Abstract
The molecular epidemiology of human caliciviruses (HuCVs) causing sporadic cases and outbreaks of acute gastroenteritis around eastern Spain (Catalonia and the Valencian Community) was studied by reverse transcription-PCR (RT-PCR) and by sequencing part of the RNA polymerase gene in open reading frame 1. HuCVs were detected in 44 of 310 stool specimens (14.19%) negative for other enteric pathogens obtained from children with acute gastroenteritis. Norwalk-like viruses (NLVs) were the most common cause of the gastroenteritis outbreaks investigated here. They were detected in 14 out of 25 (56%) outbreaks with an identified pathogen. Genotypes producing both sporadic cases and outbreaks were diverse, with a predominance of GGII strains related to genotypes Melksham and Lordsdale. Five strains clustered with a “new variant” designated GGIIb, which was detected circulating throughout quite a few European countries in the years 2000 and 2001. The emergence mechanism of these strains might be the occurrence of intertypic recombinations between different viruses. The nucleotide sequence of part of the capsid gene (ORF2) from three of these strains demonstrated their relationship with Mexico virus.
Human caliciviruses (HuCVs) are considered the most common cause of nonbacterial gastroenteritis outbreaks in persons of all ages worldwide (14, 17). These viruses can phylogenetically be divided into two genera, Norwalk-like viruses (NLVs), with genogroups I and II, and Sapporo-like viruses (SLVs), a group of typical human caliciviruses with distinctive morphology by electron microscopy. NLVs are the viruses most commonly associated with food- and waterborne outbreaks of gastroenteritis (9, 22). The course of the disease is usually mild and self-limiting, but the viruses are highly infectious and gastroenteritis outbreaks can involve small family groups or hundreds of individuals (19). Several studies have found human caliciviruses second only to rotaviruses as a cause of viral gastroenteritis in young children, and seroprevalence studies suggest that childhood infections in both developing and developed countries are common (4, 21, 30). However, their relative importance in mild or severe infantile gastroenteritis compared to other viruses has seldom been evaluated.
Molecular characterization of NLV strains are now an essential tool in our attempts to understand the epidemiology of this group of viruses. Genomic analysis of HuCVs causing outbreaks and sporadic cases of acute gastroenteritis reveals diversity, even in the RNA polymerase gene that is considered to be quite well conserved (1, 13, 37). Less sequence diversity in the polymerase gene has led to the widespread use of this genomic region as a target to detect and conduct molecular epidemiological studies of calicivirus infections (2, 3, 39, 40). According to this sequence divergence, NLVs from the two genogroups have been grouped so far into 15 clusters or genotypes and SLV into four genotypes (2, 12, 20, 37). NLV genogroup I (GGI) includes Norwalk, Southampton, Desert Shield, Queens Arms, and Winchester viruses, whereas NLV GGII includes Hawaii, Mexico, Lordsdale, Melksham, Hillingdon, and Grimsby viruses, among others (7, 25). Criteria for defining those strains belonging to the same genotype have been (i) more than 80% amino acid sequence identity in the complete capsid gene sequence and (ii) nucleotide sequence similarities of more than 85% (GGI) or 90% (GGII) when considering the polymerase gene in ORF1 (37).
The aim of this study was to investigate the role of HuCVs, both NLVs and SLVs, which cause sporadic cases of infantile diarrhea and outbreaks of acute gastroenteritis throughout eastern Spain, in particular Catalonia and the Valencian Community, and to characterize the genotypes of the calicivirus strains involved.
MATERIALS AND METHODS
Stool samples.
A total of 310 fecal samples obtained from children under 5 years of age with sporadic acute gastroenteritis were submitted to the laboratory between January 2000 and December 2001. Another 91 specimens were collected from patients involved in 30 outbreaks which occurred in different places during 2001. All the samples had already been proved negative for intestinal pathogenic parasites.
Bacteriological studies.
Salmonella, Shigella, Yersinia, and Campylobacter species were investigated by conventional bacterial culture procedures (10). The presence of enteropathogenic, enterotoxigenic, and enterohemorrhagic categories of diarrheagenic Escherichia coli was studied during outbreak investigations by PCR-based methods (8, 16, 31). When it was suggested by the clinical data, staphylococcal enterotoxin and Bacillus cereus enterotoxin were also investigated by enzyme immunoassays (EIAs) (TECRA Diagnostics, New South Wales, Australia).
