Abstract
Background
Metabolic profiling of blood metabolites, particularly in plasma and serum, is vital for studying human diseases, human conditions, drug interventions and toxicology. The clinical significance of blood arises from its close ties to all human cells and facile accessibility. However, patient-specific variables such as age, sex, diet, lifestyle and health status, along with pre-analytical conditions (sample handling, storage, etc.), can significantly affect metabolomic measurements in whole blood, plasma, or serum studies. These factors, referred to as confounders, must be mitigated to reveal genuine metabolic changes due to illness or intervention onset.
Review objective
This review aims to aid metabolomics researchers in collecting reliable, standardized datasets for NMR-based blood (whole/serum/plasma) metabolomics. The goal is to reduce the impact of confounding factors and enhance inter-laboratory comparability, enabling more meaningful outcomes in metabolomics studies.
Key concepts
This review outlines the main factors affecting blood metabolite levels and offers practical suggestions for what to measure and expect, how to mitigate confounding factors, how to properly prepare, handle and store blood, plasma and serum biosamples and how to report data in targeted NMR-based metabolomics studies of blood, plasma and serum.
Supplementary Information
The online version contains supplementary material available at 10.1007/s11306-025-02259-7.
Keywords: Metabolomics, Standardization, Blood, Serum, Plasma, NMR, Metabolites
Introduction
Metabolomics offers a powerful approach to monitor the chemical response of biological systems to internal and external perturbations. In particular, the comprehensive measurement of metabolite changes in selected tissues or biofluids can provide a detailed, molecular snapshot of the down-stream effects of physiological, environmental, pathological or genetic changes and exposures. In this regard, metabolomics offers a very powerful route to measure the chemical phenotype of an organism (Wishart, 2016). In addition to serving as a useful vehicle to perform rapid and inexpensive molecular phenotyping, metabolomics is increasingly being integrated with genomics, transcriptomics, and proteomics to provide a more complete molecular understanding of biological systems (Jendoubi, 2021; Patt et al., 2019; Wishart, 2022).
Metabolomic analyses can be either untargeted or targeted. Untargeted methods attempt to measure all detectable features in the investigated samples, typically without absolute metabolite quantification. In targeted metabolomics, a pre-selected set of metabolites is identified and (often) absolutely quantified in each sample. Multivariate statistics are then used to determine a set ofmetabolites, either feature-based in the untargeted approach or based on their absolute concentrations in the targeted approach, significantly associated with the response of interest, e.g., a phenotype (Barba et al., 2009; DeSilva et al., 2009; Jung et al., 2013; H. S. Kim et al., 2009; Moussallieh et al., 2014; Porzel et al., 2014; Schicho et al., 2010; Van Doorn et al., 2007). Untargeted metabolomics is ideal for exploratory analysis and novel metabolite discovery, while targeted metabolomics is most suited for clinical applications and biomarker discovery (Patti et al., 2012; Roberts et al., 2012; Schrimpe-Rutledge et al., 2016; Wishart, 2016).
Both targeted and untargeted metabolomics can be done on a wide range of analytical platforms, including liquid chromatography coupled with mass spectrometry (LC–MS), gas chromatography coupled with mass spectrometry (GC–MS), inductively coupled plasma mass spectrometry (ICP–MS) and nuclear magnetic resonance (NMR) spectroscopy. NMR and MS methods are the most commonly used analytical approaches with each method having its own strengths and weaknesses. In terms of strengths, MS is highly sensitive, offers broad metabolite coverage and requires only micrograms or nanograms of sample (Letertre et al., 2021). However, MS is an inherently destructive technique, requiring substantial sample preparation with limited ability to perform accurate quantification (X. Liu & Locasale, 2017). On the other hand, NMR is a non-destructive technique that is highly reproducible, requires little-to-no sample preparation, is very amenable to full automation and offers facile, accurate quantification (Wishart et al., 2022a, 2022b, 2022c). However, NMR is much less sensitive than MS, has much more limited metabolite coverage and often needs milligrams of material (Edison et al., 2021).
Regardless of the choice of method (targeted or untargeted) or analytical platform (NMR, LC–MS or GC–MS), the selection, collection and handling of the biological samples is of paramount importance in metabolomic studies. Traditionally, most metabolomic studies have focused on the analysis of biofluids (blood, saliva, cerebrospinal fluid) or excreta (urine or feces), as these are more easily collected. The most popular and most important biofluid in clinical metabolomics is blood.
Blood is a specialized body fluid that has four main components: plasma, red blood cells, white blood cells, and platelets. It is the carrier by which oxygen, nutrients, metabolites and drugs are transported throughout the body and it directly or indirectly (via interstitial fluids between various blood/organ barriers) reaches all living cells. Blood maintains homeostasis in the body via several continuous regulatory mechanisms. Because it touches every organ and collects and delivers both essential and waste products, blood has played a pivotal role in many areas of biomedical science, including the assessment of human health, the search for new disease biomarkers, the exploration of the effects of dietary intake on health, the monitoring of drug or surgical interventions, and the investigation of drug toxicity (Angioni et al., 2022; Farley et al., 2013; Krewski et al., 2010; Schwedes et al., 2002; Ubaida-Mohien et al., 2023).
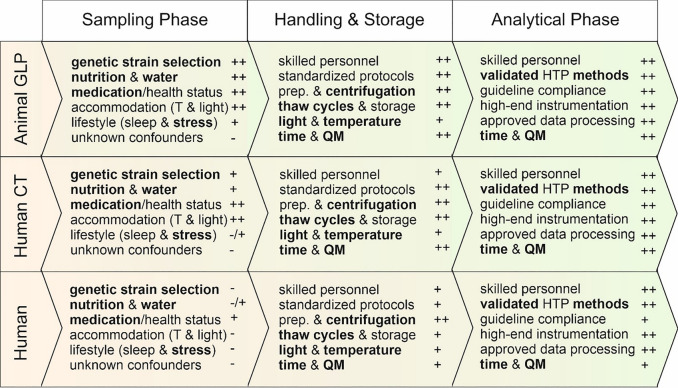
While blood is perhaps the ideal biofluid to monitor health via metabolomics, the challenges of storing and preserving blood for metabolomic studies have forced most researchers to use cell-free versions of blood—namely plasma and serum. Serum is the straw-colored liquid portion that remains after the blood has clotted, while plasma is the liquid portion that remains when clotting is prevented with the addition of an anticoagulant. Both serum and plasma contain metabolites and proteins, and both are easily prepared via centrifugation. While plasma and serum are widely thought to be very uniform and very similar in chemical composition (certainly much more so than urine), many different factors, including genetics, age, sex, diet, physical activities and lifestyle, can affect their metabolic composition. Additionally, the collection and storage conditions of serum or plasma specimens—referred to as pre-analytical factors—can significantly affect metabolite concentrations. These include donor diurnal variations, emotional or physical stress, collection temperature, collection methods, collection tubes, processing times, storage temperatures and storage time. These confounders can complicate the interpretation of metabolomic data, the assessment of health status and the discovery of novel biomarkers. Some of these factors can be easily controlled during the sampling, pre-analytical and analytical phases of a metabolomics study while others are often beyond the control of the researcher (Fig. 1).
Fig. 1.
Operative phases, confounding factors* and associated uncertainties (−)** for in-vitro metabolome studies in humans and animals. Animal testing as part of good laboratory practice (GLP) provide the highest level of controllability (+ +)**. Similar favorable conditions can be found in human clinical trials (CT) where the most important confounding variables can be well controlled. In human field studies, important parameters are placed in the test subjects' confidence range. This leads to uncertainties within a study, which can no longer be fully compensated for. Thus, good planning, extensive standardization, efficient quality management, and detailed collection and reporting of potential confounding variables is a guarantee for high-quality metabolome studies. There is currently no global standard for conducting metabolome studies. *the most critical factors are in bold; **rough estimate, whereby (−) indicates little controllable and ( +) well controllable; prep. = sample preparation; QM = quality management; HTP = high throughput
Over the past 15 years, a number of excellent reviews on blood, plasma and serum metabolomics (and metabolomes) have been published (Bar et al., 2020; James & Parkinson, 2015; Kalantari & Nafar, 2019; Kondoh et al., 2020; Likhitweerawong et al., 2021; Psychogios et al., 2011; Ren et al., 2021; Serkova et al., 2011; Trabado et al., 2017; A. Zhang et al., 2012). Most have focused on the application of MS-based approaches to analyze the metabolome of blood/serum/plasma for specific diseases or conditions such as autism, various cancers, dementia, chronic kidney disease, sepsis or aging (Kalantari & Nafar, 2019; Kondoh et al., 2020; Likhitweerawong et al., 2021; Ren et al., 2021; Serkova et al., 2011; A. Zhang et al., 2012). A smaller number of papers have focused on NMR-based approaches to analyze the metabolome of blood/serum/plasma (Nagana Gowda & Raftery, 2017, 2019; Nagana Gowda et al., 2022; Schicho et al., 2012; Silva et al., 2020; Soininen et al., 2015; Würtz et al., 2017). An even smaller number reviewed the analytical aspects of blood/serum/plasma metabolomics (sample preparation, sample processing, metabolite extraction/filtering, separation methods, sample referencing, batch corrections, and data processing) (Beckonert et al., 2007; Lipfert et al., 2019; Madrid-Gambin et al., 2023; Snytnikova et al., 2019; Wishart et al., 2022a, 2022b, 2022c). Recent advancements, particularly the ISO 23118: 2021- “Molecular in vitro diagnostic examinations—specifications for pre-examination processes in metabolomics in urine, venous blood serum and plasma” standard, provide guidelines for pre-examination processes in urine, venous blood plasma, and serum for metabolomics (IOS, 2021). This standard outlines detailed recommendations for handling, documentation, and processing to ensure the reliability of metabolomics studies. The latest additional guidelines particularly focus on the pre-examination phases of multi-center metabolomic studies (Ghini et al., 2022) and on the pre-analytical factors influencing metabolomic studies (Thachil et al., 2024) without considering patient associated factors. However, there is a lack of comprehensive guidelines on the best practices for the preparation of these blood samples, or on the expected compounds in targeted NMR-based metabolomics studies. Moreover, increased awareness of the impact of diet, sex, gender, lifestyle, and gut microbiota on metabolite levels in blood highlights the significance of considering and controlling these pre-analytical factors. These variables are crucial for the accurate interpretation of metabolite data in blood, serum, or plasma, as they can lead to potential misinterpretations if not properly accounted for.
The main purpose of this review is to assist metabolomics researchers in conducting well-designed, well-controlled studies so that reliable, standardized, shareable datasets from targeted or untargeted, NMR-based metabolomics of blood, plasma or serum can be obtained. This document is divided into five parts. The first part of this review focuses on what is known about the blood, serum and plasma metabolomes as measured by various NMR techniques. This is important for understanding the subsequent topics and caveats discussed in this review. In part two of this review, we will focus on the participant or patient-specific confounders and highlight how significant these effects are especially when it comes to biomarker discovery. The third part of this review discusses the effects of sample handling, sample storage and sample preparation on NMR-detectable blood, plasma and serum metabolites. The fourth part discusses the results of a literature review conducted to assess the common practices among labs conducting NMR-based metabolomics studies of blood, serum or plasma. A number of shortcomings in current practices and reporting methods are highlighted. The fifth part of this review focuses on providing recommendations regarding the use of appropriate methods to reduce pre-analytic variability and improve reporting practices. This includes recommendations for optimizing study design, cohort/subject data collection, sample selection, sample collection, and sample preparation of blood, serum or plasma for NMR-based metabolomics. We anticipate that these descriptions, recommendations, and assessments will enhance the quality of NMR-based metabolomics studies performed on human blood, plasma and serum.
Defining the whole blood, serum and plasma metabolomes as measured by NMR
As highlighted earlier, whole blood, plasma, and serum are fundamentally different in their preparation methods, with detectable differences in their composition. Whereas large scale, multi-platform (NMR, GC–MS, LC–MS, ICP-MS) studies have described or reviewed the metabolite content of human plasma and serum (Lawton et al., 2008; Psychogios et al., 2011; Trabado et al., 2017; Wishart, Guo, et al. 2022), there has been no detailed summary of the NMR-measurable metabolite content of serum, plasma or blood. Given this paucity of information, we believe a short summary of what is known about the NMR-measurable metabolite content of serum, plasma or blood would be quite useful. The detection limit for biomolecules using NMR methods (typically in the micromolar range) is generally one to two orders of magnitude (10- to 100-fold) less sensitive than commonly applied LC–MS metabolomics methods. Thus, in NMR metabolomics of blood, typically only about 50–100 metabolites are detected, whereas LC/GC–MS methods can detect far more than 1000 metabolites (Mandal et al., 2025). Likewise, the number of detectable compounds also depends on the sample preparation or sample pre-treatment protocols applied to each biofluid, the NMR pulse sequences applied, the NMR spectrometer frequency and the target(s) of the chemical analysis.
