Abstract
Erythromycin is currently being used for both prophylaxis and treatment of pertussis infections. Erythromycin resistance was first recognized in Bordetella pertussis in Arizona in 1994, and since then, three additional resistant isolates have been identified in the United States. To better assess the potential public health impact of erythromycin-resistant B. pertussis, we used the disk diffusion assay to evaluate the frequency of erythromycin resistance among 1,030 recently circulating U.S. isolates and found the rate of occurrence to be <1%. We also describe a novel heterogeneous phenotype, with erythromycin-resistant colonies appearing only after a 7-day incubation period. To optimize patient management, we recommend that clinicians be alert to potential treatment failures and that laboratorians use a 7-day incubation period when screening for resistance. Our ongoing national surveillance will continue to monitor for resistant B. pertussis isolates and their potential association with changing pertussis epidemiology.
Erythromycin has been used in the United States for over 45 years for treatment of patients with pertussis and is currently recommended for prophylaxis as well (1). To date, erythromycin resistance in Bordetella pertussis has been reported in only the United States, with the first resistant isolate detected in Arizona in 1994 (7). Three additional resistant isolates have since been reported from patients in Minnesota, Utah, and California (5, 6; J. Bartkus, B. A. Juni, K. Ehresman, C. Miller, G. N. Sanden, P. K. Cassiday, M. Saubolle, B. Lee, J. Long, and J. M. Besser, Abstr. 41st Intersci. Conf. Antimicrob. Agents Chemother., abstr. C1-1817, p. 102, 2001). Due to the low frequency of resistance and the lack of a standardized screening method, very few clinical laboratories currently perform susceptibility testing of B. pertussis (3, 8). Reference laboratories that do perform susceptibility testing primarily use the disk diffusion assay.
To assess the frequency of erythromycin resistance in the United States, we tested B. pertussis isolates collected through an enhanced pertussis surveillance program, as well as a subset of those already in our strain collection. The enhanced surveillance program involves several states (California, New York, Minnesota, Massachusetts, Illinois, Arizona, and Georgia) and aims to associate variations in the circulating B. pertussis population with relevant changes in pertussis epidemiology, such as overall burden of disease, disease severity, and treatment or vaccine failures (2). This evaluation of over 1,000 B. pertussis isolates resulted in the detection of a novel erythromycin-resistant phenotype in B. pertussis that is unlikely to be recognized by the current disk diffusion protocols used to screen for erythromycin resistance.
MATERIALS AND METHODS
B. pertussis strains.
We used two previously reported erythromycin-resistant isolates from Arizona (A228) and California (C310) as resistant controls and B. pertussis ATCC 9797 as the erythromycin-susceptible control (6, 7). We tested 1,030 B. pertussis isolates for erythromycin resistance: 776 were obtained from 1994 to 2000 through our enhanced pertussis surveillance program in California, New York, Minnesota, Massachusetts, Illinois, Arizona, and Georgia; the remaining 254 isolates were a convenience sample from the B. pertussis collection at the Centers for Disease Control and Prevention and were isolated from 1958 to 2001.
Erythromycin disk diffusion assay.
Since no universally standardized and validated macrolide susceptibility assay exists for B. pertussis, we used a disk diffusion assay adapted from previously published guidelines specific for B. pertussis as follows (3, 4, 8). Isolates were grown at 37°C under high humidity for 3 to 5 days on plates of charcoal agar (Oxoid, Unipath, Ltd., Basingstoke, Hampshire, England) supplemented with 10% defibrinated horse blood (Lampire Biological Laboratories, Pipersville, Pa.). The cells were harvested and suspended in 2 ml of phosphate-buffered saline (pH 7.4), and the turbidity was adjusted to the 0.5 McFarland standard. The cell suspension was applied with a cotton swab over an entire charcoal-horse blood agar plate surface (100 by 15 mm), and a 15-μg erythromycin disk (Becton Dickinson, Baltimore, Md.) was placed in the center of the plate. The plates were incubated at 37°C under high humidity, and zones of growth inhibition were measured on days 3, 5, and 7.
Definitions.
A susceptible isolate exhibited a zone of growth inhibition of >40 mm in diameter. A resistant isolate showed no zone of growth inhibition, i.e., growth was observed all the way up to the disk. An isolate with the novel heterogeneous phenotype exhibited an initial zone of growth inhibition of >40 mm in diameter after 3 days of incubation; resistant colonies appeared within the initial zone of inhibition after the extended 5- to 7-day incubation.
Testing heterogeneous phenotype isolates.
The confluent growth outside the zone of inhibition was harvested with an inoculating loop and streaked on a charcoal-horse blood agar plate for isolation. Following a 3-day incubation at 37°C under humid conditions, four individual colonies were subcultured and retested by the disk diffusion assay described herein. The same 0.5 McFarland suspension used in the disk diffusion assay was also used to perform the Etest according to the manufacturer's instructions. Etest MICs were recorded on days 3 and 7.
