Abstract
The qualitative Cobas Amplicor hepatitis C virus (HCV) version 2.0 assay (HCV PCR) and the Bayer Reference Testing Laboratory HCV RNA transcription-mediated amplification assay (HCV TMA) were compared for analytical sensitivity, clinical performance, and workflow. Limits of detection were determined by testing dilutions of the World Health Organization HCV standard in replicates of 15 at concentrations of from 1.0 to 70 IU/ml. The limit of detection of the HCV PCR assay was calculated to be 45 IU/ml on initial testing and 32 IU/ml after resolution of gray zone results. The calculated limit of detection for HCV TMA was 6 IU/ml. To compare clinical performance, 300 specimens, grouped as follows, were evaluated: 112 samples that were indeterminate in an anti-HCV enzyme immunoassay (EIA) and for which HCV RNA was not detected by HCV PCR; 79 samples that were EIA positive and for which HCV RNA was not detected by HCV PCR; and 105 samples that were both EIA and HCV PCR positive. For these groups, interassay concordance ranged from 96.2% to 100%. In addition, three HCV PCR gray zone specimens and one neonatal specimen were also evaluated. A 64-sample run (full run, 91 specimens) required 5 h for testing by HCV TMA, whereas almost 8 h were required to test a full run of 22 specimens by HCV PCR. HCV TMA demonstrated excellent concordance with HCV PCR when clinical samples were tested. However, HCV TMA was more sensitive than HCV PCR, required less time for test result completion, and had a greater throughput.
It is estimated that 170 million people worldwide are chronically infected with hepatitis C virus (HCV), a major cause of cirrhosis and liver cancer (10, 20). Given the burden of the disease and the current potential for cure, there is a compelling need for diagnosis of active HCV infection (10, 20). Although diagnostic tests that employ enzyme immunoassays (EIA) and recombinant immunoblot assays are used for serological screening and confirmation, respectively, nucleic acid testing and HCV antigen detection methods are required to demonstrate active infection (3, 9). As HCV antigen tests are still under development, qualitative nucleic acid testing is presently the method of choice to confirm active infection and to assess virologic clearance in response to therapy (3, 9, 22). Qualitative nucleic acid tests are also 10- to 100-fold more sensitive than currently available quantitative tests (9).
The qualitative Roche Cobas Amplicor HCV version 2.0 assay (HCV PCR), a reverse transcription-PCR assay, has a reported limit of detection of 50 IU/ml (11) and has been used to confirm viremia and to measure treatment response (10). Recent studies reported that another nucleic acid test based on transcription-mediated amplification (TMA), the HCV RNA transcription-mediated amplification qualitative test (HCV TMA), was able to detect HCV RNA in some patients who had no detectable HCV RNA (as determined by the first and second versions of HCV PCR) at the end of treatment and subsequently experienced virologic relapse (4, 18). These results suggested that HCV TMA is more sensitive than HCV PCR and that test sensitivity may be important for accurate assessment of antiviral response especially when measuring end-of-treatment viral clearance (6, 12, 23). We therefore assessed the limits of detection of the two assays with a dilution panel of the World Health Organization (WHO) international HCV standard, analyzed the interassay concordance with clinical specimens, and assessed assay throughput and workflow.
MATERIALS AND METHODS
Cobas Amplicor HCV PCR test version 2.0 (HCV PCR).
HCV PCR is a semiautomated assay. After manual extraction of HCV RNA from serum or plasma, subsequent steps, including reverse transcription, DNA amplification, and detection, are performed on the Cobas instrument. RNA detection is achieved by hybridizing the denatured, biotinylated PCR products to complementary HCV DNA and to internal control probes. There are three possible outcomes for this test: HCV RNA detected, HCV RNA not detected, and gray zone (i.e., indeterminate). The Amplicor Cobas analyzer automatically interprets specimens with weak signals (A660 range of 0.15 to 1.0) as gray zone. Specimens whose signals fall within this range are to be retested in duplicate. On retesting, the final result is considered to be HCV RNA positive if all three tests yield an A660 equal to or greater than 0.15. The result is HCV RNA not detected if the A660 value of any one of the three tests is less than 0.15 and the corresponding internal control is amplified and detected (11). Maximum throughput for a single run is 22 reportable specimens.