Blood, serum, and plasma consist of a complex mixture of both large molecules (proteins, lipoprotein complexes, and lipid vesicles) and numerous small molecules. In a given NMR spectrum, the signals from all these components are superimposed on top of each other. In particular, the signals from the large molecules tend to be much more intense than the small molecule metabolites. NMR samples are usually analyzed without chromatographic separation (such as GC or LC), which results in complex, highly overlapped spectra. The analytical complexity is further increased by the presence of both large molecules (proteins, lipoprotein complexes, lipid vesicles) and small molecules (metabolites). Without signal suppression or separation of the large molecules, the NMR signals from these macromolecules interferes with the small molecule signals. This interference leads to difficulties in identifying and quantifying the small molecule metabolites (or to properly measure large molecules—if desired). Removal of the proteins (deproteinization) as well as the lipids and lipoproteins (delipidization) is commonly done to enhance metabolite signals in NMR. This can be done chemically by solvent (methanol) extraction (Nagana Gowda & Raftery, 2014; Nagana Gowda et al., 2015) or mechanically by ultrafiltration (Psychogios et al., 2011; Rout et al., 2023; Wevers et al., 1994). It can also be done spectroscopically using the Carr-Purcell-Meiboom-Gill (CPMG) pulse sequence which takes advantage of the T2 relaxation time differences between large molecules and small molecules in NMR (Beckonert et al., 2007; Nagana Gowda et al., 2015). Because of its simplicity, the use of the CPMG “spectroscopic filtering” method in blood metabolomics has been and still is widely used, in particular, for semi-quantitative, untargeted metabolomics studies. In comparison to chemical or mechanical deproteinization and delipidization, the CPMG method can be employed on blood/serum/plasma specimens without any additional sample preparation steps, typically lowering consumable costs, workload, and sources of unwanted pre-analytical variability. Therefore, the CPMG method is particularly advantageous for the analysis of large-scale metabolomics studies involving hundreds to thousands of specimens. However, the CPMG method does not completely remove large molecule signals, and, because of various uncontrollable signal suppression effects, it makes the quantification of metabolite levels very difficult and inconsistent (Bliziotis et al., 2020; Nagana Gowda et al., 2015).
Nevertheless, over the past 10 years, it has been recognized that NMR of intact serum and plasma (i.e. with all intrinsic proteins and lipids) can provide very useful quantitative information about lipid and lipoprotein content—as well as some number of small molecules (Cacciatore et al., 2021; Funderburg et al. 2017; Jeyarajah et al., 2006; Julkunen et al., 2023; Lodge et al., 2021; Robinson et al., 2020; Sliz et al., 2018; Zeleznik et al., 2023). Therefore, we provide a list of small molecules measured by traditional NMR methods employing ultrafiltration or methanol extraction/lyophilization, as well as metabolites, lipids, proteins and lipoproteins identified in intact plasma via the Nightingale (See Table 1, “Serum concentrations” or “Plasma concentrations” (Julkunen et al., 2023)) as well as the Bruker IVDr Quantification Engine for Human Plasma & Serum (B.I. Quant-PS) (38 metabolites; see Table 1, “Whole serum” column) (Jiménez et al., 2018; Trautwein, 2025)) approaches, currently the most popular approaches for serum/plasma characterization by NMR. It is also worth noting that LabCorp (LipoScience) has developed a similar assay or platform for measuring lipids and lipoproteins in intact serum and plasma (Funderburg et al. 2017; Jeyarajah et al., 2006).
Table 1.
List of metabolites which can be measured using 1D 1H NMR in whole blood, plasma or serum, after various sample preparation methods
| Name | HMDB | Ultra-filtration (Rout et al., 2023) | Deproteinization (Nagana Gowda & Raftery, 2019; Nagana Gowda et al., 2015) | Cell Lysisa (Nagana Gowda & Raftery, 2017; Nagana Gowda et al., 2022) | Whole Serum (Julkunen et al., 2023; Zeleznik et al., 2023) | Lipid Extraction (Julkunen et al., 2023; Zeleznik et al., 2023) | Serum concentrations (µmol/L)b,c | Plasma concentrations (µmol/L)b,c |
|---|---|---|---|---|---|---|---|---|
| 1-Methylhistidine | HMDB0000001 | * | * |
7.1 ± 1.3 (4.6–9.1) |
– | |||
| 2-Hydroxybutyrate | HMDB0000008 | * | * | *d |
48.1 ± 16.6 (25.0–81.8) |
14.5 | ||
| 3-Hydroxybutyrate | HMDB0000011 | * | * | *d,e |
60.5 ± 62.1 (0.0–3843.4) |
215.9 ± 127.3 (59.1–1570.0) | ||
| 2-Oxoisovalerate | HMDB0000019 | * | 3.2 | – | ||||
| Acetate | HMDB0000042 | * | * | *d,e |
18.7 ± 33.0 (0.0–1800.6) |
44.8 ± 31.2 (19.4–940.0) | ||
| Betaine | HMDB0000043 | * | * |
43.5 ± 11.9 (18.7–59.8) |
7.4 | |||
| AMP | HMDB0000045 | * | – | – | ||||
| Carnitine | HMDB0000062 | * | * |
39.5 ± 5.8 (29.1–45.3) |
18.7 | |||
| Creatine | HMDB0000064 | * | * | *d |
34.1 ± 21.2 (12.3–70.5) |
17.1 | ||
| Glycerophosphocholine | HMDB0000086 | * | – | – | ||||
| Dimethylamine | HMDB0000087 | * | – | – | ||||
| Dimethylglycine | HMDB0000092 | * | * | *d |
2.7 ± 1.0 (1.4–4.7) |
0.8 | ||
| Citrate | HMDB0000094 | * | * | *d,e |
65.6 ± 13.2 (3.4–572.0) |
111.6 ± 18.0 (60.4–245.0) | ||
| Choline | HMDB0000097 | * | * | *d |
8.6 ± 2.1 (5.8–12.6) |
6.1 | ||
| Ethanol | HMDB0000108 | * | *d |
17.7 ± 11.1 (7.1–42.0) |
69 | |||
| Galactose | HMDB0000143 | *d | ||||||
| Glucose | HMDB0000122 | * | * | *d,e |
3747.8 ± 1192.1 (289.5–41,833.0) |
4437.8 ± 1430.3 (2120.0–22,700.0) | ||
| Glycine | HMDB0000123 | * | * | *d,e |
171.0 ± 66.7 (0.0–793.2) |
257.4 ± 37.6 (159.0–555.0) | ||
| Glutathione | HMDB0000125 | * | – | – | ||||
| Glycerol | HMDB0000131 | * | * | *d,e |
252.1 ± 76.6 (193.3–423.5) |
88.9 ± 23.2 (20.6–212.0) | ||
| Fumarate | HMDB0000134 | * | * | – | – | |||
| Formate | HMDB0000142 | * | * | *d |
42.1 ± 3.5 (35.2–48.6) |
– | ||
| Glutamate | HMDB0000148 | * | * | *d |
35.1 ± 27.7 (12.5–96.7) |
– | ||
| Hypoxanthine | HMDB0000157 | * | * |
5.2 ± 1.6 (3.4–7.9) |
– | |||
| Tyrosine | HMDB0000158 | * | * | *d,e |
63.0 ± 14.5 (5.5–389.4) |
49.7 ± 9.7 (22.9–120.0) |
||
| Phenylalanine | HMDB0000159 | * | * | *d,e |
47.2 ± 11.6 (5.2–1309.2) |
72.7 ± 10.9 (44.3–355.0) | ||
| Alanine | HMDB0000161 | * | * | *d,e |
296.6 ± 78.5 (61.3–1272.6) |
382.5 ± 56.9 (232.0–739.0) | ||
| Proline | HMDB0000162 | * | * | *d |
140.4 ± 40.6 (72.3–198.3) |
84.5 | ||
| Threonine | HMDB0000167 | * | * | *d |
126.4 ± 21.6 (95.2–160.3) |
86 | ||
| Asparagine | HMDB0000168 | * | * | *d |
55.8 ± 11.7 (37.4–78.0) |
44.7 | ||
| Mannose | HMDB0000169 | * | * |
41.3 ± 6.3 (31.9–53.8) |
13.7 | |||
| Isoleucine | HMDB0000172 | * | * | *d,e |
51.1 ± 18.2 (0.0–240.5) |
56.8 ± 17.8 (20.5–235.0) | ||
| Inosine monophosphate | HMDB0000175 | * | – | – | ||||
| Histidine | HMDB0000177 | * | * | *d,e |
65.6 ± 11.4 (21.2–657.4) |
58.9 ± 8.9 (24.0–109.0) |
||
| Lysine | HMDB0000182 | * | * | *d |
195.5 ± 37.9 (132.7–236.1) |
101.4 | ||
| Serine | HMDB0000187 | * | * |
109.1 ± 24.5 (79.1–152.9) |
72.1 | |||
| Lactate | HMDB0000190 | * | * | *d,e | 3873.7 ± 1141.1 (722.2–21,248.0) | 1454.8 ± 489.0 (543.0–3940.0) | ||
| Aspartate | HMDB0000191 | * | * |
5.0 ± 2.2 (0.0–8.4) |
– | |||
| Acetylcarnitine | HMDB0000201 | * | * |
10.8 ± 6.1 (5.5–27.0) |
6.3 | |||
| Oxoglutarate | HMDB0000208 | * | *d |
7.8 ± 2.4 (4.2–12.3) |
– | |||
| Myoinositol | HMDB0000211 | * | 22.4 | 27.2 | ||||
| Ornithine | HMDB0000214 | * | * | *d |
78.7 ± 17.9 (49.2–113.7) |
27.2 | ||
| NADPH | HMDB0000221 | * | – | – | ||||
| Pyruvate | HMDB0000243 | * | *d,e |
79.8 ± 32.5 (1.0–1847.8) |
87.9 ± 24.4 (34.1–232.0) | |||
| Succinate | HMDB0000254 | * | * | *d |
3.6 ± 1.5 (0.0–5.4) |
4.8 | ||
| Sucrose | HMDB0000258 | * | – | – | ||||
| Phosphoenolpyruvate | HMDB0000263 | * | – | – | ||||
| Pyroglutamate | HMDB0000267 | * | – | – | ||||
| Sarcosine | HMDB0000271 | * | * | *d |
1.9 ± 1.0 (0.7–4.1) |
– | ||
| Trimethylamine-N-oxide | HMDB0000925 | *d | ||||||
| Uridine diphosphate-glucose | HMDB0000286 | * | – | – | ||||
| Uridine monophosphate | HMDB0000288 | * | – | – | ||||
| Uridine diphosphate–N–acetylglucose | HMDB0000290 | * | – | – | ||||
| Xanthine | HMDB0000292 | * | – | – | ||||
| Urea | HMDB0000294 | * | 1716.9 ± 556.8 (944.7–2776.9) | – | ||||
| Uridine | HMDB0000296 | * | – | – | ||||
| 2–Hydroxyisovalerate | HMDB0000407 | * | * |
4.7 ± 2.4 (2.8–11.1) |
– | |||
| 2-Aminobutyrate | HMDB0000452 | * | * | *d |
19.7 ± 6.7 (5.3–27.4) |
– | ||
| Allantoin | HMDB0000462 | * | – | – | ||||
| 3-Methylhistidine | HMDB0000479 | * | – | – | ||||
| 3-Methyl-2-oxovalerate | HMDB0000491 | * | * |
2.6 ± 1.9 (0.0–5.2) |
16.4 | |||
| Arginine | HMDB0000517 | * | * |
78.1 ± 22.5 (29.6–121.3) |
– | |||
| N-Acetylglycine | HMDB0000532 | * | – | – | ||||
| ATP | HMDB0000538 | * | – | – | ||||
| Creatinine | HMDB0000562 | * | * | *e |
67.4 ± 15.5 (15.1–940.8) |
71.4 ± 18.3 (26.4–438.0) | ||
| Glutamine | HMDB0000641 | * | * | *d,e |
554.4 ± 85.3 (84.4–1881.8) |
469.2 ± 67.7 (96.5–781.0) | ||
| Leucine | HMDB0000687 | * | * | *d,e |
103.8 ± 28.9 (13.7–447.7) |
77.5 ± 17.0 (32.1–260.0) | ||
| Malonate | HMDB0000691 | * |
7.8 ± 3.4 (2.9–12.6) |
– | ||||
| Ketoleucine | HMDB0000695 | * | * |
4.6 ± 1.2 (2.6–6.8) |
– | |||
| Methionine | HMDB0000696 | * | * | *d |
27.4 ± 3.5 (20.0–30.9) |
15 | ||
| Hippurate | HMDB0000714 | * | – | – | ||||
| Isovalerate | HMDB0000718 | * | – | – | ||||
| 3-Hydroxyisovalerate | HMDB0000754 | * | * |
1.5 ± 0.7 (0.0–2.8) |
– | |||
| Isopropanol | HMDB0000863 | * |
1.9 ± 1.2 (0.9–4.9) |
– | ||||
| Valine | HMDB0000883 | * | * | *d,e |
210.3 ± 43.5 (67.4–847.7) |
157.9 ± 27.6 (62.8–372.0) | ||
| NAD + | HMDB0000902 | * | – | – | ||||
| Tryptophan | HMDB0000929 | * | * |
2.8 ± 1.5 (0.3–5.1) |
27 | |||
| 2,3-diphosphoglycerate | HMDB0001294 | * | – | – | ||||
| ADP | HMDB0001341 | * | – | – | ||||
| Guanosine monophosphate | HMDB0001397 | * | – | – | ||||
| Nicotinamide | HMDB0001406 | * | – | – | ||||
| NADH | HMDB0001487 | * | – | – | ||||
| Phosphocholine | HMDB0001565 | * | – | – | ||||
| Acetone | HMDB0001659 | * | *d,e |
14.2 ± 5.6 (2.3–421.2) |
– | |||
| Benzoate | HMDB0001870 | * | – | – | ||||
| Isobutyrate | HMDB0001873 | * | * |
7.1 ± 2.8 (3.7–11.4) |
5.7 | |||
| Methanol | HMDB0001875 | * |
39.4 ± 17.6 (21.8–62.9) |
161 | ||||
| Propylene glycol | HMDB0001881 | * | * |
57.9 ± 24.0 (22.5–97.7) |
– | |||
| Glutathione disulfide | HMDB0003337 | * | – | – | ||||
| α-D-Glucose-1,6-biphosphate | HMDB0003514 | * | – | – | ||||
| Dimethyl sulfone | HMDB0004983 | * | *d |
10.1 ± 5.5 (4.7–19.5) |
– | |||
