic and have a ground-glass appearance. The colonies of isolate F show tailing along the lines of inoculation, which is a characteristic suggestive of B. anthracis.
DISCUSSION
To date, few diagnostic laboratories have had to contend with a bioterrorism episode. In such an episode, level A laboratories might have to play a major role in screening for organisms that potentially would need to be referred for definitive testing by reference laboratories. Nasal screening for anthrax is extremely time-consuming and labor-intensive during the specimen accessioning and planting stages. It is critical that labeling of specimens and culture plates, and entry of data into computers, be done rigorously to prevent mislabeling of cultures and subsequent confusion in patient identification. The evaluation of culture plates is relatively straightforward, as most nasal swabs contain colonies of normal flora that are easily differentiated from the larger, flat Bacillus colonies. We took a conservative approach, in that any colony that was indeterminate by colony morphology was Gram stained to determine if the organisms were microscopically consistent with a Bacillus sp. We believe that use of experienced personnel to screen cultures enhanced both the speed and sensitivity of the screening process.
From 3,936 cultures, 6 isolates needed to be referred for specific B. anthracis confirmatory testing; all of the isolates were subsequently determined not to be B. anthracis. By determining biochemical characteristics and 16S rRNA sequences, we identified four of these isolates as B. megaterium and one as B. cereus; the sixth was an unidentifiable Bacillus sp. B. anthracis can be identified and differentiated from other Bacillus spp. by certain characteristics which are only determinable in specialized reference laboratories (11, 16). However, 16S rRNA sequencing cannot distinguish between B. cereus and B. anthracis (1); isolate B is illustrative of this problem. Although definitive testing by the AFIP determined that isolate E was positive for the B. anthracis capsule by direct fluorescent-antibody staining, it was negative by phage sensitivity as well as by PCR for five different markers and thus was identified as not B. anthracis.
The recommendations for utilizing nasal swabs to screen for exposure to aerosolized B. anthracis spores are based on data from an aerosol exposure study performed using rhesus monkeys (8). The data from this study indicated that spore counts from nasal swabs decreased over time after primary exposure and that the rate of decrease was directly related to the initial dose of inhaled spores. In addition, the authors reported that spores were more difficult to retrieve and were retrieved in lower numbers from a swab transport container with a moist transport medium. We found B. anthracis-like isolates in 0.4% of the CH specimens, which were transported in containers with a moist holding medium, but we found such isolates in only 0.1% of the PO specimens, which were transported in plastic bags containing no transport medium. However, as we recovered no B. anthracis isolates from either site, we cannot determine if differences in specimen collection procedures affected the results. According to the available animal data (8), positive nasal cultures for B. anthracis could be reliably demonstrated from specimens obtained within 12 to 24 h after spore exposure. We suspect that the time interval between exposure and specimen collection is one of the most important factors affecting the sensitivity of the procedure, as the clearance of spores from the nose occurs progressively over time. To the best of our knowledge, all the positive cultures from the CH site that were processed elsewhere had been obtained very shortly after the exposure occurred. Other factors affecting sensitivity may include the extent of spore exposure and culture methodology. Despite the fact that all of the cultures we processed were negative, it was subsequently recommended that all the potentially exposed postal workers receive antimicrobial prophylaxis.
Acknowledgments
We thank the technologists of the Microbiology Service for all their efforts in coping with the large and unexpected workload that these screening cultures occasioned. We are grateful for the help provided by the National Institutes of Health administrative and security personnel, the staff of the NNMC, and the District of Columbia Department of Health. We thank Ted L. Hadfield for his helpful comments and the Division of Microbiology of the AFIP for definitive testing for B. anthracis.