Viral assays.
EIAs for group A rotaviruses and adenoviruses 40 and 41 (Rotaclone and Adenoclone; Meridian Diagnostics, Cincinnati, Ohio) and astroviruses (IDEIA Astrovirus; Dako Diagnostics, Cambridgeshire, United Kingdom) were performed.
RNA extraction.
Viral RNA was extracted by binding to size-fractionated silica particles (RNaid; Q-Biogene, Carlsbad, Calif.) in the presence of guanidinium isothiocyanate (5), eluted in diethyl pyrocarbonate-treated water containing RNasin (Promega, Madison, Wis.) and was either reverse transcribed immediately or stored at −70°C.
RT-PCR.
Stool samples were analyzed by RT-PCR for HuCVs (NLVs and SLVs). Primers NVp110/NI and NVp69, which allow the detection of both GGI and GGII NLV strains (24), and primer pair JV33 and SR80 for SLV detection (38) were used to amplify sequences of the RNA polymerase region as previously described (22, 38). A 326-nucleotide sequence of the same region was amplified using the JV12 and JV13 primer pair to conduct phylogenetic analysis of the NLV strains. When needed, capsid gene sequences were amplified by using primers JV21 (5′-CCN RCM YAA CCA TTR TAC AT-3′) and JV24 (5′-GTG AAT GAA GAT GGC GTC GA-3′).
Gel electrophoresis and Southern blotting of PCR products.
The PCR products were analyzed by electrophoresis on a 1.5% agarose gel containing ethidium bromide and visualized with UV light. After denaturation with 0.5 M NaOH and neutralization in Tris-buffered saline (0.5 M Tris-HCl, 1.5 M NaCl) the PCR products in the agarose gel were transferred overnight with 20× SSC buffer to a positive charged nylon membrane (NEN Dupont) by the capillary transfer method (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate).
Hybridization of PCR products.
The DNA transferred to the membrane was cross-linked in a UV Cross-linker (Hoefer). Hybridization was performed with a set of three different NLV-specific oligonucleotide probes labeled at the 5′end with digoxigenin (Roche Molecular Biochemicals). The probes used were NVp116 (24) and SR47D and SR61D (1). Hybridization and chromogenic detection of the hybrids with nitroblue tetrazolium (NBT) and BCIP (5-bromo-4-chloro-3-indolylphosphate) were carried out according to the protocols recommended by the manufacturer.
Nucleotide sequencing.
The 326-bp products from the RT-PCR were excised from the gel, extracted, and purified by using the Concert rapid gel extraction system (Life Technologies). Sequencing was carried out in both directions with the JV12 and JV13 primers using the ABI PRISM Big Dye Terminator Cycle Sequencing kit (Applied Byosistems) and an automated sequencer (Applied Biosystems ABI PRISM 377).
Phylogenetic analyses.
Prior to any phylogenetic analyses, translated amino acid sequences were aligned using CLUSTAL-X program (34) with the default parameters. Using this alignment as a template, nucleotide sequences were aligned with the DAMBE program (41). Only positions determined without uncertainty for at least 90% of the isolates were included in the following analyses. The phylogenetic content of this data set was assessed using the four-cluster likelihood-mapping method (32) as implemented in the program TREEPUZZLE. The method is based on an analysis of the maximum likelihood for the three fully resolved tree topologies that can be computed from four taxa (individual sequences or groups of sequences). Finally, phylogenetic reconstructions used the minimum evolution and the maximum parsimony methods implemented in the MEGA2 program (23). For the minimum evolution method, the nucleotide substitution model employed was the p-distance. We used the first and second positions of the protein-coding regions because the substitutions in the third codon positions are likely to be saturated among highly diverged taxa. Gaps were removed in pairwise comparisons when we used the minimum evolution method but employed as a fifth character when the tree reconstruction method was maximum parsimony. Statistical confidence for the evolutionary trees was assessed by bootstrap (1,000 replicates). The phylogenetic tree was drawn using the MEGA2 program.