| Acetoacetate | HMDB0304256 | * | *d,e |
13.2 ± 12.5 (0.0–758.2) |
90.1 ± 62.7 (0.0–860.0) |
|||
| NADP + | HMDB0304435 | * | – | – | ||||
| Glycoprotein acetylation (GlycA) | NA | *e |
812.3 ± 120.3 (274.8–1884.7) |
1332.8 ± 259.9 (717.0–4920.0) | ||||
| Chylomicrons/VLDL (6 subclasses) | NA | *f,g |
0.1 ± 0.0 (0.0–0.6) |
0.1 ± 0.0 (0.0–0.5) |
||||
| LDL (3 subclasses) | NA | *f,g |
1.2 ± 0.3 (0.1–3.5) |
0.5 ± 0.1 (0.0–1.2) |
||||
| IDL | NA | *f,g |
0.3 ± 0.1 (0.1–0.9) |
0.1 ± 0.0 (0.0–0.3) |
||||
| HDL (4 subclasses) | NA | *f,g |
15.3 ± 2.5 (2.4–36.7) |
7.8 ± 0.8 (0.0–14.5) |
||||
| Apolipoprotein A1 | NA | *h |
52.1 ± 8.8 (10.8–123.5) |
49.7 ± 5.8 (29.1–74.7) |
||||
| Apolipoprotein B | NA | *h |
1.6 ± 0.4 (0.3–4.8) |
1.9 ± 0.4 (0.7–5.4) |
||||
| Free cholesterol | HMDB0000067 | * | 1266.7 ± 271.9 (230.1–3430.8) | 1346.2 ± 267.0 (477.0–3030.0) | ||||
| Cholesterol esters | NA | * |
3368.2 ± 685.4 (512.8–8405.8) |
3165.0 ± 669.3 (791.0–6040.0) | ||||
| Average fatty acid chain length | NA | * | – |
17.5 ± 0.3 (16.5–19.0) |
||||
| Average fatty acid saturation degree | NA | * |
1.4 ± 0.1 (0.9–3.0) |
1.2 ± 0.1 (0.9–1.5) |
||||
| Omega-3 fatty acids | NA | * |
532.9 ± 223.1 (1.0–4156.4) |
449.9 ± 152.5 (145.0–2070.0) | ||||
| Omega-6 fatty acids | NA | * |
4512.4 ± 692.4 (1005.3–16,411.0) |
3645.4 ± 730.1 (1060.0–10,800.0) | ||||
| Lineolic acid | HMDB0000673 | * |
3463.5 ± 694.7 (4.9–16,411.0) |
2980.6 ± 657.7 (792.0–9810.0) | ||||
| Docosahexaenoic acid | HMDB0002183 | * |
237.1 ± 84.2 (0.3–1789.8) |
157.2 ± 59.1 (38.1–678.0) |
||||
| Triglycerides | NA | * |
1321.0 ± 588.8 (179.2–8401.5) |
1568.9 ± 749.5 (421.0–9450.0) | ||||
| Phosphatidylcholines | NA | * |
2110.0 ± 381.1 (0.0–5697.4) |
1780.8 ± 311.5 (687.0–3440.0) | ||||
| Phosphoglycerides | NA | * |
2296.4 ± 405.0 (109.4–6112.4) |
1781.7 ± 328.9 (625.0–3540.0) | ||||
| Sphingomyelins | NA | * |
451.9 ± 73.3 (115.2–979.2) |
435.6 ± 81.8 (162.0–822.0) |
||||
| Albumin | NA | * |
587.7 ± 50.7 (0.0–1074.8) |
– | ||||
| Monounsaturated Fatty Acids | NA | * |
2904.2 ± 837.3 (0.0–13,077.0) |
3033.4 ± 904.6 (1110.0–15,000.0) | ||||
| Polyunsaturated Fatty Acids | NA | * |
5045.3 ± 815.0 (1006.3–19,399.0) |
4095.0 ± 811.6 (1210.0–11,700.0) | ||||
| Saturated Fatty Acids | NA | * |
4128.0 ± 969.5 (0.0–17,514.0) |
4175.5 ± 921.4 (1620.0–14,600.0) |
a—listing only uniquely identified compounds. b—mean values, standard deviations, and ranges, when available. c—references: (Madrid-Gambin et al., 2023; Nagana Gowda & Raftery, 2014; Rout et al., 2023); https://biobank.ndph.ox.ac.uk/ukb/label.cgi?id=220; https://view.officeapps.live.com/op/view.aspx?src=https%3A%2F%2Fwww.ucl.ac.uk%2Fepidemiology-health-care%2Fsites%2Fepidemiology_health_care%2Ffiles%2Fbrhs_1998-2000_q20_20_year_follow-up_metabolites_data_dictionary_v1.xlsx&wdOrigin=BROWSELINK. d—metabolites quantified by B.I. Quant-PS (Trautwein, 2025). e—using CPMG pulse sequence. f—methods include using lineshape fitting (Jeyarajah et al., 2006), statistical methods/machine learning (Ala-Korpela et al., 1995) or diffusion spectroscopy (Mallol et al., 2015). g—number and naming of subclasses may differ between tests (Balling et al., 2020; Rief et al., 2022). h—measured using partial least squared (PLS) or neural network analysis (Bathen et al., 2000)
As can be seen from this table, the number of metabolites identifiable by NMR in deproteinized/delipidated serum and plasma as prepared via ultrafiltration is identical for both biofluids and includes typically 57–58 compounds (Bahado-Singh et al., 2018; Mandal et al., 2025; Rout et al., 2023). The number of metabolites identifiable by NMR in deproteinized/delipidated serum and plasma prepared by methanol extraction/lyophilization is also identical and includes typically 67–71 compounds (Nagana Gowda & Raftery, 2019; Nagana Gowda et al., 2015). The compounds that are most different between samples prepared via the methanol vs. ultrafiltration methods include acetone, dimethylamine, ethanol, hippurate, isopropanol, methanol, sucrose and urea (Table 1). These differences are primarily due to the volatility of some compounds (which are lost due to lyophilization) as well as the concentration effects enabled by methanol extraction. Furthermore, compounds showing strong protein binding can only be assessed following methanol extraction. On the other hand, we would like to point out that, due to the necessary drying step, methanol extraction is not suitable for the analysis of volatile compounds, and traces of exogenous methanol, even after careful drying, will be found in the corresponding NMR spectra. Interestingly, the number of compounds identifiable by NMR in deproteinized/delipidated whole blood, as prepared via methanol extraction/lyophilization is ~ 90 (Nagana Gowda et al., 2022). This is nearly 30% more than seen with plasma or serum prepared in the same manner. The reason for this difference is due to the release of red blood cell contents (which include many cofactors and redox molecules) into the medium during the blood sample work-up (Nagana Gowda et al., 2022).
In contrast to the deproteinized/delipidated samples, when “intact” plasma is analyzed by NMR, the number of metabolites and lipoprotein components that can be identified can be as high as 168, including 150 lipid, protein and lipoprotein components and 18 small molecules when employing the Nightingale assay (Julkunen et al., 2023; Zeleznik et al., 2023), and up to a total of 112 different lipoprotein subclasses and 38 individual metabolites when employing the Bruker IVDr system (Jiménez et al., 2018; Trautwein, 2025). Note that the number of small molecules identifiable in intact plasma is significantly lower than what is achievable in deproteinized/delipidated plasma. This is primarily due to the extensive overlap of lipid or lipoprotein signals, leading to the loss of identifiable peaks associated with small molecule metabolites. On the other hand, the ability to identify many lipoprotein or cholesterol components including high-density lipoprotein (HDL), low-density lipoprotein (LDL) and very low-density lipoprotein (VLDL) fractions in intact plasma provides a very informative picture of many key cardiovascular disease markers (Patrizia Bernini et al., 2011a, 2011b; Khakimov et al., 2022).
The fact that different components of blood (i.e., whole blood, serum or plasma), prepared or processed in different ways and analyzed with different analytical techniques, can lead to different chemical compositions is important to remember. Likewise, this list of compounds is useful to recall when discussing the pre-analytical or environmental factors (such as diet, microbiome, collection or storage conditions) that influence NMR-measurable metabolite levels in human plasma, serum or whole blood.
As Table 1 shows, if the sample preparation method is identical, the chemical composition of serum and plasma are essentially identical. However, the concentrations of specific metabolites will certainly differ between plasma and serum. This is evident in the concentration ranges reported for ultracentrifuged serum/plasma as measured by NMR (Table 1). This NMR-derived information agrees with the study by Liu et al. (L. Liu et al., 2010) who used GC–MS to investigate the metabolome differences between serum and plasma. They measured the concentration levels of 72 metabolites in human plasma and compared them to the corresponding serum samples (L. Liu et al., 2010). They found that 29 metabolites were at higher concentrations in serum than plasma, while 7 metabolites in plasma were at higher concentrations than serum. Of these, only 24 metabolites are relevant to NMR studies. Of the NMR-relevant metabolites, most amino acids were at higher levels in serum, while citrate and pyruvate were more elevated in plasma samples (L. Liu et al., 2010). Similar results were also found in another publication that used LC–MS to quantify metabolites in serum and plasma, and higher levels of several amino acids such as glycine, serine, arginine and phenylalanine were reported in serum samples compared to plasma (Yu et al., 2011).
In addition to these modest metabolite concentration differences, there are other differences between serum and plasma that are worth noting. The incubation time effect on metabolite concentration levels has generally been found to be more dominant in plasma when compared to serum samples (Ferreira et al., 2019; L. Liu et al., 2010). That is, the longer that plasma is left unfrozen, the more likely metabolite levels will change due to enzyme-mediated events. Additionally, the effect of freeze–thaw cycles on metabolite levels has also been found to be more prominent in plasma than serum (Saito et al., 2014). Likewise, the presence of additional signals caused by anticoagulants in plasma can reduce the number of compounds identified by NMR (Barton et al., 2009; Kennedy et al., 2021). On the other hand, the use of EDTA as an anticoagulant can facilitate the identification and absolute quantification of bivalent cations such as Mg2+ and Ca2+, otherwise invisible in NMR experiments, which form EDTA-complexes with distinct NMR signal shifts (Barton et al., 2009; Somashekar et al., 2006).
Patient-specific factors influencing metabolite levels in human blood, plasma and serum
In this section, we will focus on participant or patient-specific effects that can significantly influence NMR-detectable metabolite levels in blood, plasma and serum. Researchers need to be mindful of these effects, especially in studies focusing on metabolite-based biomarker identification. Here, we briefly review the influence of different “intrinsic” confounders on serum and plasma metabolite composition and possible ways to compensate or minimize these perturbations.
Effects of food intake
As the old adage goes: “we are what we eat”. In other words, the metabolites found in the human metabolome, including the blood, serum or plasma metabolome, are primarily derived from metabolites in our diet. While the human body itself can produce thousands of metabolites, many other compounds, such as essential amino acids, vitamins, and essential fatty acids, must come from external dietary sources. Likewise, many other food-derived xenobiotics such as polyphenols, phytosterols and alkaloids, as well as a variety of food additives, food preservatives or food colorants, are digested and transformed into a host of other compounds that can also become part of the blood, serum or plasma metabolome. Furthermore, the presence of some food components or food-derived chemicals can have significant downstream effects that change the metabolism, the metabolic pathways or abundance of seemingly unrelated metabolites.