HCV RNA TMA qualitative test (HCV TMA).
Following a lysis step, target HCV RNA is captured to magnetic particles coated with oligonucleotides complementary to the 5′ untranslated region of the HCV genome. Monitoring of target capture and amplification is achieved by adding an internal control RNA to each sample. The target RNA is then amplified with an isothermal TMA process that requires the addition of primers, reverse transcriptase, and T7 RNA polymerase. Detection of the amplified product is based on hybrid protection and the dual kinetic assays (18, 19). Each tube produces a chemiluminescent signal that is read as relative light units (RLU). Data are reported both as calculated RLU and as signal-to-cutoff ratios. When the signal-to-cutoff ratio is >1, the specimen is considered reactive or as having detectable HCV RNA. There are two possible outcomes of this assay; HCV RNA is either detected or not detected. Maximum throughput of HCV TMA in one run is 91 reportable specimens. A total of nine calibrators and controls are required.
WHO HCV dilution panel.
A dilution panel of the WHO standard for HCV RNA (WHO 96/790) was gravimetrically made at the British Columbia Centers for Disease Control (BCCDC). Lyophilized standard was reconstituted in 0.5 ml of distilled water as per the package insert (National Institute for Biological Standards and Control) to yield a solution containing 105 IU/ml. By international agreement, 100,000 IU is defined as the amount of HCV in 1 ml of this standard (16, 17). Dilutions were made with anti-HCV and HCV PCR-negative human plasma. The dilution panel consisted of 15 replicates each (panel members) at concentrations of 1.0, 2.5, 5.0, 10, 20, 30, 40, 50, 60, and 70 IU/ml. Multiple aliquots of panel members were frozen at −70°C, and all aliquots underwent one freeze-thaw cycle.
Aliquots for HCV TMA testing were shipped on dry ice to the Bayer Reference Testing Laboratory (BRTL) in Emeryville, Calif. Specimens were assayed randomly with each of the panel members typically represented at least once in each run. Additional negative controls were added to each run of both HCV PCR and HCV TMA. HCV PCR was performed as directed by the manufacturer. For HCV PCR, the BCCDC employed the run validity criteria described in the package insert. BRTL assay procedures were followed for HCV TMA, a home-brew assay. Standard run validity criteria developed by GenProbe, the manufacturer of the assay, were employed by the BRTL. An assay run was considered valid if the controls gave correct results and a minimum of 90% of specimens had internal controls whose RLUs were above the value for the internal control cutoff. Specimen results were considered invalid if the internal control RLU value was below the internal control cutoff value.
Qualitative Cobas Amplicor PCR assay testing of the WHO panel.
Over a period of 7 working days, six full runs and one partial run were carried out on a single Cobas instrument by the same BCCDC technologist, who was blinded to sample concentrations tested. Samples yielding gray zone results were retested in duplicate according to the manufacturer's directions.
Qualitative TMA-based testing of the WHO panel.
Fifteen replicates of most of the dilution panel members (containing 1 to 50 IU/ml) were analyzed in two runs of 64 samples each. The 60-IU/ml and 70-IU/ml replicates were tested randomly on a subsequent assay run. In addition to the standard negative calibrators, four negative control samples were included in each run. Validity criteria for the run are described in the previous section on the WHO panel. All HCV TMA assays were performed on the same instrument and the same kit lot.
Assessment of assay limit of detection.
The limit of detection, or analyte detection sensitivity, is defined as the minimum analyte concentration at which an assay can distinguish a positive specimen from negative specimens 95% of the time (13). A logistic regression model was used to estimate the detection rate as a function of concentration via the maximum-likelihood method. The limit of detection was estimated with the inverse of the estimated logistic equation to calculate the concentration corresponding to a 95% detection rate (8).