Nucleotide sequence accession numbers. The nucleotide sequences determined in this study have been deposited in GenBank under accession numbers AJ487474 and AJ487789 to AJ487811.
RESULTS
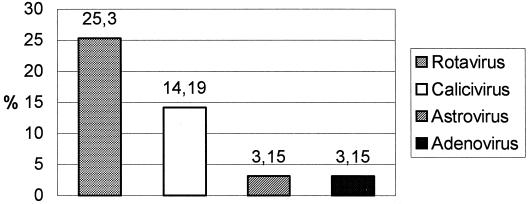
HuCVs were detected in 44 cases out of 310 cases of sporadic acute gastroenteritis in children (14.19%) (Table 1). A clear predominance of NLV GGII (33 out of 40 NLV-positive samples) was found. SLVs were detected in only four cases. Other enteric viruses detected were group A rotaviruses (25.3%), astroviruses (3.15%), and adenovirus type 40 or 41 (3.15%) (Fig. 1). The etiologic agent was identified in 25 of 30 gastroenteritis outbreaks investigated in this study (Table 2). Bacterial pathogens were detected in specimens obtained from patients involved in 11 outbreaks. Salmonella enterica was the bacteria most frequently found (seven outbreaks), followed by S. aureus (two outbreaks), Campylobacter jejuni, and B. cereus (one outbreak caused by each bacterial species). No diarrheagenic E. coli strains were detected.
TABLE 1.
Detection of human caliciviruses by RT-PCRa
| Test group | No. of samples | No. (%) RT-PCR positive | No. of NLV | No. of GGI | No. of GGII | No. of SLV |
|---|---|---|---|---|---|---|
| Sporadic cases | 310 | 44 (14.19) | 40 | 7 | 33 | 4 |
| Patients in outbreaks | 91 | 71 (78) | 71 | 23 | 48 | 0 |
| Total | 401 | 115 (28.67) | 111 | 30 | 81 | 4 |
Fecal specimens from sporadic cases of infantile acute gastroenteritis and from 14 gastroenteritis outbreaks were collected from January 2000 to December 2001.
FIG. 1.
Enteric viruses detected among 310 fecal specimens obtained from children under 5 years of age with acute gastroenteritis in Spain (2000 to 2001).
TABLE 2.
Enteropathogenic agents identified in outbreaks of acute gastroenteritis during 2001
| Outbreak characteristic | No. of outbreaks |
|---|---|
| Total | 30 |
| Unknown etiology | 5 (16.6%) |
| Identified pathogen | 25 (83.3%) |
| Bacteria | 11 |
| Salmonella enterica | 7 |
| Staphylococcus aureus | 2 |
| Campylobacter jejuni | 1 |
| Bacillus cereus | 1 |
| Viruses | 14 |
| Norwalk-like viruses | 14 |
| Parasites | 0 |
NLVs were identified as the etiologic agents in 14 gastroenteritis outbreaks (56%). Eleven outbreaks were caused by NLV GGII strains and three were caused by NLV GGI strains (Table 3).
TABLE 3.
Outbreaks of acute gastroenteritis caused by NLVs during 2001 in different sites in Catalonia and in the Valencian Community (Spain)
| Location | No. of cases | Setting | No. of samples | No. of positives | Genogroup | Genotype |
|---|---|---|---|---|---|---|
| Castell (Alicante) | 20 | Rural village | 4 | 4 | NLV GGII | GGIIb Mexico |
| Meliana (Valencia) | 32 | School | 3 | 2 | NLV GGII | Hillingdon |
| La Bisbal (Tarragona) | 14 | Nursing home | 4 | 3 | NLV GGII | GGIIb Mexico |
| Figueres (Girona) | 25 | Nursing home | 4 | 3 | NLV GGI | NDa |
| Lleida (Lleida) | 6 | Catering service | 6 | 3 | NLV GGI | ND |
| Sant Feliu (Girona) | 21 | Vacation camp | 3 | 3 | NLV GGII | Melksham |
| Vilafranca (Barcelona) | 17 | School | 2 | 1 | NLV GGII | Melksham |
| Ullastrell (Barcelona) | 37 | School | 21 | 17 | NLV GGI | ND |
| Montblanc (Tarragona) | 34 | Nursing home | 4 | 3 | NLV GGII | Lordsdale |
| Suria (Barcelona) | 75 | School | 15 | 15 | NLV GGII | GGIIb |
| Les Borges (Lleida) | 96 | School | 6 | 5 | NLV GGII | Melksham |
| Esteni d'Aneu (Lleida) | ? | Rural village | 9 | 6 | NLV GGII | Melksham |
| Utiel (Valencia) | 7 | Private home | 2 | 1 | NLV GGII | Lordsdale |
| Barcelona (Barcelona) | 80 | School | 8 | 5 | NLV GGII | ND |
ND, not determined.