There are numerous published NMR metabolomic studies that have explored these food or diet-related impacts on the blood, serum or plasma metabolome. One study described by Hefni et al. in 2018 investigated the effect of folate supplements on plasma metabolite levels by NMR spectroscopy (Hefni et al., 2018). These authors observed an increase in six different metabolites: glycine, choline, betaine, formate, histidine, and threonine, all of which are known to be affected by folate mediated one-carbon metabolism. This result suggests that folate supplementation should be monitored, or at least noted, in plasma/serum metabolomic studies. Another study conducted in 2013 by Gregory et al. (Gregory et al., 2013) explored the effect of vitamin B6 restriction on plasma samples for 23 different participants. They found the ratios of glutamine/glutamate and 2-oxoglutarate/glutamate increased. This is consistent with the role that vitamin B6 plays in transaminase activity. They also noted an increase in the plasma concentration of acetate, pyruvate (reflecting the role of vitamin B6 in glucose metabolism), and trimethylamine-N-oxide. Given that vitamin B6 deficiencies can range from 13 to 50% of the population (Ho et al., 2016; Kjeldby et al., 2013) (depending on the age of the population), these effects on the serum or plasma metabolome need to be taken seriously.
Habitual diets also influence the human serum/plasma metabolome in significant ways. Fotiou et al. in 2018 explored the effects of habitual dietary patterns on the chemical composition of serum (as well as amniotic fluid, serum and urine) in pregnant women in their second trimester (Fotiou et al., 2018). The authors found that women who consumed a less healthy diet consisting of refined cereals, yellow cheese, red meat, poultry, and ready-to-eat foods had moderately higher serum levels of glucose, alanine, tyrosine, valine, citrate, cis-aconitate, and formate compared to women who ate a healthier diet consisting of whole grain cereals, vegetables, fruits, legumes, and nuts. Whether these metabolite levels reflect the habitual diet per se, or the metabolic consequence of the habitual diet, is still to be determined.
Similar effects on the serum or plasma metabolome have also been observed with other kinds of habitual or modified habitual diets. In 2011, Moazzami et al. (Moazzami et al., 2011) studied the effect of a diet rich in whole grain rye products on plasma metabolites of prostate cancer patients. These patients were given a diet rich in whole grain (WG) rye and rye bran products (RP) or refined white wheat products (WP) for six weeks followed by a two-week washout period. At the end of the intervention period, patient plasma samples were collected and studied by 1H NMR. A modest increase in plasma levels of 3-hydroxy butyric acid, acetone, betaine, N,N-dimethylglycine, and dimethyl sulfone after RP intake was observed (Moazzami et al., 2011). Elevated levels of betaine and N,N-dimethylglycine could be attributed to the higher levels of these two compounds in WG foods. On the other hand, changes in 3-hydroxybutyric acid and acetone (ketone bodies) appear to reflect long-term, endogenous metabolic responses in the subjects that were induced by the WG diet.
Habitual consumption of meat or an omnivorous diet versus a vegetarian or vegan diet can also lead to NMR-detectable serum metabolome differences. A study by Lindqvist et al. in 2019 (Lindqvist et al., 2019) involving 40 omnivores, 37 vegetarian and 43 vegan dieters demonstrated that all three groups could be distinguished by their 1H NMR serum metabolomic profiles. Serum levels of branched-chain amino acids, creatine, creatinine, 3-hydroxyisobutyrate, lysine and 2-aminobutyrate (tentative ID) were all elevated in the meat eaters/omnivores while glutamine, glycine and trimethylamine were elevated in the vegans and vegetarians.
The Mediterranean diet (MD), because of its clear health benefits, is becoming increasingly popular (Vázquez-Fresno et al., 2015). As a result, the effects of a habitual MD on the plasma metabolome of 58 subjects were studied by Macias et al. in 2019 using 1H NMR-based metabolomics (Macias et al., 2019). Based on an established MD adherence scoring scheme, subjects were classified into two groups (“low” and “high”). This study found that those who scored high on the MD scale had moderately-elevated plasma levels of citric acid, betaine and acetic acid, but lower levels of pyruvate, mannose and myo-inositol relative to those who scored low on the MD scale (Macias et al., 2019). Citrate was found to be an excellent marker of habitual fruit intake. As with other diet-metabolome studies, certain metabolite changes could be attributed directly to food components while others appeared to arise from general shifts in the subject’s metabolism.
While habitual food intake or habitual diets can modestly change an individual’s serum or plasma metabolome, so too can acute consumption of certain foods. An acute intervention study conducted by Radjursoga et al. in 2019 (Rådjursöga et al., 2019) investigated the serum metabolic responses to two different kinds of breakfast: a ham and egg breakfast, and a cereal breakfast. The postprandial serum samples (n = 24) were subjected to 1H NMR-based metabolomics and it was found that concentrations of proline, tyrosine, and N-acetylated amino acids increased after consumption of the cereal breakfast, while concentrations of creatine, methanol and isoleucine were increased after the ham and egg breakfast (Rådjursöga et al., 2019).
A similar acute intervention study by Trimigno et al., 2018 (Trimigno et al., 2018) looked at the serum from 11 healthy individuals who consumed milk, cheese, or a soy drink. Using 1H NMR-based metabolomics, they found that levels of tyrosine, isoleucine, valine and 3-hydroxyisobutyrate were elevated in the serum of cheese consumers relative to milk and soy drink consumers (Trimigno et al., 2018). While the effects are relatively modest, these examples show that serum/plasma metabolite concentration levels can be modified by both habitual and acute food intake.
Effects of extended fasting
Periods of fasting have been, and still are, very common in human culture. Fasting has been followed by Muslims, Christians, Jews, Buddhists, and others as religious practice for centuries. For example, during the Ramadan, intermittent fasting is practiced with a daytime (12–14 h) cessation of food consumption for nearly a month (Ang et al., 2012; Townsend et al., 2016). Fasting affects energy production of the human body and will trigger catabolic and anabolic reactions. Glucose and its short-term storage form glycogen comprise the major energy sources of the body. During fasting, glycogen stores are rapidly exhausted forcing the body to activate gluconeogenesis. For fasting subjects, it was shown by Rothman et al. using 13C NMR that gluconeogenesis accounts for a substantial fraction of total blood glucose during the first 22 h of a fast (Rothman et al., 1991). Likewise, Teruya et al. showed that during fasting, various non-carbohydrate metabolites such as lipids and branched chain amino acids (BCAAs) (Teruya et al., 2019) are used as additional energy sources leading to a substantial increase in blood ketone bodies such as β-hydroxybutyrate and acetoacetate (Nicholson et al., 1984). As a result, samples collected from fasted individuals will tend to have higher levels of ketone bodies along with lower levels of glucose, lipids and BCAAs than those samples collected from individuals who have just consumed a meal (Bermingham et al., 2023; Shrestha et al., 2017).
The preference for fasted samples for plasma or serum in clinical studies originally arose from a study conducted by Cohn et al. nearly 40 years ago (Cohn et al., 1988) which showed that the lipoprotein cholesterol concentration measured in plasma sampled from probands fed a high-fat diet differed significantly from those measured in the fasted control group. As a result, most plasma samples being analyzed for lipids, lipoproteins and apolipoproteins—whether by NMR or by other means—are usually measured in the fasting state (De Backer et al., 2003; Remaley et al., 2006). Likewise, for the analysis of type 2 diabetes mellitus and diabetic microangiopathy, it is also recommended that plasma and serum samples should be obtained after an at least 8 h fast to ensure stable fasting blood glucose levels (Lin et al., 2019). Therefore, for the analysis of metabolites, lipids and lipoproteins, it is generally recommended to collect human blood samples after an overnight fast.
Effects of age and gender
The impact of age and sex on plasma/serum composition is well established (Pinto et al., 2014). Age-related changes include alterations in the levels of amino acids that are involved in muscle mass and protein synthesis as well as metabolites associated with inflammation, oxidative stress, energy metabolism and lipid metabolism. For example, van den Akker et al. demonstrated that it is possible to use NMR-derived metabolite concentrations to predict an individual’s chronological age with a median error of 7.3 years (van den Akker et al., 2020). Specifically, they used concentrations of 52 metabolites to develop a linear model that correlated with chronological age with a Pearson correlation coefficient of 0.64. The metabolites that correlated negatively with chronological age included histidine, leucine, isoleucine, alanine, linoleic acid, docosahexaenoic acid, phosphatidylcholine and total choline. Conversely, the metabolites that correlated positively with chronological age included glutamine, citrate, phenylalanine, creatinine, tyrosine, lactate, valine, acetate and glucose.
In agreement with this study, Lau et al. (Lau et al., 2023) reported an age-related decrease in the concentration of the amino acids leucine and histidine (as well as the protein albumin, small HDL phospholipids, and VLDL cholesterol). The authors also observed age-related increases in levels of tyrosine, phenylalanine, glutamine, creatinine, citrate, glucose, omega-3 fatty acids, triglycerides, and the degree of unsaturation of fatty acids. In a separate NMR-based metabolomics study, Castro and co-authors (Castro et al., 2022) found that concentrations of leucine, isoleucine, asparagine, threonine, and 3-hydroxybutyrate correlate negatively, while aspartate, succinate, valine, dimethylsulfone and ornithine correlate positively with age in a statistically significant manner. In other words, a significant number of blood, serum or plasma metabolites will either decrease or increase with age. Therefore, if the confounding effects of age are unaccounted for in disease-related metabolomics studies, the variation due to age could be misinterpreted as being disease related or could hide the presence of other more subtle changes in truly meaningful biomarkers.
Similar to age differences, biological sex differences have a significant impact on metabolite concentrations measured by NMR. The sex-specific differences in metabolite levels originate from well-known differences in hormone levels, body composition, and metabolic processes between males and females. For instance, estrogen and testosterone have been shown to modulate lipid metabolism, which can be detected as variations in lipid-related metabolites in serum/plasma samples analyzed by the Nightingale or B.I. LISA NMR assays. Additionally, sex-specific differences in muscle mass and fat distribution can influence concentrations of metabolites, particularly those that are related to energy metabolism and amino acid turnover. For example, Barba et al. (Barba et al., 2019) identified and quantified 19 serum metabolites from methanol deproteinized serum samples and found that lactate, glucose, valine and glycine were notably elevated in women, compared to men. The study involved 39 healthy individuals (22 males and 17 females) aged between 55 and 70 years, who underwent stress tests and were considered negative for any sign of coronary artery disease.
A significant sex-related difference in metabolite concentrations was also found by Bell et al. in a study that looked at the sex differences in metabolites across four life stages (Bell et al., 2021). This study used data from the Avon Longitudinal Study of Parents and Children, which involved the analysis of 7727 offsprings (49% male/51% female) and 6500 parents. This study examined over 200 serum or plasma components (metabolites, lipids, proteins and lipoproteins) quantified through targeted NMR metabolomics using the Nightingale assay. The researchers found that at age 8, males had lower levels of total lipids in VLDL compared to females. As males aged, VLDL levels increased, particularly in medium-or-larger subclasses of VLDL triglycerides. By 18 years, males had significantly higher levels of VLDL triglycerides, a trend that continued into early adulthood (25 years) and middle adulthood (50 years). While males showed increasing levels of VLDL triglycerides with age, females had generally higher levels of LDL cholesterol, apolipoprotein B, and acetylated glycoproteins across all ages.
In agreement with this study, Ellul et al. (Ellul et al., 2020), who also used the Nightingale NMR assay, reported higher average serum levels of lipids in small and very-small VLDL particles in their study of sex differences in serum metabolites of infants (Ellul et al., 2020). The Ellul et al. study was part of the Barwon Infant Study and involved a population of 485 infants. The authors found that several serum cholesterol measures (i.e., total serum, and remnant, esterified and free cholesterol) were higher in girls. Likewise, serum docosahexaenoic acid, omega-3, omega-6, and monounsaturated fatty acids were higher in female infants. Other serum metabolites that were elevated in girls included intermediate density lipoproteins, LDL-related measures, inflammation marker acetylated glycoproteins, total serum albumin, glycerol and glycine.
A higher plasma glycine concentration in female participants was also found in the Karlsruhe Metabolomics and Nutrition (KarMeN) study conducted by Rist and colleagues (Rist et al., 2017). This cross-sectional study included 301 healthy men and women aged 18–80 years. The KarMeN study aimed to investigate how these sex- and age-related physiological conditions such as body composition and physical fitness are reflected in the metabolome and whether sex and age can be predicted based on plasma and urine metabolite profiles. This study employed several experimental techniques, including 1H NMR-based metabolomics. Overall, it was discovered that men had higher plasma concentrations of creatinine, leucine and valine, while women had higher plasma levels of creatine and glycine. Overall, these data indicate a significant number of blood, serum or plasma metabolites that are affected by sex. Therefore, if sex effects or their confounding influence are not properly accounted for in disease-related NMR metabolomic studies, one’s ability to discover important metabolite markers, pathways or trends will be compromised.
Effects of physical activity
The involvement in exercise and sports has long been known to cause substantial changes in human physiology and in the human metabolism (Jaguri et al., 2023). Exercise typically brings about an immediate metabolic response that influences the rate of synthesis or generation of many different metabolites that are linked to aerobic and anaerobic glucose metabolism, lipid and amino acid metabolism, muscle damage and liver function (Coelho et al., 2016).