Resistant colonies appearing within the zone of growth inhibition were counted after 7 days. Four resistant colonies from each culture were selected and subcultured onto a charcoal-horse blood agar plate; these colonies were retested by the disk diffusion assay described above. The same 0.5 McFarland suspension used in the disk diffusion assay was also used to perform the Etest. Etest MICs were recorded on days 3 and 7.
Testing for degradation of erythromycin in the disk diffusion assay.
Erythromycin disks were placed in the center of 45 fresh charcoal-horse blood agar plates. These plates were incubated at 37°C under high humidity for up to 14 days before testing. On days 0, 3, 5, 7, 10, and 14, 0.5 McFarland suspensions of the susceptible (ATCC 9797), resistant (C310), and heterogeneous phenotype (B715) isolates were prepared from 3-day cultures and 100 μl of each suspension was inoculated in triplicate around the erythromycin disks on the previously incubated charcoal-horse blood agar plates. These plates were incubated for an additional 10 days, and zones of inhibition were measured after 3, 5, 7, and 10 days. As a control, a disk diffusion assay using a fresh charcoal-horse blood agar plate and erythromycin disk was performed with suspensions of strains representing each erythromycin susceptibility phenotype on days 0, 3, and 7.
RESULTS
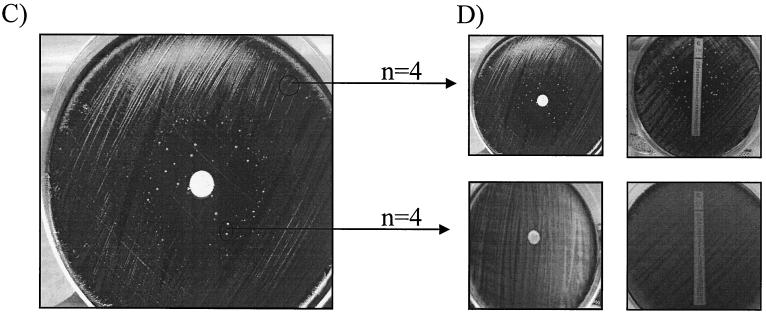
All 1,030 test isolates showed zones of inhibition of >40 mm by the disk diffusion assay after a 3-day incubation; accordingly, they were all initially defined as susceptible. However, after 5 to 7 days of incubation, 5 of the 1,030 isolates (0.5%) demonstrated the heterogeneous phenotype, as isolated colonies appeared inside the initial zone of growth inhibition (Fig. 1). All five isolates were collected through the enhanced surveillance program; four were from Arizona (1994 to 1995) and one was from Georgia (1999).
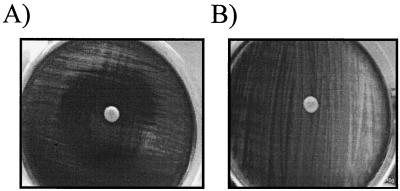
FIG. 1.
Disk diffusion assay and Etest for screening of B. pertussis for erythromycin resistance. (A) B. pertussis ATCC 9797 susceptible control displaying a zone of inhibition of >40 mm. (B) B. pertussis C310 resistant control (Calif., 1999) showing growth confluent to the disk. (C) B. pertussis B715 (Ariz., 1994) displaying a heterogeneous phenotype after 7 days of incubation. (D) B. pertussis B715. Four colonies from both inside and outside the zone of growth inhibition were subcultured and retested by the disk diffusion assay and Etest. Colonies outside the zone of inhibition were reproducibly of the heterogeneous phenotype, while colonies inside the zone were resistant (MIC > 256 μg/ml).
The number of resistant colonies appearing within the zone differed for each of these five strains, ranging from approximately 10 for each of the four Arizona strains to 100 for the Georgia strain. When these resistant colonies were tested by the disk diffusion assay, all displayed only the resistant phenotype within the standard 3- to 5-day incubation and MICs of >256 μg/ml were obtained by the Etest. When the colonies outside of the initial zone of inhibition for the same five strains were subcultured and retested by the disk diffusion assay, the heterogeneous phenotype was consistently reproduced. Comparable results were seen by using the Etest, e.g., MICs for the colonies outside the initial zone in the disk diffusion assay were <0.06 μg/ml, while the resistant colonies growing within the initial zone of inhibition in the disk diffusion test also grew inside the Etest inhibition zone (Fig. 1). No decreases in the size of the zone of inhibition were observed between the plates that contained fresh and preincubated erythromycin disks.
For the five heterogeneous phenotype strains, all tested colonies, both within and outside of the initial zone of inhibition, were confirmed to be B. pertussis by direct fluorescent-antibody testing (8).
DISCUSSION
We report here for the first time on five Bordetella pertussis isolates that exhibit a heterogeneous phenotype for erythromycin resistance. These isolates have a zone of inhibition characteristic of erythromycin susceptibility (>40 mm) after 3 days of incubation, but after extended incubation, resistant colonies appeared inside this zone with Etest MICs of >256 μg/ml. An incubation period greater than the commonly used 3 to 5 days is necessary for the detection of this heterogeneous phenotype; this extended incubation period might be a reason that no such B. pertussis isolates have been reported to date.