Some epidemiological characteristics of the calicivirus-associated outbreaks and the viral genogroups and genotypes identified as the causative agents are also shown in Table 3. The majority of these outbreaks occurred in either schools (42.8%) or nursing homes (21.4%). Genotypes detected producing both sporadic cases and outbreaks were diverse, with GGII strains belonging to genotypes Melksham (eight strains), Lordsdale (five strains), Hillingdon (one strain), Mexico (one strain), and five viral strains that clustered with a “new variant” designated GGIIb, which has been detected circulating throughout several European countries during 2000 and 2001 (M. Koopmans, personal communication).
Three of the five GGIIb strains were detected in patients involved in outbreaks (Tarragona/238/01, Suria/312/01, and Castell/217/01), whereas the remaining two strains caused sporadic cases. In order to identify these strains more accurately, the capsid gene (ORF2) from three of them was sequenced and appeared to be related to Mexico genotype (results not shown).
Viral sequences determined in samples from patients in the same outbreak were highly homogeneous. However, the sequence of the virus involved in the Barcelona outbreak could not be established because the PCR product was not pure enough, presumably due to the presence of more than one single strain.
Six NLV GGI strains could not be clearly grouped with any of the previously described genotypes based on the sequence of the polymerase gene, although two of them (Lleida/235/01 and Sagunt/338/01) were related to Norwalk and Southampton viruses.
Prior to the phylogenetic inference, we applied the four-cluster likelihood-mapping method, which strongly confirmed the tree-likeness of the data set (81.8%) (i.e., the likelihood of the data set generating a true phylogenetic tree). Because a tree-likeness value of well above 50% can apparently be trusted (33), we may conclude that despite the high heterogeneity observed among GGI and GGII, the quality of the data still allows us to estimate the relationship between all calicivirus isolates.
Figure 2 shows a phylogenetic tree of sequences of a 326-nucleotide region of the RNA polymerase (ORF1) of 27 NLV strains isolated in Spain during 2000 to 2001 and 14 reference strains available either in GenBank or in the database of the European Union-funded project “Foodborne viruses in Europe.” Information about this database is available upon request. The GenBank accession numbers for the calicivirus reference strains are as follows: Lordsdale, x86557; Hawaii, HCU07611; Melksham, x81879; Hillingdon, AB 020558; Norwalk, M87661; Southampton, L07418; and Desert Shield, DSU04469.
FIG. 2.
Phylogenetic tree of sequence data of the RNA polymerase gene (ORF1) of 27 Norwalk-like virus strains collected in Spain during 2000 to 2001 and of reference strains. Phylogenetic reconstructions were done using the minimum evolution and the maximum parsimony methods implemented in the MEGA2 program (23). Statistical confidence for the evolutionary tree was assessed by bootstrap (1,000 replicates). Reference strains are shown in boxes, and those strains isolated from gastroenteritis outbreaks are underlined. The GenBank accession numbers for the calicivirus reference strains are as follows: Lordsdale, x86557; Hawaii, HCU07611; Melksham, x81879; Hillingdon, AB 020558; Norwalk, M87661; Southampton, L07418; Desert Shield, DSU04469.
Our results confirm a significant incidence of calicivirus infections, mainly NLV, among young children, causing gastroenteritis which needs medical attention. Only rotaviruses were found more frequently (25.3%) than caliciviruses. Mixed viral or viral-bacterial infections were not investigated in our study, and this may reduce to some extent the final figures of prevalent pathogens.