1H NMR metabolomics studies, such as those by Brugnara et al. (Brugnara et al., 2012), have shown that immediately after an intense workout, an increase in serum tricarboxylic acid (TCA) cycle intermediates (succinate, citrate, and glycerol) and products of anaerobic glycolysis (lactate and pyruvate) are often observed, reflecting the increased energy demands during exercise (Brugnara et al., 2012). Gluconeogenic precursors, which includes alanine and lactate, also increased in serum post-workout (Brugnara et al., 2012). 1H NMR metabolomics studies have also shown that after exercise of longer durations, such as running long distances (i.e., a marathon), several additional changes in energy metabolism occur as the body responds to this massive energy demand. Bester et al. (Bester et al., 2021) showed that for serum lactate and pyruvate, post-marathon levels doubled compared to pre-marathon levels, suggesting increased anaerobic glycolysis. Serum amino acid levels (lysine, proline, leucine, isoleucine, valine) decreased post-marathon suggesting that amino acids were catabolized for energy generation. An increase in ketone bodies (3-hydroxybutyric acid, acetone, acetoacetic acid) was also seen in post-marathon serum which is strongly indicative of upregulated lipid catabolism. Increased serum creatine and creatinine were also observed, suggesting that muscle damage may be occurring (Bester et al., 2021). Similar changes in serum TCA and anaerobic glycolysis metabolites were also seen by Pechlivanis et al., who studied effects of sprint interval training, suggesting that these are global changes seen with exercise (Pechlivanis et al., 2012).
Additional changes to the serum/plasma metabolome have been detected via NMR-based metabolomics over the long term with regular exercise. Overall, changes of metabolites related to protein and fatty acid metabolic pathways are typically observed (Castro et al., 2023; Pechlivanis et al., 2012; Sardeli et al., 2022). These studies found that serum lipid levels are decreased, while other changes to serum metabolites related to fatty acid metabolism, such as 3-hydroxybutyrate and acetoacetate, are also observed. With moderate and high-level inspiratory muscle training, Castro et al. (Castro et al., 2023) observed higher serum levels of proline and methionine, suggesting upregulation of amino acid metabolism. The above 1H NMR study, focussed on sprint interval training by Pechlivanis et al. (Pechlivanis et al., 2012), also detected a decrease in serum glycoprotein acetylation, which has been previously identified as a biomarker for inflammation and cardiovascular disease (Otvos et al., 2015). Pechlivanis et al. also noted increased serum methylguanidine with increased training (Pechlivanis et al., 2012). Methylguanidine, derived from protein catabolism, has been reported as a biomarker of oxidative stress (Ienaga et al., 2007) and exhibits anti-inflammatory activity (Marzocco et al., 2004). Collectively, these results point to the need to be aware of the effects of both acute and long-term exercise on serum/plasma metabolomes and that mitigation of exercise effects or providing guidance about when blood should be collected after exercise is important in the design of 1H NMR metabolomics studies.
Effects of obesity
Several studies have reported a significant association of body-mass index (BMI) and obesity measures, such as waist–hip ratio (WHR) and android/gynoid fat ratio (AGR) with changes in serum metabolites. For example, in a 2019 paper, Wulaningsih and colleagues used 1H NMR and the Nightingale assay to investigate the relationship between adiposity measures (from childhood through adulthood), BMI and serum metabolites for 900 British individuals (Wulaningsih et al., 2019). They quantified 233 serum metabolites, lipids, and lipoproteins and found significant associations between BMI, WHR, AGR, and numerous serum metabolite concentrations and features. Specifically, 168, 126, and 133 compounds were associated with BMI, WHR, and AGR at age 60–64, respectively. The study found strong associations for HDL, particularly HDL particle size, with a notable decrease in HDL diameter with each unit increase in BMI. The study also identified inverse associations between BMI at age 7 and levels of glucose and glycoprotein at age 60–64, suggesting early-life BMI influences on metabolic profiles in later life.
In a similar study, Saner et al. (Saner et al., 2019) applied the Nightingale NMR assay to investigate the associations between adiposity measures and serum metabolomic profiles in youth with obesity. This study utilized the Childhood Overweight BioRepository of Australia (COBRA) cohort and looked at 214 participants. Positive associations were found between BMI z-score and serum phenylalanine and tyrosine levels, as well as in medium HDL levels. Negative associations with BMI were detected with the ratio of serum docosahexaenoic acid/total fatty acids and histidine.
Discovering the association between metabolite concentrations and BMI as well as modifiable life factors (smoking status, alcohol consumption, physical activity, diet, etc.) was also the subject of a study by Hamaya et al. (Hamaya et al., 2022). These authors focused on measuring plasma BCAAs (isoleucine, leucine, and valine) by 1H NMR (using the LipoScience/LabCorp assay) because of their known association with an increased risk of type 2 diabetes. The study included a cross-sectional analysis among 18,897 women from the Women's Health Study to identify modifiable lifestyle factors that influence BCAA concentrations. BMI was found to be significantly associated with BCAA levels. Compared to women with a BMI of less than 25.0, plasma BCAAs were 8.6%, 15.3%, and 21.0% higher for women with a BMI of 25.0–29.9, 30.0–39.9, and ≥ 40.0, respectively. Diet, physical activity, and other lifestyle factors had a smaller impact.
This connection between BMI and serum BCAA concentrations appears to be in agreement with the results of the Shanghai Changfeng Study, published in 2021 by Wu et al. (Wu et al., 2021). This study, which looked at 1078 individuals, was aimed at identifying serum metabolites that could indicate future development of metabolic disorders in individuals who are initially metabolically healthy. Participants were categorized based on their BMI and metabolic health into metabolically-healthy overweight/obese and metabolically-healthy normal weight groups. Their serum metabolic profiles were analyzed using 1H NMR spectroscopy via the B.I. LISA assay (which detected 112 lipids and lipoproteins) and the B.I.Quant-PS system (which detected 38 small-molecule metabolites). This study found that, after a follow-up of 4 years, a higher proportion of healthy overweight participants transitioned to a metabolically unhealthy status compared to normal-weight healthy participants. It was also found that healthy overweight individuals had higher concentrations of BCAAs, alanine and tyrosine as well as lower concentrations of HDL components and glycine when compared with normal-weight individuals. Overall, it is clear that obesity or higher BMI has a significant influence on NMR measurable metabolites. Therefore, if obesity or BMI-related metabolite effects are not included in NMR-based metabolomic studies, one’s ability to discover important metabolite markers, pathways or trends may be compromised.
Effects of smoking
While smoking or smoke-exposure compounds such as cotinine are detectable in certain biofluids via GC–MS and LC–MS studies (Lehtovirta et al., 2023; Thomas et al., 2020), no smoking or smoke-derived compounds have yet been detected by NMR of serum or plasma. However, because smoking (like obesity) can lead to negative, long-term health effects, the effects of smoking are typically detected through secondary effects on metabolism or general health. One study by Lehtovirta et al. used the Nightingale NMR assay on serum to assess the “smoker’s metabolome” (Lehtovirta et al., 2023). This study found that serum lipid levels, particularly unsaturated fatty acids, triglycerides, and VLDL particle concentrations and size were increased in smokers. Interestingly, these biomarkers are also associated with increased risk of cardiovascular disease. Another NMR study by Bernini et al. (Patrizia Bernini et al., 2011a, 2011b) found that changes to metabolites related to fatty acid metabolism (such as 3-hydroxybutyrate, α-ketoglutarate, threonine, and dimethylglycine) can be correlated with blood lipid levels. Other NMR studies appear to show weaker correlations between the serum metabolome and smoking. However, effects were detected including reduced serum amino acid concentrations (Labaki et al., 2019), increased serum tryptophan levels (Aguilar et al., 2021), decreased BAAAs in plasma (Hamaya et al., 2022; Jang et al., 2022), and decreased levels of plasma citrate and glycerol (Jang et al., 2022). Collectively, these data indicate that smoking does influence the NMR measurable serum/plasma metabolome and that these confounding effects should be modeled into any cohort measured via NMR-based metabolomics.
Effects of alcohol
Alcohol consumption has a strong and multifaceted effect on NMR-detectable metabolites in serum and plasma. These effects depend on the subject’s age, sex, genetics, physiology and the amount of consumed alcohol. A large study by Würtz et al. (Würtz et al., 2016) applied an early version of the Nightingale NMR assay to investigate the associations between alcohol intake and 86 serum metabolites, lipids and lipoproteins in a cross-sectional study from three population-based cohorts from Finland. In addition, these authors also examined the serum metabolic changes associated with changes in alcohol intake in 1,466 individuals during a 6-year follow-up. This study found that increased alcohol intake was associated with higher serum HDL levels and smaller LDL particle size, increased monounsaturated fatty acids, decreased omega-6 fatty acids, and lower serum concentrations of glutamine and citrate. Many serum metabolic biomarkers were found to have U-shaped associations with alcohol consumption, indicating differential effects depending on the quantity of alcohol intake.
The U-shaped dependence on alcohol consumption was also detected for several other serum metabolites (e.g., lipoprotein lipids in VLDL subclasses, VLDL triglycerides) using the more extensive Nightingale-based NMR assay as reported by Du and co-authors (Du et al., 2020). Alcohol intake was significantly associated with changes in 23 out of 37 lipids, 12 out of 16 fatty acids, and six out of 20 low-molecular-weight metabolites, independent of confounding factors. These effects were similar for total alcohol consumption and different types of alcohol (beer, wine, and spirits). Many metabolites, including those in several HDL subclasses, HDL cholesterol, apolipoprotein A-1, phosphotriglycerides, total fatty acids, monounsaturated fatty acids, and omega-3 fatty acids, had positive correlations with alcohol intake. Conversely, LDL particle size, omega-6 fatty acids ratio to total fatty acids, and citrate had negative linear associations with alcohol consumption. The findings indicate that alcohol intake can be a significant confounding variable that should be taken into consideration in NMR metabolomics studies.
Effects of gut microbiota
The gut microbiome (GM) is a complex ecosystem that comprises bacteria, protozoa, archaea, viruses, and fungi, which symbiotically interact with each other and their human host and the food that their host consumes. The gut microbiome plays important physiological roles, participating in digestion, immunomodulation, cardiovascular health, gut health, and a number of different pathological disorders (Amedei & Morbidelli, 2019).
The metabolites generated by gut microbes can easily pass into the circulatory system, including amino acids, lipids, sugars, biogenic amines, organic acids, peptides, glycolipids, oligosaccharides, terpenoids or secondary bile products, and volatile small molecules. These compounds can influence both the health and the metabolic state of the host (Amedei & Morbidelli, 2019). Gut microbes can chemically cleave dietary compounds containing tertiary amines such as choline, phosphatidylcholine, glycerophosphocholine, carnitine and betaine leading to the release of trimethylamine (TMA). These can be further oxidized by liver flavin monooxygenase (FMO) to produce trimethylamine-N-oxide (TMAO) (Fan et al., 2015; Koeth et al., 2013). TMAO is a known uremic toxin and has been implicated in the formation of atherosclerotic plaques. Short-chain fatty acids or SCFAs (such as acetic acid, propionic acid, butyric acid, and the less abundant, valeric acid and caproic acid) are synthesized by bacteria from the glycolysis of glucose to pyruvate, to acetyl-CoA, and finally to the SCFAs acetic acid, propionic acid, and butyric acid (Miller & Wolin, 1996). Unlike TMAO, SCFAs are widely recognized to have beneficial effects (Abdul Rahim et al., 2019).
The diversity and composition of the gut microbiome can influence the metabolites found in serum. An NMR study by Org et al. (Org et al., 2017) found associations between gut microbes and the fasting serum levels of several metabolites, including fatty acids, amino acids, lipids, and glucose (Org et al., 2017). They found a strong association of microbial diversity with serum levels of, glutamine, glycated haemoglobin and acetate levels. Partula et al. (Partula et al., 2021) found that higher gut microbial diversity was correlated with higher concentrations of amino acids, glucose, and citrate, and with lower concentrations of metabolites involved in fatty acid metabolism.
Bacterial infections in the gastrointestinal tract can also influence the serum metabolome. Using 1H NMR, Fang et al. (Fang et al., 2020) found that serum TMAO and lactate levels were affected by Helicobacter pylori infection in the stomach, and that these compounds could be potential biomarkers for monitoring the course of antibiotic treatment. Kato et al. (Kato et al., 2018) used 1H NMR to investigate the effects of oral administration of Porphyromonas gingivalis on the gut microbiome and serum metabolome in mice. While the mechanism by which P. gingivalis alters the gut microbiome is not clear, these authors found that the gut microbiome composition was altered and serum amino acids, notably the aromatic amino acids, were also increased.
Interventions targeting the gastrointestinal tract or altering the gut microbiome can affect the serum metabolome. Gralka et al. (Gralka et al., 2015) used 1H NMR to explore the effects of bariatric surgery on the serum metabolome. They found that obese subjects, prior to surgery, had higher levels of methanol and isopropanol and that these levels dropped to healthy, normal levels after bariatric surgery. The fact that methanol and isopropanol are gut-derived metabolites suggests that bariatric surgery induced changes in their gut microbiota. Ghini et al. (2020) used 1H NMR to measure the effect of probiotics on gut microbiota and serum metabolome. They found that serum pyruvate, phenylalanine and proline levels were increased over the course of probiotic treatment in healthy subjects. Overall, these data clearly show that individual differences in gut microbiota as well as interventions, such as surgical procedures or dietary modifications that alter the gut microbiota, can significantly affect the serum or plasma metabolome. In this regard, the gut microbiome must be considered as a potential confounding factor in the interpretation of NMR-based metabolomic studies.