Clinical samples.
Between December 1999 and February 2000, approximately 19,000 clinical specimens were received at the BCCDC for HCV testing. A subset of 300 specimens, as described below, were chosen for clinical evaluation of the nucleic acid test assays. Separate aliquots of these samples, whose serum had been removed from the clot within 4 h of the draw and stored at −70°C, had been used previously for HCV PCR testing. HCV PCR was initially performed because nucleic acid testing was requested or the specimen was anti-HCV indeterminate. Aliquots of all 300 samples were shipped on dry ice to the BRTL, where they were stored at −80°C prior to HCV TMA testing.
As per standard BCCDC procedures, specimens submitted for HCV testing are screened for anti-HCV by third-generation enzyme immunoassay (EIA) on an AxSYM instrument (Abbott, North Chicago, Ill.). Specimens with signals above the assay cutoff undergo secondary serological testing on an Ortho Vitros Eci instrument (Ortho-Clinical Diagnostics). Specimens whose EIA test signals are greater than 2 to 3 standard deviations (SD) above the cutoff of both assays have been shown to be serologically HCV positive >98% of the time (1, 2, 7, 9). When the results of one or both EIA tests are greater than the assay cutoff but the signal intensity is less than 2 SD above the assay cutoff, the specimen is considered serologically indeterminate.
Specimens were divided into three groups: 112 samples that were EIA indeterminate (signals < 2 SD above the cutoff) and for which HCV RNA was not detected by HCV PCR; 79 samples that were anti-HCV positive by both EIAs (> 2 SD above the assay cutoff) and for which HCV RNA was not detected by HCV PCR; and 105 samples that were positive by both EIAs (>2 SD above the cutoff) and for which HCV RNA was detected by HCV PCR. Three HCV PCR gray zone and one neonatal clinical specimen were also tested by HCV TMA. The PCR gray zone samples had insufficient volume to be retested in duplicate, as required by the package insert. The neonatal specimen had detectable HCV RNA by HCV PCR but had not been tested by EIA because antibody in neonates is passively acquired. Aliquots of all 300 samples were shipped on dry ice to the BRTL for HCV TMA testing.
Assessment of workflow.
A data collection worksheet was developed for the HCV TMA and HCV PCR assays. One run of each assay was timed during the WHO panel testing. The HCV PCR workflow measurements were performed at the BCCDC, where one BCCDC technologist timed another BCCDC technologist performing one full run of 22 reportable specimens. The HCV TMA workflow assessment was performed at the BRTL, where one BRTL technologist timed a BCCDC technologist performing the first test run of 64 specimens as part of the WHO panel evaluation.
RESULTS
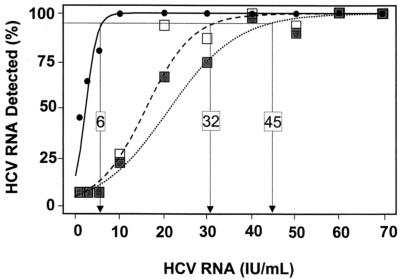
The WHO international standard HCV dilution panel was used to assess the sensitivity of HCV PCR and HCV TMA (Fig. 1). After testing 15 replicates at each of the 10 dilutions by HCV PCR, 20 of 150 (13.3%) specimens with concentrations between 1.0 and 50 IU/ml yielded gray zone signals (Fig. 2). Three specimens contained either 1.0 or 2.5 IU/ml; 13 of the 20 ranged in concentration between 5 and 20 IU/ml; and four specimens had concentrations of 30 to 50 IU/ml. On retesting in duplicate, all gray zone specimens at concentrations between 1.0 and 10 IU/ml resolved as HCV RNA not detected, while those with concentrations between 20 and 50 IU/ml resolved as HCV RNA positive (Fig. 2). As there are no established guidelines for determining the limit of detection of a test with more than two outcomes (13), the limit of detection of HCV PCR was calculated to be 45 IU/ml before and 32 IU/ml after resolution of specimens yielding gray zone values, respectively.