DISCUSSION
Molecular detection of HuCVs in stool specimens has become a more common diagnostic procedure of acute gastroenteritis in clinical laboratories (3). The application of PCR-based methods to screen stool specimens has not only shown that the overwhelming majority of outbreaks of acute gastroenteritis are attributable to NLVs but also that HuCVs also cause numerous cases of sporadic gastroenteritis (35, 39).
Our results show that human caliciviruses, mainly GGII of the NLVs, are indeed a major cause of both outbreaks and sporadic cases of acute gastroenteritis in infants and young children in eastern Spain. The prevalence rate detected (14.19%) is similar to that reported by other authors, 14% in Dijon, France (4), 9% in Melbourne, Australia (21), and 6.5% in the United Kingdom (35).
To our knowledge, this is the first Spanish study in which genogroups and genotypes of Norwalk-like viruses have been investigated as etiologic agents of sporadic gastroenteritis cases in children and in gastroenteritis outbreaks. We found that among viral pathogens rotaviruses are the most frequent agents causing sporadic gastroenteritis cases (25.3%), followed by HuCVs (14.19%). In previous studies, we characterized the genotypes of group A rotavirus and the serotypes of astrovirus detected in our geographical area (6, 15) as well as the presence of astroviruses in wastewater (28). Our study confirms that NLVs are the main cause of gastroenteritis outbreaks in Spain, as in other EU countries (4, 26, 29, 36). Interestingly, although it is well known that group A rotaviruses are the primary cause of sporadic cases of gastroenteritis in children, they are not frequently found as the causative agents of gastroenteritis outbreaks.
The predominance of NLV GGII strains during this period of study agrees with previous reports of a higher prevalence of GGII strains than of GGI (4, 21). A notable difference has also been found in antibody acquisition to Mexico virus (GGII), to which over 70% of children older than 2 years are seropositive, whereas only 12% of these children had been infected with Norwalk virus (GGI) (27). Molecular epidemiological studies of outbreaks have revealed that GGII strains dominate GGI strains (11, 39). The reason for this is unknown, although differences in biological properties, such as virulence, routes of transmission, or stability of the virus in the environment, are possible explanations.
The alignments of the RNA polymerase nucleotide sequences of the seven GGI strains isolated in this study with reference strains of the accepted GGI genotypes showed similarities of less than 85%. This, together with the results of the phylogenetic analysis, suggests that some of these isolates might represent new variants of GGI or even new genotypes. This possibility awaits confirmation by capsid gene sequence analysis.
In our study, SLV strains were rarely detected (only four cases [1.29%]) as a cause of severe gastroenteritis in young children, perhaps reflecting the idea that SLVs cause less severe symptoms than NLVs. Similar results were obtained by Kirkwood and Bishop in Melbourne, Australia (21), who also consider that SLV-associated gastroenteritis may be clinically milder, not requiring hospitalization.
The number of gastroenteritis outbreaks included in this study does not reflect the real figure of outbreaks which occurred during the period of study, because many of them are not reported to the Regional Health Services. Even so, not all outbreaks are tested for a viral etiology and, consequently, calicivirus infections are underdiagnosed.
Strains isolated from particular outbreaks where highly homogenous and no sequence differences among specimens investigated within one outbreak were found. However, in some specimens, a mixture of sequences was detected, both from patients involved in an outbreak and from sporadic cases of infantile diarrhea. This finding was presumably due to mixed infections with more than one single viral strain. We have not detected any remarkable difference in the viral genotypes producing outbreaks or sporadic cases in the community. It has been suggested that there are two distinct epidemiological patterns of NLV strains, one producing endemic infections (i.e., Grimsby virus strains considered endemic in the United Kingdom) and another causing epidemic infections (Norwalk and Mexico viruses) (17). Our data do not support such hypotheses, although the number of strains sequenced is limited. The same genotypes were found causing both sporadic cases and gastroenteritis outbreaks in 2000 to 2001, and it seems that multiple viral genotypes are circulating at any given time. Continued NLV surveillance across Europe and investigations of the biological differences between strains are needed in order to clarify their molecular epidemiology.
The genetic relatedness of caliciviruses does not always correspond when regions in different open reading frames are sequenced, this may originate by recombinations in the evolution of caliciviruses (29, 36). The detection of differences in phylogenetic tree topology between different parts of the genome, like the RNA polymerase and the capsid genes, suggests the existence of the recombination process among HuCVs (18). To identify recombinants more clearly, one must compare the sequence of different regions of the genome against a panel of well-characterized representative sequences in a database, such as the one created by the EU-funded project on “Foodborne viruses in Europe” to search for matching strains.