Effects of health status
An individual’s health status can significantly influence their NMR-detectable serum, plasma or whole blood metabolome. These health-status effects can be broadly distinguished according to whether the condition is acute or chronic. Acute diseases, such as viral infections, including influenza (Banoei et al., 2017), and SARS-CoV-2 (Bruzzone et al., 2020, 2023), sepsis (Mickiewicz et al., 2018) as well as acute lung diseases (Dasgupta et al., 2023) have been shown to cause broad alterations in the composition of NMR-detectable blood, serum and plasma metabolomes. Similar metabolome-wide effects have been shown for acute organ injuries, including acute damage to the liver (Amathieu et al., 2016) and kidney (Zacharias et al., 2015). Numerous chronic diseases have likewise been associated with significant changes in the NMR-detectable blood, serum and plasma metabolomes, including cardiovascular diseases (Ussher et al., 2016), diabetes (Roberts et al., 2014), chronic kidney disease (Schultheiss et al., 2021), chronic liver diseases (Amathieu et al., 2016), chronic lung diseases (Moitra et al., 2023), chronic gastrointestinal disorders (Bjerrum et al., 2021; Y. Zhang et al., 2013), neurodegeneration (Zacharias et al., 2022), chronic viral infections, e.g. HIV (Sitole et al., 2013), gout (Li et al., 2023), psychiatric disorders (Pedrini et al., 2019), various cancers (Nannini et al., 2020; Vignoli et al., 2021), and even hereditary metabolic diseases, e.g., phenylketonuria (Cannet et al., 2020). These findings have also been reaffirmed with large-scale epidemiological association studies using NMR-derived metabolomics data from the UK Biobank for many common diseases (Buergel et al., 2022; Julkunen et al., 2023). A detailed accounting of the individual serum/plasma metabolite changes associated with these many health conditions is beyond the scope of this review, but suffice to say, these NMR-detectable metabolite changes are often highly significant and their confounding effects cannot be ignored.
Given the large influence of health status on the blood, serum and plasma metabolomes (A.-H. M. Emwas et al., 2013), researchers should try to carefully assess the individual health states of their study population (and, if applicable, their healthy control population) by appropriate medical exams, patient/treating physician questionnaires, and/or information retrieval from medical records. Further recommendations for mitigating the confounding effects of health status on serum/plasma/blood metabolomics studies are provided in the Recommendations Section (Sect. 8).
Effects of drug intake
Besides the individual’s health status, the intake of drugs can significantly influence a patient’s blood metabolome. NMR-based metabolomics is able to accurately detect and quantify both small molecule drugs, e.g., paracetamol or D-mannitol, as well as downstream drug metabolites, such as propofol-glucuronide (J W Kim et al., 2013; Zacharias et al., 2015). The reflection of a patient’s medication intake in the NMR metabolic fingerprint of his/her blood specimen can have serious consequences for subsequent analyses, as large drug NMR signals—for example the large multiplets of D-mannitol administered during surgery—might obscure signals of small molecules of interest (Pertinhez et al., 2014) and/or drugs and their metabolites might act as confounders, inducing, e.g., spurious differences between patients and healthy controls, which only reflect the fact that the patients received a specific medication, while the healthy controls did not. Moreover, drugs themselves can have drastic effects on the endogenous metabolism of a patient, thereby potentially obscuring metabolic effects of diseases, which are oppressed by the drug treatment, or inducing metabolic differences between patients and healthy controls, which do not reflect the pathomechanisms of disease, but of the treatment itself (Preuss & Burris, 1996).
On the other hand, drugs and drug metabolites reflected in NMR metabolic fingerprints can also be used to study pharmacokinetics, drug metabolism, drug response as well as toxicity and their relation to the patient’s disease status (A-H Emwas et al., 2021). Zacharias et al., for example, utilized increased levels of propofol metabolites as well as the antifibrinolytic agent tranexamic acid in plasma NMR spectra to diagnose acute kidney injury in patients undergoing cardiac surgery, potentially reflecting both prolonged drug administration and delayed excretion in patients with renal impairment (Zacharias et al., 2015). Likewise, drug effects on the endogenous metabolome can be studied by NMR-based metabolomics, providing insights into, e.g., hepatotoxicity induced in humans by acetaminophen (J W Kim et al., 2013).
In conclusion, each metabolomics study of blood, plasma, and/or serum should carefully consider potential drug effects directly being reflected in the NMR spectra as well as indirectly by influencing the endogenous metabolome. Therefore, we recommend, analogously to assessing the health states of investigated patients, to also assess the drug intake, including individual timing and dosage, of the patients under study by employing appropriate patient/treating physician questionnaires, and/or information retrieval from medical records. We would like to point out that patients undergoing anesthesia are particularly exposed to high-dosage drug administration, and researchers should, in this setting, be especially aware of NMR signals arising from drugs or metabolites thereof in the investigated metabolic fingerprints.
Summary of participant-specific factors
It should be clear from the NMR-based metabolomic studies that the whole blood, serum and plasma metabolomes are affected significantly by diet, fasting status, sex, age, exercise, obesity or weight, smoking, alcohol consumption, gut microbiota, health status, and drug administration. The magnitude of these effects varies considerably, and in unfortunate cases, may lead to a significant misinterpretation of a typical metabolomics study. Their influence or the added metabolic “noise” from these effects can lead to a loss of signal or the diminishment of potentially interesting findings. Whereas most of these confounders can be controlled and adjusted through suitable patient selection, well-defined sample collection protocols, careful tracking or the use of questionnaires, it is important to remember that some confounders (such as the microbiome) cannot be as easily controlled. Being aware that these confounders exist and being knowledgeable about their likely contributions to a given study can certainly help improve the quality and the robustness of the findings of any NMR-based metabolomic study of serum, plasma or whole blood.
Effects of sample processing time
In addition to the participant- or patient-specific confounding factors discussed in the previous section, another critical variable is the blood sample processing time, i.e., the amount of time it takes to prepare plasma or serum from blood. Blood is a “living” biofluid containing cells and enzymes that carry out a range of chemical functions, including glycolysis, oxidation, transamination, hydroxylation and others. Therefore, the time between blood collection and the processing of the blood samples (involving removing the cellular components for plasma and removing cells and clotting components in the case of serum) is important because it directly affects measured metabolite concentrations. Extended delays before and during processing can alter the final concentrations of several “time-sensitive” metabolites measured by 1H NMR spectroscopy. A thorough study of this phenomena was performed by Bernini et al. (P Bernini et al., 2011a, 2011b). This study found that delays in processing blood caused a significant decrease in glucose concentration and an increase in lactate concentration, likely due to glycolysis in erythrocytes. These glucose and lactate changes were also observed in another study that also noted relatively small changes in lipid components’ levels with delays in sample processing (Debik et al., 2022a, 2022b). Changes in pyruvate concentration, an intermediate metabolite of glycolysis, depended on the type of blood derivative (serum or plasma) and on temperature. Specifically, the pyruvate concentration slightly decreased in plasma at 4 °C and increased at 25 °C, while in serum, it decreased at 4 °C and was almost constant at 25 °C. After blood processing, concentrations of glucose, lactate, and pyruvate remained stable over time. However, the presence of oxygen led to changes in several other metabolites and proteins, including albumin, triglycerides, LDL/VLDL, proline, citrate, and histidine. Histidine signals showed a shift consistent with a pH variation increase of about 0.1 pH units.
Debik et al. (Debik et al., 2022a, 2022b) proposed that the lactate/glucose ratio can be used as a measure for compliance with the sample collection protocols, specifically, the expected time between blood collection and blood centrifugation (to obtain either plasma or serum). However, as detailed above it should be noted that both glucose and lactate may be affected by numerous other factors. Debik et al. also found that several NMR-measured metabolites other than lactate, glucose and pyruvate can be affected by delays in sample processing. Specifically, an 8-h delay resulted in 147% increase in ornithine levels in plasma, along with 15.0 and 52.5% increases in phenylalanine concentrations in plasma and serum, respectively. Likewise, a 10.4 and 13.7% increase was seen in alanine levels in plasma and serum, respectively, and a 32.9% increase was seen in glycine concentration in serum (Debik et al., 2022a, 2022b).
Bervoets et al. (Bervoets et al., 2015) found that the impact of sample processing delays can be minimized if blood is stored at 4 °C before centrifugation. Specifically, they found that changes in the concentrations of lactate, glucose and pyruvate after storing blood at 4 °C for 8 h are significantly reduced and much smaller than the inter-individual variation in the study (Bervoets et al., 2015).
An alternative approach to minimize the impact of time delays in blood processing was proposed by Brunius et al. (Brunius et al., 2017). After observing similar trends in changes in metabolite concentrations, they developed an algorithm, called sampleDrift, to predict pre-centrifugation time and temperature from metabolite concentrations. This method also predicts the initial concentrations of time-sensitive metabolites such as lactate, glucose, pyruvate, hypoxanthine, acetate, ornithine and histidine.
More recently, based on comprehensive investigations of whole blood, plasma, and serum, robust biomarkers to assess or validate the preanalytical quality of plasma and serum for metabolomics have been discovered (Nagana Gowda et al., 2024). In cold human blood, the anomalous dynamics (Nagana Gowda et al., 2023) of adenosine triphosphate (ATP) result in the progressive accumulation of adenosine diphosphate (ADP), adenosine monophosphate (AMP), inosine monophosphate (IMP), inosine, and hypoxanthine. While the ATP, ADP, AMP, and IMP are confined to red blood cells (RBCs), inosine and hypoxanthine are excreted into plasma/serum. The plasma and serum levels of inosine and hypoxanthine depend on the temperature of blood and the plasma and serum contact time with the RBCs and hence they represent robust biomarkers for evaluating the preanalytical quality of plasma and serum. These biomarkers are highly specific since they are generally not present or at very low levels in fresh plasma and serum and are highly sensitive since they are derived from ATP, one of the most abundant metabolites in blood. Further, whether blood was kept at room temperature or on ice could be predicted based on inosine levels. An analysis of more than 2000 plasma/serum samples processed for metabolomics-centric analyses showed alarmingly high levels of inosine and hypoxanthine. The results highlight the gravity of sample quality challenges with high risk of grossly inaccurate measurements and incorrect study outcomes. Overall, these data suggest that particular care should be taken in noting the time delays and processing methods for serum and plasma preparation to reduce the effects of systematic errors in metabolite measurements.
Effects of hemolysis
Hemolysis involves the rupturing of RBCs and the release of their contents into blood plasma. Hemolysis can be visually detected and graded by looking for a pink or (in serious cases) red color in selected plasma or serum samples. It is a major issue of concern in clinical studies involving plasma and serum samples. Most hemolysis occurs because of procedural errors during the pre-analytical phase of blood collection, processing (during centrifugation to separate serum or plasma) and transport (Lippi et al., 2006). Hemolysis is of particular concern for blood metabolomics studies because the metabolic contents of RBCs are released into the surrounding fluid leading to significant alterations in “normal” serum or plasma metabolite levels. These altered metabolite levels typically have nothing to do with the condition being studied. While the number of studies looking at the effects of hemolysis on metabolite levels is relatively small, one study of note used NMR to characterise how hemolysis affected the metabolite levels in serum samples from umbilical cord (Lippi et al., 2006). The authors found that the concentrations of acetate, citrate, formate, glycerol, ornithine, phenylalanine, and succinate were reduced in hemolyzed samples while the concentration of glucose was increased (Nagana Gowda et al., 2024).
Effects of sample storage time and conditions
After the serum or plasma preparation process is completed, there may be further delays before the serum/plasma samples are either analyzed or placed into frozen storage. If serum and plasma samples are left at room temperature, significant metabolite changes in the contents of the sample can occur. These changes typically arise from residual enzyme activity from cells damaged during processing or from prior tissue (primarily muscle) injury. Using 1H NMR to evaluate the effects of sample “neglect”, Bernini et al. (P Bernini et al., 2011a, 2011b) found that changes in serum were larger than those of plasma, when these biofluids were left at room temperature over 24 h. They observed changes in triglycerides, proline, choline, citrate, histidine, and VLDL/LDL. In a related study, Ghini et al. (Ghini et al., 2019) observed changes to serum citrate levels, even when samples were kept at 4 ºC. In the case of long-term storage, Pinto et al. (Pinto et al., 2014) found that changes to proline and glucose levels in plasma became significant after 1 week at − 20 ºC. Encouragingly, they found that changes in plasma and serum were negligible after 30 months at − 80 ºC. These data suggest that serum and plasma samples should be frozen quickly (within 1 h) after preparation and stored at − 80 ºC to ensure long term stability. While minimal changes in lipoprotein profiles were seen with multiple freeze–thaw cycles (thawed from − 80 ºC) (Loo et al., 2020) and no significant changes in NMR-relevant metabolites were seen with four freeze–thaw cycles, slightly increased concentrations of amino acids (glycine, methionine, phenylalanine, tryptophan and tyrosine) were seen with increasing freeze–thaw cycles (Anton et al., 2015). Effects of repeated freezing and thawing on lipoprotein levels have been reported (Feng Wang et al., 2019). Therefore, repeated freezing and thawing should be omitted as far as possible.
