Abstract
Three clinical strains of enterohemorrhagic Escherichia coli O157:H7 which were subcultured repeatedly or stored at room temperature over a 25-week period showed appreciable variations in their pulsed-field gel electrophoresis fragment patterns. The variations could be explained by a couple of spontaneous genetic events at most and thus did not invalidate the genetic lineage of the strains.
Enterohemorrhagic Escherichia coli (EHEC) O157:H7 has been increasingly recognized as a major food-borne pathogen that causes hemorrhagic colitis and hemolytic-uremic syndrome (7, 9). In an attempt to identify the possible source and route of infection of the pathogen, pulsed-field gel electrophoresis (PFGE) (1) has been employed in many epidemiological investigations of both sporadic and multiple outbreaks almost worldwide, including Germany (4), the United States (2), and Japan (8, 15). The PFGE analysis (8) performed on the Japanese isolates derived from the major outbreaks in 1996 revealed that the isolates were genotypically classified into at least five groups, but there were appreciable minor variations in the PFGE profiles within each PFGE type. Recently, Murase et al. (10) have reported that PFGE fragment patterns of EHEC O157:H7 isolates were changed by the loss of single fragments during maintenance or subculturing, possibly due to curing of Shiga toxin (Stx)-converting bacteriophages. Variations in PFGE fragment patterns in a single strain that had been cultured repeatedly over time were reported for several bacterial species (3, 11, 12). For example, an extensive genotypic change occurred in a strain of Campylobacter coli after extended subculturing under standard in vitro culture conditions (12). However, to date EHEC O157:H7 has hardly been evaluated in this regard. We here describe the variability of PFGE patterns of three clinical isolates of EHEC O157:H7 through repeated subculturing and prolonged storage in vitro.
One of the clinical isolates, EDL933 (= ATCC 43895), used in the present study was kindly provided by the Centers for Disease Control and Prevention (Atlanta, Ga.). The strain was responsible for a hamburger-associated food poisoning episode in 1982. Two other isolates, NIID2 and NIID220, were from patients involved in the major outbreaks that occurred in Japan during 1996 (15). All isolates were found to possess both Stx 1- and Stx 2-encoding genes, and their PFGE profiles were different from one another as described elsewhere (13).
In the present study, PFGE was performed essentially according to the procedure described by Izumiya et al. (8) with minor modifications. Briefly, bacterial cells on heart infusion agar (HIA) (Difco Laboratories, Detroit, Mich.) were directly embedded in low-melting-temperature agarose (FMC BioProducts, Rockland, Maine). After appropriate preparations for restriction endonuclease digestion were made, the DNAs in each plug were digested with 30 U of XbaI (Takara Shuzo, Tokyo, Japan) at 37°C for 4 h. PFGE was performed with a 1% agarose gel by using a contour-clamped homogeneous electric field DRII apparatus (Bio-Rad Laboratories, Richmond, Calif.) in 0.5× Tris-borate-EDTA buffer at 12°C at 200 V. For separation of a whole genome, a linearly ramped switching time from 4 to 8 s was applied for 12 h and then a linearly ramped switching time from 8 to 50 s was applied for 10 h. After PFGE, the gels were stained with ethidium bromide (0.2 μg/ml) and were photographed under UV transillumination.
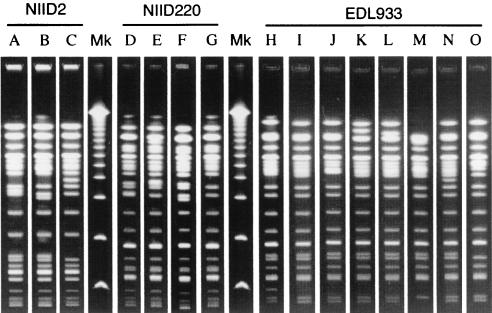
The strains were continually subcultured on a slant medium of HIA at a rate of twice per week at 37°C, and the subculturing was repeated 50 times (for 25 weeks). It should be noted that each subculturing would have created approximately 200 to 300 generations of the cells if they divided every 20 min. After every 10 subcultures, a loopful of the culture was streaked onto a separate HIA plate and incubated at 37°C for 18 h. After incubation, 14 well-isolated colonies were randomly selected and subjected to the PFGE analysis as described above. The results are presented in Table 1. The PFGE fragment patterns for the original NIID2, NIID220, and EDL933 strains were designated patterns A, D, and H, respectively. The 14 randomly selected colonies of EDL933 did not show any variation in PFGE fragment pattern for the first 10 subcultures but did start to show variation after 20 subcultures. A total of seven patterns (I, J, K, L, M, N, and O) different from the original PFGE pattern (Table 1 and Fig. 1) were observed through the repeated subculturing. The fragment patterns (L, M, N, and O) shown by the strain at the 50th subculturing were different from the original pattern by five fragments at most. By contrast, majorities of the subcultures of NIID2 and NIID220 seem to have retained their original fragment pattern during the treatment, in which sporadic variations (patterns B and C for NIID2 and patterns E, F, and G for NIID220) from the original pattern by one to three fragments were observed (Table 1 and Fig. 1). The evidence points to the possibility that E. coli O157:H7 strains differ in genomic stability.