Capsid gene sequence analysis can be used to verify the viral genotyping based on the sequence of the polymerase gene fragment produced by the diagnostic PCR. There is a correlation between antigenic grouping based on the use of virus-like particles generated using baculovirus expression systems and genomic grouping based on capsid amino acid sequences (12, 18). Further characterization of the genetic and antigenic relationships will help the classification of human caliciviruses.
Understanding the real prevalence of these viruses, the mechanisms that lead to the emergence of new viral variants and how they spread within populations will improve the prevention of the diarrheal diseases that they cause.
Acknowledgments
This study was supported by the EU grant QLK1-CT-00594 (EU 5th Framework Program “Quality of life and management of living resources”).
We thank Marion Koopmans, Jan Vinjé, and Harry Vennema (RIVM, National Institute of Public Health and the Environment, Bilthoven, The Netherlands) for providing reference strains and viral sequences and Santiago F. Elena for assistance with phylogenetic analyses.
REFERENCES
- 1.Ando, T., S. S. Monroe, J. R. Gentsch, Q. Jin, D. C. Lewis, and R. I. Glass. 1995. Detection and differentiation of antigenically distinct small round-structured viruses (Norwalk-like viruses) by reverse transcription-PCR and Southern hybridization. J. Clin. Microbiol. 33:64-71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ando, T., J. S. Noel, and R. L. Fankhauser. 2000. Genetic classification of “Norwalk-like viruses.” J. Infect. Dis. 181:S336-S348. [DOI] [PubMed] [Google Scholar]
- 3.Atmar, R. L., and M. K. Estes. 2001. Diagnosis of noncultivatable gastroenteritis viruses, the human caliciviruses. Clin. Microbiol. Rev. 14:15-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bon, F., P. Fascia, M. Dauvergne, D. Tenenbaum, H. Planson, A. M. Petion, P. Pothier, and E. Kohli. 1999. Prevalence of group A rotavirus, human calicivirus, astrovirus, and adenovirus type 40 and 41 infections among children with acute gastroenteritis in Dijon, France. J. Clin. Microbiol. 37:3055-3058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Boom, R., C. J. A. Sol, M. M. M. Salimans, C. L. Jansen, P. M. E. Wertheim-van Dillen, and J. van der Noordaa. 1990. Rapid and simple method for purification of nucleic acids. J. Clin. Microbiol. 28:495-503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Buesa, J., C. O. de Souza, M. Asensi, C. Martínez, J. Prat, and M. T. Gil. 2000. VP7 and VP4 genotypes among rotavirus strains recovered from children with gastroenteritis over a 3-year period in Valencia, Spain. Eur. J. Epidemiol. 16:501-506. [DOI] [PubMed] [Google Scholar]
- 7.Dingle, K. E., P. R. Lamden, E. O. Caul, and I. N. Clarke. 1995. Human enteric Caliciviridae: the complete genome sequence and expression of virus-like particles from a genetic group II small round structured virus. J. Gen. Virol. 76:2349-2355. [DOI] [PubMed] [Google Scholar]
- 8.Feng, P., and S. R. Monday. 2000. Multiplex PCR for detection of traid and virulence factors in enterohemorrhagic Escherichia coli serotypes. Mol. Cell. Probes 14:333-337. [DOI] [PubMed] [Google Scholar]
- 9.Frankhauser, R. L., J. S. Noel, S. S. Monroe, T. Ando, and R. I. Glass. 1998. Molecular epidemiology of “Norwalk-like viruses” in outbreaks of gastroenteritis in the United States. J. Infect. Dis. 178:1571-1578. [DOI] [PubMed] [Google Scholar]
- 10.Gilligran, P. H., J. M. Janda, M. A. Karmali, and J. M. Miller. 1992. Laboratory diagnosis of bacterial diarrhea. American Society for Microbiology, Washington, D.C.