FIG. 1.
HCV TMA and HCV PCR detection sensitivity as a function of HCV RNA concentration as estimated by logistic regression. Fifteen replicates of each sample at various concentrations were tested by both assays (except for the 60-IU/ml specimens, for which 14 replicates were tested by HCV TMA). The percentage giving positive HCV PCR results at each concentration when gray zone results were not resolved is illustrated by solid squares (▪), and the dotted line shows the regression curve. The percentage giving positive HCV PCR results at each concentration when gray zone results were resolved by retesting in duplicate is shown by open squares (□), and the dashed line shows the corresponding regression curve. The percentage giving positive results in HCV TMA at each concentration is shown by solid circles (•), with the regression curve denoted by the black line. The horizontal line indicates the 95% detection value. The x coordinate where each calculated curve intercepts the 95% line represents the limits of detection of each assay.
FIG. 2.
Distribution of 20 HCV PCR gray zone results according to HCV RNA concentration based on testing replicates of 15. The symbol above the bar (− or +) represents the resolved status, negative or positive, respectively, after retesting in duplicate.
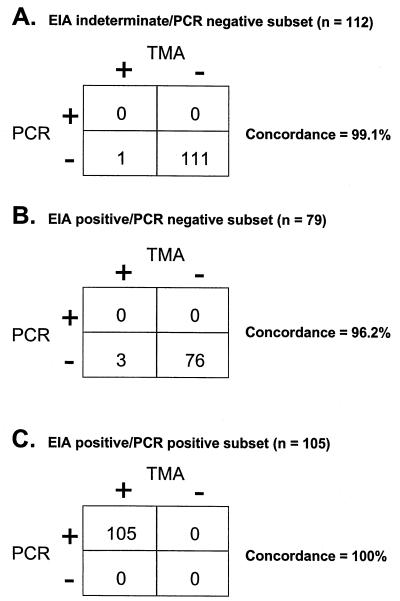
In contrast, the limit of detection of HCV TMA was determined to be 6 IU/ml. At concentrations of 10 IU/ml and higher, 100% of the samples were HCV RNA positive, and at 2.5 IU/ml, 9 of 15 (60%) of the replicates contained detectable HCV RNA by HCV TMA. Therefore, HCV TMA was 5.3- to 7.5-fold (6 IU/ml versus 32 to 45 IU/ml) more sensitive than HCV PCR. To assess the concordance of the two assays, 300 clinical specimens were tested by both assays. The data for 296 of 300 of the specimens are presented in Fig. 3, grouped according to the three patient subsets. Among the first group of anti-HCV-indeterminate/PCR-negative specimens (Fig. 3A), HCV RNA was detected by HCV TMA in 1 of 112 samples (0.9%), giving a concordance of 99.1%. This single positive specimen was obtained from a patient with a history of substance abuse. In the second group of anti-HCV-positive/HCV PCR-negative specimens (Fig. 3B), HCV TMA detected HCV RNA in 3 of 79 samples (3.8%), resulting in a concordance rate of 96.2%. One of the three specimens came from a patient with a history of substance abuse, and the other two were from patients undergoing Rebetron therapy for HCV infection. These four discrepant samples had low signal-to-cutoff ratios of 1.13, 1.22, 2.59, and 6.39 by HCV TMA, as might be expected for samples with low levels of virus.
FIG. 3.
Overall performance of HCV PCR and HCV TMA in detecting HCV RNA in three distinct subgroups.
The concordance was 100% in the third group of 105 anti-HCV-positive/HCV PCR-positive specimens (Fig. 3C). Four additional specimens that did not fall into the three patient groups gave the following results: three of four (approximately 1% of the 300 samples) gave gray zone results on initial testing by HCV PCR but had undetectable HCV RNA by HCV TMA. The HCV PCR-positive neonatal specimen was also positive by HCV TMA.