TABLE 1.
Occurrence of different PFGE fragment patterns in three strains of EHEC O157:H7 during a 25-week period of repeated subculturing at 3- to 4-day intervals
| Strain and PFGE fragment pattern | Typical no. of fragment differences from original pattern | No. of colonies (n = 14) showing difference in PFGE pattern after number of subculturing:
|
|||||
|---|---|---|---|---|---|---|---|
| 1 | 10 | 20 | 30 | 40 | 50 | ||
| NIID2 | |||||||
| A | Original pattern | 14 | 14 | 14 | 14 | 13 | 11 |
| B | 2 | 1 | 2 | ||||
| C | 3 | 1 | |||||
| NIID220 | |||||||
| D | Original pattern | 14 | 12 | 13 | 14 | 14 | 10 |
| E | 2 | 1 | 1 | 1 | |||
| F | 1 | 1 | |||||
| G | 1 | 3 | |||||
| EDL933 | |||||||
| H | Original pattern | 14 | 14 | 11 | 12 | 1 | |
| I | 1 | 3 | 1 | 8 | 1 | ||
| J | 2 | 1 | 2 | ||||
| K | 2 | 2 | |||||
| L | 2 | 1 | 10 | ||||
| M | 5 | 1 | |||||
| N | 2 | 1 | |||||
| O | 4 | 1 | |||||
FIG. 1.
XbaI-derived PFGE DNA profiles of repeatedly subcultured EHEC O157:H7 strains. Capital letters above the lanes correspond to fragment patterns in Table 1. Lanes Mk, molecular size markers (Lambda Ladder PFG Marker; New England BioLabs, Beverly, Mass.).
Meanwhile, the initial cultures of the three strains, which were maintained in screw-cap test tubes containing a slant of HIA medium, were stored in the dark at room temperature (23°C) for a period of 25 weeks. At the end of every 5 weeks of storage, a loopful of the cultures on the medium was streaked onto a separate HIA plate and incubated at 37°C for 18 h. After incubation, 14 well-isolated colonies were randomly selected and also subjected to PFGE analysis. The results are presented in Table 2. Occurrence of fragment patterns (pattern P for NIID2; Q, R, and S for NIID220; and T for EDL933) different from the original patterns was sporadic and limited to three fragments at most in all EHEC O157:H7 strains during the treatment (Table 2 and Fig. 2).
TABLE 2.
Occurrence of different PFGE fragment patterns in three strains of EHEC O157:H7 through prolonged room temperature storage for up to 25 weeks
| Strain and PFGE fragment pattern | Typical no. of fragment differences from original pattern | No. of colonies (n = 14) showing difference of PFGE pattern, after storage for no. of wks:
|
|||||
|---|---|---|---|---|---|---|---|
| 0 | 5 | 10 | 15 | 20 | 25 | ||
| NIID2 | |||||||
| A | Original pattern | 14 | 14 | 14 | 14 | 13 | 14 |
| P | 2 | 1 | |||||
| NIID220 | |||||||
| D | Original pattern | 14 | 13 | 14 | 14 | 11 | 14 |
| Q | 2 | 1 | 1 | ||||
| R | 1 | 1 | |||||
| S | 1 | 1 | |||||
| EDL933 | |||||||
| H | Original pattern | 14 | 14 | 14 | 14 | 13 | 14 |
| T | 3 | 1 | |||||
FIG. 2.
XbaI-derived PFGE DNA profiles of EHEC O157:H7 strains during prolonged storage at room temperature. Capital letters above the lanes correspond to fragment patterns in Table 2. Lanes Mk, molecular size markers (Lambda Ladder PFG Marker).
After the above treatments, the strains that showed fragment patterns different from the originals were tested for any loss of Stx 1 and Stx 2 genes by the PCR assay described elsewhere (6). The assay revealed that both genes were retained in all strains except one EDL933 strain that had been stored for 20 weeks. The evidence suggests that the observed variations in the fragment pattern were due mostly to other spontaneous genetic alterations during storage.
Recently, electronic storage of digitized PFGE patterns has become common for comparing historical and present isolate patterns (5). According to the criteria for interpreting PFGE patterns as proposed by Tenover et al. (14), two strains in question are closely related if they differ by two to three fragments, implying a single genetic event (i.e., point mutation, insertion, or deletion of DNA). The strains are possibly related if they differ by four to six fragments, implying two independent genetic events. The results of the present study suggest that the criteria are applicable to strain typing of EHEC O157:H7. Repeated subculturing or long-term storage can thus cause variations in PFGE profiles of the strains, but the variations will not invalidate epidemiological lineage.