- 11.Green, J., C. I. Gallimore, J. P. Norcott, D. Lewis, and D. W. G. Brown. 1995. Broadly reactive reverse transcriptase polymerase chain reaction in the diagnosis of SRSV-associated gastroenteritis. J. Med. Virol. 47:392-398. [DOI] [PubMed] [Google Scholar]
- 12.Green, J., J. Vinjé, C. Gallimore, M. P. G. Koopmans, A. Hale, and D. W. G. Brown. 2000. Capsid protein diversity among Norwalk-like viruses. Virus Genes 20:227-236. [DOI] [PubMed] [Google Scholar]
- 13.Green, J., J. P. Norcott, D. Lewis, C. Arnold, and D. W. G. Brown. 1993. Norwalk-like viruses: demonstration of genomic diversity by polymerase chain reaction. J. Clin. Microbiol. 31:3007-3012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Green, K. Y. 1997. The role of human caliciviruses in epidemic gastroenteritis. Arch. Virol. 13(Suppl.):153-165. [DOI] [PubMed] [Google Scholar]
- 15.Guix, S., S. Caballero, C. Villena, R. Bartolomé, C. Latorre, N. Rabella, M. Simó, A. Bosch, and R. M. Pintó. 2002. Molecular epidemiology of astrovirus infection in Barcelona, Spain. J. Clin. Microbiol. 40:133-139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gunzburg, S. T., N. G. Tornieporth, and L. W. Riley. 1995. Identification of enteropathogenic Escherichia coli by PCR-based detection of the bundle-forming pilus gene. J. Clin. Microbiol. 33:1375-1377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hale, A., K. Mattik, D. Lewis, M. Estes, X. Jiang, J. Green, R. Eglin, and D. Brown. 2000. Distinct epidemiological patterns of Norwalk-like virus infection. J. Med. Virol. 62:99-103. [DOI] [PubMed] [Google Scholar]
- 18.Hardy, M. E., S. F. Kramer, J. J. Treanor, and M. K. Estes. 1997. Human calicivirus genogroup II capsid diversity revealed by analysis of the prototype Snow Mountain agent. Arch. Virol. 142:1469-1479. [DOI] [PubMed] [Google Scholar]
- 19.Inouye, S., K. Yamashita, S. Yamadera, M. Yoshikawa, N. Kato, and N. Okabe. 2000. Surveillance of viral gastroenteritis in Japan: pediatric cases and outbreak incidents. J. Infect. Dis. 181:S270-S274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jiang, X., W. D. Cubbit, T. Berke, W. Zhong, X. Dai, S. Nakata, L. K. Pickering, L. K., and D. O. Matson. 1997. Sapporo-like human caliciviruses are genetically and antigenically diverse. Arch. Virol. 142:1813-1827. [DOI] [PubMed] [Google Scholar]
- 21.Kirkwood, C. D., and R. Bishop. 2001. Molecular detection of human calicivirus in young children hospitalized with acute gastroenteritis in Melbourne, Australia, during 1999. J. Clin. Microbiol. 39:2722-2724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Koopmans, M., J. Vinjé, M. de Wit, I. Leenen, W. van der Poel, and Y. van Duynhoven. 2000. Molecular epidemiology of human enteric caliciviruses in The Netherlands. J. Infect. Dis. 181(Suppl. 2):S262-S269. [DOI] [PubMed] [Google Scholar]
- 23.Kumar, S., K. Tamura, I. B. Jacobsen, and M. Nei. 2001. MEGA2: molecular evolutionary genetics analysis software. Arizona State University, Tempe. [DOI] [PubMed]
- 24.Le Guyader, F., M. K. Estes, M. E. Hardy, F. H. Neill, J. Green, D. W. G. Brown, and R. L. Atmar. 1996. Evaluation of a degenerate primer for the PCR detection of human caliciviruses. Arch. Virol. 141:2225-2235. [DOI] [PubMed] [Google Scholar]
- 25.Lew, J. F., A. Z. Kapikian, J. Valdesuso, and K. Y. Green. 1994. Molecular characterization of Hawaii virus and other Norwalk-like viruses: evidence for genetic polymorphism among human caliciviruses. J. Infect. Dis. 170:535-542. [DOI] [PubMed] [Google Scholar]