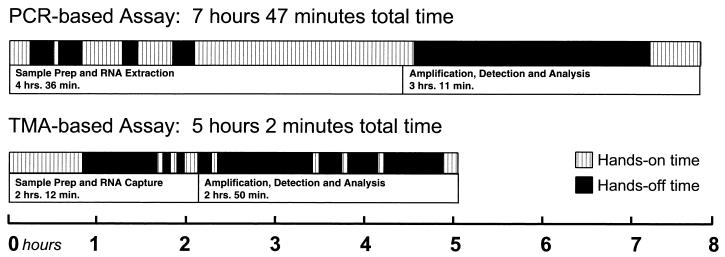
The hands-on and hands-off time as well as the total run time for one representative run of each assay were also determined (Fig. 4). The total run time was divided into time spent on (i) sample preparation, including RNA extraction and capture, and (ii) amplification, detection, and analysis. Hands-on time is defined as the time the technologist was actively manipulating the specimen or reagents, while hands-off time refers to incubation time periods when the technologist was not actively involved in assay procedures. For the HCV TMA assay run with 64 reportable results, the total run time was 5 h, 2 min, with a hands-on time of 2 h, 45 min. In contrast, for 22 reportable results, the HCV PCR assay run required 7 h, 45 min to complete, with a hands-on time of 4 h, 36 min.
FIG. 4.
HCV PCR and HCV TMA workflow analysis. The upper segments of each bar graph show total running time and hands-on time per sample. The lower segments of each bar graph illustrate the proportion of time devoted to the separate tasks of (i) sample preparation and RNA extraction and capture and (ii) amplification, detection, and analysis. A total of 22 reportable samples were run by HCV PCR; 64 samples were run by HCV TMA.
DISCUSSION
Combination therapy with interferon/ribavirin or pegylated interferon/ribavirin has dramatically increased the percentage of patients who appear to be cured of HCV infection after treatment (10, 20). From these encouraging results and given the large numbers of HCV-infected individuals worldwide, we expect that nucleic acid testing will be increasingly required for diagnosis of active HCV infection and therapeutic monitoring. Assay sensitivity, workflow, and throughput will be important criteria for laboratories selecting a nucleic acid test.
The recent availability of the WHO standard facilitates interassay comparisons and allows more standardized assessment of performance, for example, limit of detection. Our data are consistent with the stated limits of detection for both HCV PCR and HCV TMA. Lee et al. (11) reported that HCV PCR had 100% sensitivity at 50 IU/ml and 91% sensitivity at 25 IU/ml. Our data, based on 95% sensitivity, showed the limit of detection for HCV PCR to be 45 IU/ml when gray zone specimens were not resolved and 32 IU/ml after specimens yielding gray zone results were resolved by retesting in duplicate.
Of note, when testing the WHO dilution panel on the HCV PCR, 20 of the 150 panel members gave gray zone results, and 16 of the 20 occurred when testing replicates at concentrations below 20 HCV IU/ml. For a given assay, a gray zone represents the range of analyte values that generate signals where there is an overlap between positive and negative test results (5). At a limiting dilution of the analyte, the ability of any assay to detect a target becomes a statistical process that is said to resemble a Poisson distribution where approximately one-third of the test reactions produce negative results (5). HCV PCR has a recognized gray zone and an algorithm developed by the manufacturer to resolve specimen status by retesting in duplicate. This study demonstrates that the resolution of HCV PCR gray zone specimens increased the sensitivity of HCV PCR from 45 to 32 IU/ml and that retesting in duplicate provides a statistical process to enhance the limit of detection of HCV PCR. It must be understood that using duplicate or triplicate testing to resolve specimens that yield gray zone signals can increase either assay sensitivity or specificity. This statistical approach to resolving gray zone specimens is possible because HCV PCR is highly specific (11).