A survey of current methods for 1H-NMR based blood processing & pre-analysis
Much of this review has focused on summarizing what is known about the metabolic effects and impacts of sample type (serum vs. plasma vs. blood), sample processing (deproteinized vs. intact) and various pre-analytical variables (patient-specific vs. general) on NMR-identifiable metabolites in human serum, plasma and blood. To better understand how the NMR community prepares, handles and reports on the metabolomic analysis of serum, plasma and blood, we decided to conduct a comprehensive literature survey. Specifically, we analyzed data from 100 scientific papers published in the past 15 years that reported metabolomic analyses of blood, serum or plasma by NMR. The papers were selected via PubMed using the following key words: NMR, metabolomics, human disease, blood, serum and plasma (Supplementary Table S1). The intent of this survey was to gather information on what is typically done or reported in terms of sample types, sample processing methods, patient information and other pre-analytical variables. It also allowed us to determine what are the most common methods or approaches for handling blood, plasma or serum for metabolomic analysis by NMR. Specifically, we assessed the following:
1) What fraction of papers analyzed serum only, plasma only, serum and plasma and blood?
2) What fraction used intact serum/plasma and what fraction used processed serum/plasma?
3) What fraction of those working with processed serum/plasma studies used methanol extraction vs. ultracentrifugation?
4) What fraction used intact serum/plasma to measure lipids and lipoproteins (via Nightingale, B.I. LISA or LabCorp) and what fraction used CPMG methods?
5) What fraction used untargeted (STOCSY, etc.) methods and what fraction used targeted or quantitative methods (Chenomx, Nightingale, B.I. LISA, B.I. Quant-PS or LabCorp)?
6) What fraction reported or controlled for patient-specific variables (diet, fasting, sex, exercise, ethnicity, etc.)?
7) What fraction reported on collection time/delays, thaw times, hemolysis protocols, temperature during preparation, storage temperature and centrifugation conditions (time and temperature)?
As seen in Table S1, the most common biofluid analyzed was serum (61%), followed by plasma (41%) and finally whole blood (2%) (with 4% reporting using serum, plasma and/or whole blood). In terms of sample processing, the majority of papers reviewed (74%) analyzed intact serum/plasma rather than deproteinized serum/plasma. Among the groups analyzing intact serum/plasma, 73% used CPMG methods to spectroscopically remove lipid and protein signals, while 7% used either Nightingale or B.I. LISA. Among the papers reporting on the analysis of deproteinized/delipidated serum/plasma, 48% used methanol extraction while 44% used ultracentrifugation. These results suggest that the analysis of intact serum or plasma via the CPMG method (despite its many shortcomings) is still very popular in the NMR metabolomics community. When proteins and lipids are removed from plasma or serum, methanol precipitation and ultracentrifugation are used with similar frequency. Interestingly, 1% of the surveyed papers did not report how their samples were processed/prepared. Across all papers that we reviewed, we found that 37% reported quantitative (targeted) metabolomic data while 70% reported qualitative (untargeted) data (with 3% reporting both). This trend is not surprising, considering that untargeted metabolomics is often the first step in a metabolomics study and is often followed by targeted analysis after statistically significant differences in untargeted data have been observed.
Information on patient specific variables (sex, age, diet/fasting state, etc.) was reported in 88% of the papers, including information on exercise status, alcohol consumption, smoking status, prior/existing disease status or other patient-specific variables that are known to affect metabolite levels. The vast majority of NMR reports (98%) appears to account or adjust for patient-specific variables in the experimental design phase rather than in the statistical analysis (by confounder adjustment or stratification) phase. We found very few papers (2%) that implemented corrections for patient-specific variables or their corresponding metabolite variations.
In terms of pre-analytical information on sample processing, such as processing time, processing temperature and centrifugation conditions, this information is reported in Figs. 2 and 3 and in Supplementary Table S1. We found that information about the time between blood collection and centrifugation for serum or plasma separation was not available for 70% or 85%, respectively, of the publications. Where the time interval was reported, we found that it was < 60 min for 20/30 publications for serum and for 6/15 publications for plasma (Fig. 2A). Information on the centrifugation conditions, such as speed, time, and temperature were available in 72, 71, and 43% of papers, respectively (Fig. 2B–E).
Fig. 2.
Summary statistics of provided a clotting time, b centrifugation speed in [g], or c in [rpm], d centrifugation temperature, and e centrifugation time to obtain serum or plasma as reported in 100 selected metabolomics publications (see Supplementary Table S1). NA—not available or reported; RT—room temperature
Fig. 3.
Summary statistics of a storage temperature or plasma or serum samplees, b storage time of plasma/serum samples at their respective temperatures, c time delay before samples were processed, d temperature before samples were processed and e temperature samples were processed using methanol for protein precipitation or organic solvents for lipid extraction reported in 100 selected metabolomics publications (see Supplementary Table S1). NA—not available or reported; RT—room temperature
With regard to sample storage temperature, we found that 86% reported storage at − 80 °C while 2% reported storage at − 70 °C (Fig. 3A). However, 8% did not report storage conditions and 3% reported storage temperatures between − 25 and − 20 ºC. The storage time was only mentioned in 6% of papers, with 2% of these storing samples from 1.5 to 13 years (Fig. 3B). Centrifugation speed was not described in 28% of analyzed papers (Figs. 2C). Among those that reported rotor speeds only, there was considerable variation, ranging from 1000 to 13,000 rpm. These alarmingly high centrifugation speeds could cause hemolysis, which would certainly affect the reported metabolite concentrations. Despite high centrifugation speeds (> 2000 × g or > 10,000 rpm) being reported in 41% of the publications, only 2% reported checking for hemolysis. Likewise, the centrifugation temperature was not mentioned in 57% of the reviewed papers, whereas 33% reported a centrifugation temperature of 4 °C (Fig. 2D). The centrifugation time was not mentioned in 29% of papers (Fig. 2E). The time of centrifugation was between 10 and 15 min in 63% of papers and 5–8 min in 6% of the papers. No mention of centrifugation time was given in 31% of papers (Fig. 2E).
When we analyzed the time between sample thawing and sample processing (either for protein precipitation using methanol or for lipid extraction using organic solvents), only a few publications (4%) mentioned the time delay before processing the plasma or serum (Fig. 3C) and only few reported the thawing temperature (Fig. 3D). As noted earlier, sample thaw time and sample thawing temperature can significantly affect sample composition. When sample processing temperatures were given, most (11%) were processed at 4 °C, followed by 5% at − 20 °C, 3% on ice (or at 0 °C) and 3% at room temperature (Fig. 3E).
In conclusion, many published manuscripts in the field of NMR-based blood/plasma/serum metabolomics failed to describe the methods or experimental design considerations needed to mitigate the most significant intrinsic or pre-analytical variables impacting metabolite concentrations. While many did report patient-specific variables such as age and sex distribution, many did not report diet (controlled, monitored or not), fasting state (when blood was collected), drug intake, weight/BMI, smoking/exercise/alcohol exposure status, health status or other potential confounders. Furthermore, descriptions of the methods (either pre-analytical or post hoc statistical) used to mitigate or control for these confounders were usually not provided. In addition to the lack of information on intrinsic or patient-specific data, critical information on how the blood samples were collected, which collection tubes were used, how the serum/plasma samples were prepared, the total time required for blood preparation, the blood processing temperatures, the centrifugation times and speeds, the thaw times, the thawing conditions and the delipidation or deproteinization protocols were often not provided. Even among those papers providing such information, there is a notable lack of standardization (compare Figs. 2 and 3). Given the lack of consistent and complete reporting seen for so many NMR studies on blood, plasma and serum, we decided to provide a series of recommendations and best-practice suggestions.
Recommendations for performing 1H-NMR metabolomics of blood, plasma and serum
According to several recent studies, errors arising from pre-analytical procedures or pre-analytical variables can contribute up to 60–80% of the total error in measured analyte quantification—even in well-established clinical tests (Carraro et al., 2012; Yin et al., 2013). As we have highlighted previously, these pre-analytical variables can be separated into patient/participant-specific variables and general sample preparation or handling variables. Patient or participant-specific variables such as age, sex, diet, drug intake, body weight, ethnicity and comorbidities can either be controlled through careful experimental design or dealt with as confounding variables during the statistical analysis phase. On the other hand, sample preparation/handling variables such as blood collection procedures (tourniquet use, vacuum systems and use of tubes with additives), plasma or serum preparation protocols (clotting time, temperature, centrifugation), and even the choice of biofluid type (blood, serum or plasma) can be controlled by following well-designed and well-tested Standardized Operating Procedures (SOPs), as, for example, specified in the ISO 23118:2021- “Molecular in vitro diagnostic examinations—specifications for pre-examination processes in metabolomics in urine, venous blood serum and plasma” standard (Ellervik & Vaught, 2015; Guder, 2014; IOS, 2021; Lippi et al., 2012; Pinto et al., 2014). In the following section, we describe a number of consensus recommendations and protocols regarding best practices for handling or mitigating confounders associated with analyzing blood, plasma or serum via 1H NMR-based metabolomics.
Recommendations for choice of sample type
Serum samples can be collected in tubes without any anti-clotting agents. The large peaks in the NMR spectra resulting from standard anti-clotting agents (such as EDTA or heparin) frequently interfere with the detection of other metabolites of interest (Y. T. Liu et al., 2019). Furthermore, the higher concentrations of many metabolites in serum (over plasma) provide higher sensitivity for detecting and analyzing biomarkers. However, plasma is less sensitive to differences in processing, which may be advantageous for smaller sample sets or studies requiring higher accuracy. Whole blood, with extraction of the intracellular metabolites, may be an option if those particular metabolites are of interest. However, the field of whole blood metabolomics is still in its infancy and more studies regarding sample handling and reproducibility are needed.
Recommendations for blood collection tubes
It is well known that contaminants from blood collection tubes contribute to numerous interfering signals and the type and magnitude of such interference potentially depend on the manufacturer (Bando et al., 2010; Deprez et al., 2002; Dunn et al., 2011). Therefore, it is important to test blank tubes beforehand to determine the presence of any such chemical contamination or interference. While tubes from a particular manufacturer are anticipated to exhibit similar levels of chemical interference, ideally, tubes from different batches need to be tested for the presence of any interfering signals in the NMR spectra. To conduct such a test, the tubes may be soaked with a blank solution such as PBS (phosphate buffered saline), then the sample should be processed using similar steps as used for blood processing and the solution tested using NMR for the presence of any chemical contaminants. It is important to distinguish between the contaminants from the blood collection tubes and those coming from the sample itself.
Recommendations for blood processing temperature
As noted earlier, allowing blood to stay at room temperature for an extended period of time (> 1 h) induces significant alterations to metabolic profiles. Hence, as soon as the blood is drawn, sample tubes should be placed on ice or kept at 4 °C until the sample processing steps (preparation of serum or plasma) are completed. This lower temperature reduces the rate of enzymatic action which can lead to significant changes in time-sensitive metabolites such as glucose, lactate, pyruvate, ornithine, triglycerides, LDL/VLDL, choline, proline, citrate, and histidine.
Recommendations for blood processing time (to serum or plasma)
Blood should be processed promptly after collection, preferably, within 2 h but optimally within 1 h (P Bernini et al., 2011a, 2011b). For plasma preparation, ideally, blood should be processed immediately after collection; however, in many cases it is impractical to do this. If blood must be stored for an hour or two, it should be kept at 4 °C, except for the 30 min at room temperature which is required for blood clotting when serum is being prepared.
Recommendations for serum preparation
After blood is collected in sterile tubes, it should be allowed to clot in the tubes in an upright position for 30 min at room temperature. After the blood clot is formed, each sample should be centrifuged at 1500–2000 × g for 10–15 min at room temperature (Kaluarachchi et al., 2018). If no further deproteinization is required, the serum samples can be mixed with buffer, D2O, reference standards (e.g. DSS) and loaded into NMR tubes for data acquisition.
Recommendations for plasma preparation
Unlike serum samples, plasma samples should be prepared in the presence of blood anti-coagulants, such as EDTA, heparin, and citrate. Once collected, the sample can be immediately centrifuged at 1600 × g for 15 min at room temperature. To prepare platelet free plasma, we recommend centrifuging blood with an anti-coagulant at 2000 × g for 10 min at room temperature, aliquoting the plasma into fresh vials and then performing a second centrifugation at 15,000 × g for 7 min. The recommendations for serum NMR sample preparation, internal concentration standards, and storage are also applicable to plasma samples.
Recommendations regarding hemolysis
Hemolysis can contribute to altered metabolic profiles in both plasma and serum. It can be visually detected from the pink or red tinge in serum or plasma. Numerous factors including improper procedure during blood collection, overly vigorous mixing, exposure to excessive heat or cold, mechanical trauma during transport, improper centrifugation speed and excessive centrifugation duration can contribute to hemolysis. As it can alter metabolite profiles, it is important to avoid hemolysis and we strongly recommend that any hemolyzed samples, detected by simple visual inspection, should be excluded from analysis. We also recommend that the number or percentage of hemolyzed samples identified in a study set should be reported as this is an excellent indicator of the overall quality of sample handling and preparation.