The frequencies of resistant and heterogeneous phenotypes detected among the 1,030 B. pertussis isolates in our study were low. However, our results also suggest a significant potential for these phenotypes to negatively affect patient management through their apparent distribution and persistence in diverse B. pertussis populations. Erythromycin treatment failures have been associated with resistant phenotypes isolated from 1994 to 2000 in Arizona, California, and Minnesota (6, 7; Bartkus et al., 41st ICAAC). The Georgia heterogeneous phenotype isolate was cultured from a pediatric AIDS patient after erythromycin treatment. Unfortunately, we were unable to retrospectively evaluate the treatment outcomes of the four Arizona patients whose cultures yielded the remaining heterogeneous phenotype isolates. But because these four cultures were isolated over a period of 2 years from different infants in a single county, they were probably maintained in the local B. pertussis population and could have been transmitted during this period of time. Isolation of both the resistant and heterogeneous phenotypes from distant geographic areas and over a 5-year period may represent geographic and temporal dissemination. Alternatively, these last observations may reflect the independent emergence of resistant phenotypes in response to local selective pressures.
The appearance of resistant colonies only after extended incubation raised the possibility that they might have been selected for by the disk diffusion assay as the erythromycin concentrations decreased with degradation over time. However, we have shown that erythromycin did not degrade in the disk diffusion assay; its activity was maintained for over 14 days at 37°C. Preliminary data also suggest the molecular basis for erythromycin resistance in B. pertussis isolates with the heterogeneous phenotype, and more extensive studies are under way to fully elucidate the mechanism (Bartkus et al., 41st ICAAC; K. Wilson, P. K. Cassiday, J. M. Bartkus, F. Doucet-Populaire, and G. N. Sanden, Abstr. 41st Intersci. Conf. Antimicrob. Agents Chemother., abstr. C1-1818, p. 102, 2001).
Our study identified B. pertussis isolates with a heterogeneous phenotype comprised of both erythromycin-susceptible and -resistant colonies. Laboratorians are advised to incubate the disk diffusion assay for at least 7 days in order to identify all strains with either the erythromycin-resistant or heterogeneous phenotypes. At the same time, clinicians should be cognizant of the potential for erythromycin treatment failures due to an infection with the heterogeneous phenotype and the need to obtain posttreatment cultures in order to identify alternative antimicrobial therapies. Rapid communication between clinicians and laboratorians and subsequent therapy changes could substantially reduce transmission of these isolates.
On the national level, the ongoing enhanced surveillance program is needed to identify resistant and heterogeneous B. pertussis phenotypes in order to monitor their appearance, dissemination, and potential association with changes in pertussis epidemiology. Correlation of resistant phenotypes with genetic subtypes, including pulsed-field gel electrophoresis patterns and pertactin and pertussis toxin gene sequence types, will allow assessment of their clonality and genetic relatedness to B. pertussis isolates currently circulating in the United States.
REFERENCES
- 1.Guris, D. 2000. Treatment and chemoprophylaxis, p. 3-1-3-14. In Guidelines for the control of pertussis outbreaks. Centers for Disease Control and Prevention, Atlanta, Ga.
- 2.Hardwick, T. H., P. Cassiday, R. S. Weyant, K. M. Bisgard, and G. N. Sanden. 2002. Changes in predominance and diversity of genomic subtypes of Bordetella pertussis isolated in the United States, 1935-1999. Emerg. Infect. Dis. 8:44-49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hill, B. C., C. N. Baker, and F. C. Tenover. 2002. A simplified method for testing of Bordetella pertussis for resistance to erythromycin and other antimicrobial agents. J. Clin. Microbiol. 38:1151-1155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hoppe, J. E., and T. Paulus. 1998. Comparison of three media for agar dilution susceptibility testing of Bordetella pertussis using six antibiotics. Eur. J. Clin. Microbiol. Infect. Dis. 17:391-393. [DOI] [PubMed] [Google Scholar]
- 5.Korgenski, E. K., and J. A. Daly. 1997. Surveillance and detection of erythromycin resistance in Bordetella pertussis isolates recovered from pediatric population in the intermountain west region of the United States. J. Clin. Microbiol. 35:2989-2991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lee, B. 2000. Progressive respiratory distress in an infant treated for presumed pertussis. Pediatr. Infect. Dis. J. 19:475, 492-493. [DOI] [PubMed] [Google Scholar]
- 7.Lewis, K., M. Saubolle, F. C. Tenover, M. F. Rudinsky, S. D. Barbour, and J. D. Cherry. 1995. Pertussis caused by an erythromycin-resistant strain of Bordetella pertussis. Pediatr. Infect. Dis. J. 14:388-391. [DOI] [PubMed] [Google Scholar]
- 8.Murphy, T., K. M. Bisgard, and G. N. Sanden. 2000. Diagnosis and laboratory methods, p. 2-2-2-3. In Guidelines for the control of pertussis outbreaks. Centers for Disease Control and Prevention, Atlanta, Ga.