Our data demonstrate that the limit of detection of the HCV TMA assay was 6 IU/ml, which is consistent with the 5-IU/ml limit of detection reported by Ross et al. (15) and Sawyer and colleagues (19). Published data have shown that HCV TMA is also very specific (15, 19). HCV TMA reports specimens as being either reactive (HCV RNA detected) or nonreactive (HCV RNA not detected). No gray zone value is reported. Given the 6-IU/ml limit of detection of HCV TMA, to evaluate whether HCV TMA has a gray zone would have required a thorough evaluation of test results from multiple replicates of a dilution series containing very low levels of HCV RNA (e.g., 5 to 0.5 IU/ml) (5). In order to verify whether specimens at such low dilutions of the WHO HCV standard truly contain HCV RNA, discrepant specimens would also need to be tested by an appropriately sensitive third-party assay. At this time, no standardized commercial HCV assay with sufficient sensitivity to adequately resolve discrepant results at such low HCV RNA concentrations has been described. Homebrew nested PCR assays which may be sensitive enough are prone to amplicon contamination and difficult to standardize (24).
The impact of resolving gray zone specimens on the laboratory workload depends on the frequency of specimens with low levels of HCV RNA. Within this highly selected specimen set, 3 of 300 specimens yielded initial gray zone results. Indeed, of the last 3,000 HCV PCR tests performed at the BCCDC, 1 to 2% yielded gray zone values on initial testing (data not shown).
To compare the performance of the two assays with clinical specimens, we selected subsets of specimens that are problematic to the laboratory along with appropriate controls. Specimen selection was influenced by analysis of the previous HCV testing results at the BCCDC. This laboratory performs approximately 75,000 anti-HCV tests per year, 8 to 10% of which have some degree of anti-HCV reactivity by AxSYM on initial screening. Of all anti-HCV-reactive specimens, 15 to 18% yield weak signals (<2 SD above the assay cutoff), and the other 82 to 85% yield signals greater than 2 SD above the assay cutoff (9). Without epidemiological data, it is difficult to determine the cause of weakly anti-HCV-reactive specimens (9, 14). Possible explanations include testing too early during acute infection (i.e., prior to complete seroconversion); blunted antibody responses due to immunosuppression; waning antibody due to resolved infection (21); and a false-positive serological test (9). Nucleic acid testing can determine if HCV RNA is present in these specimens. Similarly, in individuals with strongly reactive anti-HCV serology, nucleic acid testing is required to confirm active HCV infection. Confirmation of active infection is especially important when assessing individuals who undergo treatment and those with normal serum transaminase levels (10, 20).
We therefore assessed the concordance of the nucleic acid testing assays in selected specimen subsets, i.e., HCV PCR-negative, anti-HCV-indeterminate, and anti-HCV-positive specimens, to determine if the more sensitive HCV TMA could identify HCV RNA in additional specimens. Overall, the concordance of the two assays was excellent and varied between 96 and 100% depending on the combination of anti-HCV and HCV PCR results of the subsets tested. For example, all strongly anti-HCV-positive, HCV PCR-positive specimens were also HCV TMA positive. Of the 191 specimens (of 300) that were anti-HCV indeterminate or positive, HCV PCR-negative, four were HCV TMA assay positive. These specimens came from individuals with a history of substance abuse or from patients undergoing Rebetron therapy. Unfortunately, there was insufficient volume for further testing of these samples. Of note, Ross et al. (15) also reported the detection of HCV RNA by HCV TMA in four samples that had undetectable HCV RNA by Amplicor version 2.0 PCR among serial samples from 10 patients undergoing interferon alpha therapy. These were confirmed with a nonstandardized homebrew PCR assay.
For the three clinical specimens that gave initial gray zone results by HCV PCR and were negative by HCV TMA, there was insufficient specimen volume for retesting in duplicate as required by the HCV PCR diagnostic algorithm.
Assay sensitivity, throughput, and workflow all influence laboratory assay selection. While we ran 64 samples in our workflow study, the maximum throughput for HCV TMA (91 reportable samples) is more than four times that of HCV PCR (22 reportable samples). Notwithstanding the difficulty in accurately determining workflow times for assays that are incompletely automated, HCV TMA showed considerable timesaving over HCV PCR for both hands-on time and time to final result.