- 26.Maguire, A. J., J. Green, D. W. G. Brown, U. Desselberger, and J. J. Gray. 1999. Molecular epidemiology of outbreaks of gastroenteritis associated with small round-structured viruses in East Anglia, United Kingdom, during the 1996-1997 season. J. Clin. Microbiol. 37:81-89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Parker, S. P., W. D. Cubbitt, and X. Jiang. 1995. Enzyme immunoassay using baculovirus-expressed human calicivirus (Mexico) for the measurement of IgG responses and determining its seroprevalence in London, UK. J. Med. Virol. 46:194-200. [DOI] [PubMed] [Google Scholar]
- 28.Pintó, R. M., C. Villena, F. S. Le Guyader, S. Guix, S. Caballero, S. Pommepuy, and A. Bosch. 2001. Astrovirus detection in wastewater samples. Water Sci. Technol. 12:73-76. [PubMed] [Google Scholar]
- 29.Schreier, E., F. Döring, and U. Künkel. 2000. Molecular epidemiology of outbreaks of gastroenteritis associated with small round structured viruses in Germany in 1997/98. Arch. Virol. 145:443-453. [DOI] [PubMed] [Google Scholar]
- 30.Smit, T. K., P. Bos, I. Peenze, X. Jiang, M. K. Estes, and A. D. Steele. 1999. Seroepidemiological study of genogroup I and II caliciviruses infections in South and Southern Africa. J. Med. Virol. 59:227-231. [DOI] [PubMed] [Google Scholar]
- 31.Stacy-Phipps, S., J. J. Mecca, and J. B. Weiss. 1995. Multiplex PCR assay and simple preparation method for stool specimens detect enterotoxigenic Escherichia coli DNA during course of infection. J. Clin. Microbiol. 33:1054-1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Strimmer, K., and A. von Haeseler. 1997. Likelihood-mapping: a simple method to visualize phylogenetic content of a sequence alignement. Proc. Natl. Acad. Sci. USA 94:6815-6819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Strimmer, K., and A. von Haeseler. 1996. Quartet puzzling: a quartet maximum-likelihood method for reconstructing tree topologies. Mol. Biol. Evol. 13:964-969. [Google Scholar]
- 34.Thompson, J. D., T. J. Gibson, F. Plewniak, F. Jeanmougin, and D. G. Higgins. 1997. The CLUSTAL X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25:4876-4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tompkins, D. S., M. J. Hudson, H. R. Smith, R. P. Eglin, J. G. Wheeler, M. M. Brett, R. J. Owen, J. S. Brazier, P. Cumberland, and P. E. Kong Vm Cooke. 1999. A study of infectious intestinal disease in England: microbiological findings in cases and controls. Comm. Dis. Public Health 2:108-113. [PubMed] [Google Scholar]
- 36.Vainio, K., K. Stene-Johansen, T. O. Jonassen, A.-L. Bruu, and B. Grinde. 2001. Molecular epidemiology of calicivirus infections in Norway. J. Med. Virol. 65:309-314. [DOI] [PubMed] [Google Scholar]
- 37.Vinjé, J., J. Green, D. C. Lewis, C. I. Gallimore, D. W. G. Brown, and M. P. G. Koopmans. 2000. Genetic polymorphism across regions of the three open reading frames of “Norwalk-like viruses.” Arch. Virol. 145:223-241. [DOI] [PubMed] [Google Scholar]
- 38.Vinjé, J., H. Deijl, R. van der Heide, D. Lewis, K.-O. Hedlund, L. Svensson, and M. P. G. Koopmans. 2000. Molecular detection and epidemiology of Sapporo-like viruses. J. Clin. Microbiol. 38:530-536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Vinjé, J., and M. P. G. Koopmans. 1996. Molecular detection and epidemiology of small round-structured viruses in outbreaks of gastroenteritis in the Netherlands. J. Infect. Dis. 174:610-615. [DOI] [PubMed] [Google Scholar]
- 40.Vinjé, J., and M. P. Koopmans. 2000. Simultaneous detection and genotyping of “Norwalk-like viruses” by oligonucleotide array in a reverse line blot hybridization format. J. Clin. Microbiol. 38:2595-2601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Xia, X. 2000. Data analysis in molecular biology and evolution. Kluwer Academic Publishers, Boston, Mass.