REFERENCES
- 1.Ash, C., J. E. Farrow, M. Dorsch, E. Stackebrandt, and M. D. Collins. 1991. Comparative analysis of Bacillus anthracis, B. cereus and related species on the basis of reverse transcriptase sequencing of the 16S-rRNA. Int. J. Syst. Bacteriol. 41:343-346. [DOI] [PubMed] [Google Scholar]
- 2.Brachman, P. S. 1980. Inhalation anthrax. Ann. N. Y. Acad. Sci. 353:83-93. [DOI] [PubMed] [Google Scholar]
- 3.Centers for Disease Control and Prevention, American Society for Microbiology, and Association of Public Health Laboratories. 18March2002, revision date. Basic diagnostic testing protocols for level A laboratories for the presumptive identification of Bacillus anthracis. [Online.] American Society for Microbiology, Washington, D.C. http://www.asmusa.org/pcsrc/ban.asm.la.cp.031802.pdf.
- 4.Centers for Disease Control and Prevention. 2000. Biological and chemical terrorism: strategic plan for preparedness and response; recommendations of the CDC Strategic Planning Workgroup 2000. Morb. Mortal. Wkly. Rep. 43:1-14. [PubMed] [Google Scholar]
- 5.Centers for Disease Control and Prevention. 1999. Bioterrorism alleging use of anthrax and interim guidelines for management—United States, 1998. Morb. Mortal. Wkly. Rep. 48:69-74. [PubMed] [Google Scholar]
- 6.Centers for Disease Control and Prevention. 2001. Update: investigation of anthrax associated with intentional exposure and interim public health guidelines, October 2001. Morb. Mortal. Wkly. Rep. 50:889-893. [PubMed] [Google Scholar]
- 7.Cieslak, T. J., and E. M. Eitzen, Jr. 1999. Clinical and epidemiologic principles of anthrax. Emerg. Infect. Dis. 5:552-555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hail, A. S., C. A. Rossi, G. V. Ludwig, B. E. Ivins, R. F. Tammariello, and E. A. Henchal. 1999. Comparison of noninvasive sampling sites for early detection of Bacillus anthracis spores from rhesus monkeys after aerosol exposure. Mil. Med. 164:833-837. [PubMed] [Google Scholar]
- 9.Inglesby, T. V., D. A. Henderson, J. G. Bartlett, M. S. Ascher, E. Eitzen, A. M. Friedlander, J. Hauer, J. McDade, M. T. Osterholm, T. O'Toole, G. Parker, T. M. Perl, P. K. Russell, K. Tonat, and the Working Group on Civilian Biodefense. 1999. Anthrax as a biological weapon: medical and public health management. JAMA 281:1735-1745. [DOI] [PubMed] [Google Scholar]
- 10.Klietmann, W. F., and K. L. Ruoff. 2001. Bioterrorism: implications for the clinical microbiologist. Clin. Microbiol. Rev. 14:364-381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Logan, N. A., and P. C. Turnbull. 1999. Bacillus and recently derived genera, p. 357-369. In P. R. Murray, E. J. Baron, M. A. Pfaller, F. C. Tenover, and R. H. Yolken (ed.), Manual of clinical microbiology, 7th ed. American Society for Microbiology, Washington, D.C.
- 12.Meselson, M., J. Guillemin, M. Hugh-Jones, A. Langmuir, I. Popova, A. Shelokov, and O. Yampolskaya. 1994. The Sverdlovsk anthrax outbreak of 1979. Science 226:1202-1208. [DOI] [PubMed] [Google Scholar]
- 13.Miller, J. M. 2001. Bioterrorism—a perspective for the community hospital. Clin. Microbiol. Newsl. 23:179-185. [Google Scholar]
- 14.Rotz, L. D., A. S. Khan, S. R. Lillibridge, S. M. Ostroff, and J. M. Hughes. 2002. Public health assessment of potential biological terrorism agents. Emerg. Infect. Dis. 8:225-230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Suffin, J., W. Carnes, and A. Kaufmann. 1978. Inhalation anthrax in a home craftsman. Hum. Pathol. 9:594-597. [DOI] [PubMed] [Google Scholar]
- 16.Turnbull, P. C. B. 1999. Definitive identification of Bacillus anthracis. J. Appl. Microbiol. 87:237-240. [DOI] [PubMed] [Google Scholar]