In summary, the HCV TMA assay demonstrated excellent concordance with HCV PCR, but it was more sensitive, required less time for test result completion, and had a greater throughput. Further studies in well-characterized populations will be required to properly assess the clinical impact of the increased HCV TMA sensitivity.
Acknowledgments
Both the BCCDC and BRTL supported the analytical testing reported herein.
We thank Kristina Whitfield (Posterdocs, Oakland, Calif.) for graphics.
REFERENCES
- 1.Allain, J. P., A. Kitchen, S. Aloysius, I. Reeves, J. Petrik, J. A. J. Barbara, and L. M. Williamson. 1996. Safety and efficacy of hepatitis C virus antibody screening of blood donors with two sequential screening assays. Transfusion 36:401-405. [DOI] [PubMed] [Google Scholar]
- 2.Anderson, S. C., T. Hathaway, I. K. Kuramoto, P. V. Holland, R. Gilcher, T. Koch, and S. Hojvat. 1995. Comparison of two second-generation anti-hepatitis C virus ELISA on 21431 US blood donor samples. J. Viral Hepatitis 2:55-61. [DOI] [PubMed] [Google Scholar]
- 3.Aoyagi, K., K. Iida, C. Ohue, Y. Matsunaga, E. Tanaka, K. Kiyosawa, and S. Yagi. 2001. Performance of a conventional enzyme immunoassay for hepatitis C virus core antigen in the early phases of hepatitis C infection. Clin. Lab. 47:119-127. [PubMed] [Google Scholar]
- 4.Comanor, L., F. Anderson, M. Ghany, R. Perrillo, E. J. Heathcote, C. Sherlock, I. Zitron, D. Hendricks, and S. C. Gordon. 2001. Transcription-mediated amplification is more sensitive than conventional PCR-based assays for detecting residual serum HCV RNA at end of treatment. Am. J. Gastroenterol. 96:2968-2972. [DOI] [PubMed] [Google Scholar]
- 5.Coupland, R. W. 1994. Quality control, analytic sensitivity, and the polymerase chain reaction. J. Clin. Immunoassay 17:237-242. [Google Scholar]
- 6.Doglio, A., C. Laffont, F. X. Caroli-Bosc, P. Rochet, and J. Lefebvre. 1999. Second generation of the automated Cobas Amplicor HCV assay improves sensitivity of hepatitis C virus RNA detection and yields results that are more clinically relevant. J. Clin. Microbiol. 37:1567-1569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Goubau, P., M. Reynders, K. Beuselinck, F. Nevens, K. Peerlinck, and J. Desmyter. 1997. Confirmatory strategy of hepatitis C serology based on two screening assays in a diagnostic setting. Acta Clin. Belg. 52:31-35. [DOI] [PubMed] [Google Scholar]
- 8.Kleinbaum, D. G. 1992. Logistic regression: a self-learning text, p. 101-119. Springer-Verlag, New York, N.Y.
- 9.Krajden, M. 2000. Hepatitis C virus diagnosis and testing. Can. J. Public Health 91(Suppl. 1):S34-39. [PubMed] [Google Scholar]
- 10.Lauer, G. M., and B. D. Walker. 2001. Hepatitis C virus infection. N. Engl. J. Med. 345:41-52. [DOI] [PubMed] [Google Scholar]
- 11.Lee, S. C., A. Antony, N. Lee, J. Leibow, J. Q. Yang, S. Soviero, K. Gutekunst, and M. Rosenstraus. 2000. Improved version 2.0 qualitative and quantitative Amplicor reverse transcription-PCR tests for hepatitis C virus RNA: calibration to international units, enhanced genotype reactivity, and performance characteristics. J. Clin. Microbiol. 38:4171-4179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.McHutchison, J. G., L. M. Blatt, R. Ponnudurai, K. Goodarzi, J. Russell, and A. Conrad. 1999. Ultracentrifugation and concentration of a large volume of serum for HCV RNA during treatment may predict sustained and relapse response in chronic HCV infection. J. Med. Virol. 57:351-355. [PubMed] [Google Scholar]
- 13.National Committee for Clinical Laboratory Standards. 2000. User protocol for evaluation of qualitative test performance, vol. 20, p. 1-3. Evaluation protocol EP12-P. National Committee for Clinical Laboratory Standards, Wayne, Pa.