REFERENCES
- 1.Arbeit, R. D., M. Arthur, R. D. Dunn, C. Kim, R. K. Selander, and R. Goldstein. 1990. Resolution of recent evolutionary divergence among Escherichia coli from related lineages: the application of pulsed field gel electrophoresis to molecular epidemiology. J. Infect. Dis. 161:230-235. [DOI] [PubMed] [Google Scholar]
- 2.Barrett, T. J., H. Lior, J. H. Green, R. Khakhria, J. G. Wells, B. P. Bell, K. D. Greene, J. Lewis, and P. M. Griffin. 1994. Laboratory investigation of a multistate food-borne outbreak of Escherichia coli O157:H7 by using pulsed-field gel electrophoresis and phage typing. J. Clin. Microbiol. 32:3013-3017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Beall, B., P. K. Cassiday, and G. N. Sanden. 1995. Analysis of Bordetella pertussis isolates from an epidemic by pulsed-field gel electrophoresis. J. Clin. Microbiol. 33:3083-3086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Böhm, H., and H. Karch. 1992. DNA fingerprinting of Escherichia coli O157:H7 strains by pulsed-field gel electrophoresis. J. Clin. Microbiol. 30:2169-2172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Carson, C. A., J. M. Keller, K. K. Mcadoo, D. Wang, B. Higgins, C. W. Bailey, J. G. Thoren, B. J. Payne, M. Skala, and A. W. Hahn. 1995. Escherichia coli O157:H7 restriction pattern recognition by artificial neural network. J. Clin. Microbiol. 33:2894-2898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cebula, T. A., W. L. Payne, and P. Feng. 1995. Simultaneous identification of strains of Escherichia coli serotype O157:H7 and their Shiga-like toxin type by mismatch amplification mutation assay-multiplex PCR. J. Clin. Microbiol. 33:248-250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Griffin, P. M. 1995. Escherichia coli O157:H7 and other enterohemorrhagic Escherichia coli, p. 739-762. In M. J. Blaser, P. D. Smith, J. I. Ravdin, H. B. Greenberg, and R. L. Guerrant (ed.), Infections of the gastrointestinal tract. Raven Press, New York, N.Y.
- 8.Izumiya, H., J. Terajima, A. Wada, Y. Inagaki, K. Ito, K. Tamura, and H. Watanabe. 1997. Molecular typing of enterohemorrhagic Escherichia coli O157:H7 isolates in Japan by using pulsed-field gel electrophoresis. J. Clin. Microbiol. 35:1675-1680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kaper, J. B., and A. D. O'Brien. 1998. Escherichia coli O157:H7 and other Shiga toxin-producing E. coli strains. ASM Press, Washington, D.C.
- 10.Murase, T., S. Yamai, and H. Watanabe. 1999. Changes in pulsed-field gel electrophoresis patterns in clinical isolates of enterohemorrhagic Escherichia coli O157:H7 associated with loss of Shiga toxin genes. Curr. Microbiol. 38:48-50. [DOI] [PubMed] [Google Scholar]
- 11.Nielsen, E. M., J. Engberg, and V. Fussing. 2001. Genotypic and serotypic stability of Campylobacter jejuni strains during in vitro and in vivo passage. Int. J. Med. Microbiol. 291:379-385. [DOI] [PubMed] [Google Scholar]
- 12.On, S. L. 1998. In vitro genotypic variation of Campylobacter coli documented by pulsed-field gel electrophoretic DNA profiling: implications for epidemiological studies. FEMS Microbiol. Lett. 165:341-346. [DOI] [PubMed] [Google Scholar]
- 13.Osawa, R., S. Iyoda, S.-I. Nakayama, A. Wada, S. Yamai, and H. Watanabe. 2000. Genotypic variations of Shiga toxin-converting phages from enterohaemorrhagic Escherichia coli O157:H7 isolates. J. Med. Microbiol. 49:565-574. [DOI] [PubMed] [Google Scholar]
- 14.Tenover, F. C., R. D. Arbeit, R. V. Goering, P. A. Mickelsen, B. E. Murray, D. H. Persing, and B. Swaminathan. 1995. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J. Clin. Microbiol. 33:2233-2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Watanabe, H., A. Wada, Y. Inagaki, K. Itoh, and K. Tamura. 1996. Outbreaks of enterohaemorrhagic Escherichia coli O157:H7 infection by two different genotypes-strains in Japan. Lancet 348:831-832. [DOI] [PubMed] [Google Scholar]