- 14.Polywka, S., M. Schroter, H. H. Feucht, B. Zollner, and R. Laufs. 2001. Relevance of reactivity in commercially available hepatitis C virus antibody assays. J. Clin. Microbiol. 39:1665-1668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ross, R. S., S. O. Viazov, S. Hoffmann, and M. Roggendorf. 2001. Performance characteristics of a transcription-mediated nucleic acid amplification assay for qualitative detection of hepatitis C virus RNA. J. Clin. Lab Anal. 15:308-313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Saldanha, J. 2001. Validation and standardisation of nucleic acid amplification technology (nucleic acid testing) assays for the detection of viral contamination of blood and blood products. J. Clin. Virol. 20:7-13. [DOI] [PubMed] [Google Scholar]
- 17.Saldanha, J., N. Lelie, and A. Heath. 1999. Establishment of the first international standard for nucleic acid amplification technology (nucleic acid testing) assays for HCV RNA. W.H.O. Collaborative Study Group. Vox Sang. 76:149-158. [DOI] [PubMed] [Google Scholar]
- 18.Sarrazin, C., G. Teuber, R. Kokka, H. Rabenau, and S. Zeuzem. 2000. Detection of residual hepatitis C virus RNA by transcription-mediated amplification in patients with complete virologic response according to polymerase chain reaction-based assays. Hepatology 32:818-823. [DOI] [PubMed] [Google Scholar]
- 19.Sawyer, L., K. Leung, M. Friesenhahn, D. Duey, M. McMorrow, and B. Eguchi. 2000. Clinical laboratory evaluation of a new sensitive and specific assay for qualitative detection of hepatitis C virus in clinical specimens. J. Hepatol. 32(Suppl. 2):116A. [Google Scholar]
- 20.Shad, J. A., and J. G. McHutchison. 2001. Current and future therapies of hepatitis C. Clin. Liver Dis. 5:335-359. [DOI] [PubMed] [Google Scholar]
- 21.Takaki, A., M. Wiese, G. Maertens, E. Depla, U. Seifert, A. Liebetrau, J. L. Miller, M. P. Manns, and B. Rehermann. 2000. Cellular immune responses persist and humoral responses decrease two decades after recovery from a single-source outbreak of hepatitis C. Nat. Med. 6:578-582. [DOI] [PubMed] [Google Scholar]
- 22.Tanaka, E., C. Ohue, K. Aoyagi, K. Yamaguchi, S. Yagi, K. Kiyosawa, and H. J. Alter. 2000. Evaluation of a new enzyme immunoassay for hepatitis C virus (HCV) core antigen with clinical sensitivity approximating that of genomic amplification of HCV RNA. Hepatology 32:388-393. [DOI] [PubMed] [Google Scholar]
- 23.Yamakawa, Y., M. Sata, H. Suzuki, K. Tanaka, E. Tanaka, S. Noguchi, K. Ono, and K. Tanikawa. 1998. Monitoring of serum levels of HCV RNA in early phase of IFN therapy; as a predictive marker of subsequent response. Hepatogastroenterology 45:133-136. [PubMed] [Google Scholar]
- 24.Zaaijer, H. L., H. T. M. Cuypers, H. W. Reesink, I. N. Winkel, G. Gerken, and P. N. Lelie. 1993. Reliability of polymerase chain reaction for detection of hepatitis C virus. Lancet 341:722-724. [DOI] [PubMed] [Google Scholar]