Recommendations for sample storage
After serum or plasma samples have been prepared, we recommend that samples should be aliquoted into 0.5–1.0 mL aliquots into suitably-labeled cryovials to minimize the number of freeze–thaw cycles required when samples are eventually sent for NMR analysis. Storing large volumes (> 5 mL) of serum/plasma in larger tubes is not recommended. Based on the weight of published evidence about serum/plasma stability, we strongly recommend storing samples at– 80 ºC. For short term storage (less than 1 week), storage at– 20 ºC is acceptable.
Recommendations for sample processing time prior to NMR analysis
The manner in which blood, plasma or serum samples are processed is critical for the accuracy and reproducibility of the measured metabolite concentrations. Frozen serum or plasma samples should be thawed rapidly at room temperature or in a 37 °C water bath such that they are thawed within 10–15 min. Serum should be separated from any remaining cellular components (via centrifugation or ultrafiltration) within 15–30 min of thawing to prevent any further enzymatic degradation. If delipidation or deproteinization is planned, these should be performed quickly, as recommended below. Fomenko et al. studied the stability of metabolite levels during sample preparation over a period of 24 h. To this end, 52 metabolites in serum, whole blood extracts and whole blood homogenates were analyzed. They found marked differences, especially for compounds prone to oxidation such as GSH and its oxidation product GSSG or ATP, ADP and AMP, whereas the majority of other compounds remained relatively stable (Fomenko et al., 2022). For NMR measurements, it is important to use rapid acquisition protocols (< 20 min) to reduce sample exposure time within the spectrometer. Ideally, samples should be stored in chilled (4 °C) autosamplers for no longer than 12 h. Study samples should be randomised, e.g. by stratified permuted block randomisation or any other suitable randomisation protocol (Burger et al., 2021) so that any confounding factors arising from the sample order during automated data acquisition affect all study groups similarly, and quality control (QC) samples should be inserted at regular intervals in between the randomised study samples (about 2–3 pooled study QC samples per 100 study samples and about one long-term QC sample per 100 study samples) to monitor for metabolite degradation during sample storage in NMR autosamplers and any other confounding factors arising from the sample measurement order (Dona et al., 2014).
Choice of sample processing method prior to NMR (deproteinization or no deproteinization)
For deproteinization of serum and plasma prior to NMR analysis, we recommend either methanol extraction (followed by lyophilization and reconstitution) or ultrafiltration (with 3 kDa molecular-weight cut-off (MWCO) filters). Both are acceptable but the choice and processing details of the methods must be clearly indicated and both will give different results. Some metabolites such as tryptophan and tyrosine show strong protein binding. By ultrafiltration, the protein bound part will be removed as well and only the free amount will be obtained. Note that spectroscopic deproteinization methods such as the use of the CPMG pulse-sequence will also only yield the free amount. Whereas, by protein precipitation, the protein bound fraction will be released, and the total metabolite amount will be obtained. Note that methods from mass spectrometry usually perform a deproteinization by protein precipitation. Therefore, if results should be compared with those from mass spectrometry, protein precipitation should be used (Wallmeier et al., 2017). Recently, the use of human serum albumin binding competitors has been successfully applied to obtain total metabolite concentrations (Barrilero et al., 2017; Derveaux et al., 2021; Vanhove et al., 2022; Z Wang et al., 2021). For example, TSP which shows considerable protein binding, may be used for this purpose. Clearly an additional internal standard such as malate must be used in this case (Vanhove et al., 2022). For ultrafiltration, we recommend the following protocol. Enough amount of unfiltered blood serum/plasma should be collected to provide sufficient amount of sample after filtration for NMR analysis (e.g. 800 μL and 400 μL for 5 mm and 3 mm NMR tubes, respectively). The 3-kDa MWCO filters (e.g. from Amicon) need to be thoroughly washed to remove glycerol present in the membranes. This can be done by adding distilled water to each filter and spinning down the filter in a centrifuge at 10,000 × g for 10 min at room temperature. Once the filters are cleaned, blood serum or plasma should be added to the filter and centrifuged at 10,000 × g at 4 °C for 20–30 min. The filtrate can then be mixed with buffer, D2O, and an internal standard (e.g. DSS) and loaded to an NMR tube. The filtrate should be clear and some liquid should remain in the filter chamber. Cloudy filtrate or a lack of residual unfiltered liquid indicates filter failure/damage.
Ultrafiltration has been shown to be more reproducible (Tiziani et al., 2008) than protein precipitation with organic solvents, which often leave trace amounts of lipid/protein that are still visible in the spectra. While ultrafiltration is an excellent method for high-throughput preparation of deproteinized NMR samples, it can systematically attenuate concentrations of metabolites that interact with filter membrane or which are bound to serum albumin (e.g. tryptophan). If accurate absolute concentrations of those metabolites are important, proteins can be removed with precipitation. In the methanol precipitation method, serum samples should be mixed with methanol in a 1:2 v/v ratio, vortexed, and incubated at − 20 °C for 20 min. The mixtures are then centrifuged at 14,000 × g for 30 min to precipitate proteins. The supernatants should be transferred to fresh tubes, dried, and dissolved in a buffer with D2O and an internal standard, and loaded to NMR tubes. Deproteinized and delipidated samples allow for the detection and quantification of 60 + small molecules in serum and plasma. We do not recommend the use of spectroscopic removal (via CPMG or CPMG-like pulse sequences) of protein/lipid signals. The CPMG method is not sufficiently robust for precise metabolite quantification (Bliziotis et al., 2020).
Intact serum and plasma are most useful for detecting and quantifying protein, lipoprotein and lipid components. Intact serum and plasma are suboptimal for detecting small molecule metabolites as typically only 10–15 small molecules can be quantified due to extensive signal overlap. As previously mentioned, the processing details for the intact serum/plasma sample must be clearly indicated to ensure reproducibility and comparability.
Recommendations for handling patient or participant-specific confounders
A good study design is crucial for minimizing or accounting for patient-specific confounders. Cohorts of participants should be as homogeneous or as comparable as possible in terms of sex, age, weight/BMI, health status, diet, drug intake, physical activity, etc. For example, when both sexes are included in the study, the percentage of male and female subjects should be approximately the same (e.g. 50%/50%) in each subject class (e.g. healthy control, disease). Confounding effects arising from comorbidities and/or drug administration can significantly distort or blur metabolome effects of the disease of interest and should be kept at a minimum. Several strategies to avoid comorbidity and/or drug intake confounders are available. First, careful study participant selection that includes interviews or questionnaires to identify (and exclude) individuals with comorbidities is recommended. As a simple example, study participants with type-1 diabetes should be excluded from the control group of an observational study investigating the effects of type-2 diabetes on the blood metabolome. Other study design approaches to minimize confounding variables include patient randomization with the goal to equally distribute confounders between different study groups and patient matching, where individuals or groups are matched in order to achieve equal distribution of confounders across individuals/groups. Finally, performing longitudinal studies on the same group of subjects can help to distinguish effects of confounding factors (e.g. BMI, health status, diet) and treatment.
Another important step in minimizing or accounting for confounding factors is metadata collection. With regard to health status, diet, BMI, medication and alcohol intake, we recommend that questionnaires (diet records, health and medication records, physical activity records) and simple phenotypic measurements or medical exams (height, weight, age, health status, drug intake) should be taken to assess and record patient-specific health states, physical activities, and diets. To minimize the effect of acute food intake, blood/serum/plasma samples should be collected after fasting for a standardized period (e.g. 8–12 h) before sample collection to minimize diet impact. To minimize the effects of habitual food intake, study participants should be encouraged to keep dietary records such as food diaries or validated food frequency questionnaires to accurately measure dietary intake. Detailed data on vitamin supplements (or vitamin status) should also be collected and the resulting metabolomic data should be appropriately adjusted based on this information. If this is not possible, then the diet of the subject group should be made as consistent as possible. As a general rule, habitual dietary effects, which randomly distribute across case/control populations, tend to obscure or reduce the significance of detectable disease biomarkers. Therefore, the better dietary information or diets are managed, controlled or corrected, the more significant the detectable biomarkers will be.
Standardization of pre-sample collection conditions and sample collection procedures can help to minimize effects of patient-specific confounders, such as diet, alcohol intake, health status, drug administration, and physical activity. In a controlled setting (e.g. hospital), participants should be provided with the same meals to standardize the dietary intake before sample collection. Likewise, participants should be asked to avoid vigorous physical activity for a specific time period (24–48 h) before sample collection. In controlled clinical settings, the physical activity of participants should be standardized in the days leading up to sample collection. Finally, the timing of individual blood sample collection can be rationally chosen to minimize certain unwanted effects of a study participant’s health status and/or drug intake, particularly with respect to acute diseases or treatments. In line with the Guidelines on Assessing Donor Suitability for Blood Donation of the World Health Organization (World Health Organization, 2012), we recommend that participants should be excluded from a study when infected with a virus (such as influenza or SARS-CoV-2 without any disease symptoms within the last seven days, or, in case of symptoms, within the last two weeks prior to blood collection).
Samples should be collected at the same time of day to account for daily or diurnal variations in metabolite levels and to standardize the timing between the sample collection and confounding factors (e.g. last meal, last physical exercise, last medication, etc.). If possible, consider collecting samples at multiple time points to capture temporal variations in metabolite concentrations related to diet, exercise, and disease status.
Finally, it may be warranted to perform additional collection and analysis of biological samples (blood, urine, fecal) to evaluate microbiome status or/and to measure biomarkers that may reflect diet, alcohol consumption, and health status in an objective manner. The collected metadata and the results of additional (non-metabolomic) tests can be then used in post-hoc sample stratification and confounder adjustment during statistical analysis.
Recommendations for experimental design
As with any other experimental study, proper experimental design is important. This includes considerations of confounding factors, as well as designing a standardized protocol for sample collection and sample preparation. One additional factor that should be considered in the earliest stages of study design is the number of samples that would be required to confirm or deny a particular hypothesis, or to observe or not observe statistically-significant differences between populations (Blaise et al., 2016; Vignoli et al., 2019). Determination of the appropriate study or cohort size must be done using power analysis (Blaise et al., 2016). However, if a power analysis calculation is not possible, it is often wise to do a small pilot study, from which statistical power analysis and a suitable follow-up study size can be determined. Due to the large natural variability between individuals, most well-powered metabolomics studies on humans often require large numbers of samples (> 100 cases and > 100 controls). If large numbers of samples are not available, one can reduce some of this natural variability by using a long-term longitudinal study, following the changes over time in a smaller cohort during the development or progression of a disease. However, longitudinal study designs can be both time consuming and expensive. Furthermore, they are not necessarily guaranteed to give sufficient numbers of samples for a clear, statistically significant conclusion to be reached. The decision on which study design is most suitable depends on many factors and therefore no single study design can be recommended. However, the rationale for a given study design and cohort size should be described in detail for every metabolomics study.
Recommendations for subject information
Metabolomics studies that involve human individuals often provide general information about the individuals, such as their sex, age, BMI, ethnicity, dietary and health status (A-H Emwas et al., 2015; Navarro et al., 2023; Fenglei Wang et al., 2023). However, there is no agreement on what essential information must be included in such studies. Healthy controls are usually described with a general statement, without providing comprehensive information. To ensure comparability, it is essential to have a uniform description of both the patients and healthy controls. Generally, healthy individuals are those who do not have underlying chronic diseases or symptoms. Factors such as food intake, health status, drug administration, physical activities, and lifestyle can all contribute to the metabolite concentration levels in human blood. Moreover, these factors are not independent; they are indeed dependent, where health status affects physical activities, food intake affects the microbial footprint, and so on (Cukkemane et al., 2020; Kelly et al., 2020; Mohr et al., 2022). Thus, it is crucial to standardize and report all relevant details of all individuals involved in any metabolomics study (González-Domínguez et al., 2020).
For comparison purposes, previously collected samples, that have been stored for a long time in biobanks, should not be used to be compared with newly collected patient samples. However, old NMR spectra acquired under the same conditions might be used. To ensure reliable comparability, healthy individuals should also share the same ethnicity, BMI, geographical location, diet, and lifestyle (e.g., level of exercise) (A-H Emwas et al., 2015). Additionally, this information should be collected at the same period as the patients’ specimens. Table 2 shows the recommended minimum information that should be collected and reported in metabolomics studies.
Table 2.
The study individual’s baseline characteristics including demographic, age, gender, and ethnicity that should be reported in the methods or results section of any metabolomics study
| Individual information | Age | Sex | Weight | Height | BMI | Ethnicity |
|---|---|---|---|---|---|---|
| Food intake | Diet | Fasting | Alcohol consumption | Vegetarian | Vegan | |
| Sample | Type | Size | ||||
| Collection | Date | Time | Collection tube | Location | Storage time | |
| Health status | Previous health issues including acute and chronic diseases |
Medication/ therapeutic treatments |
Any medication for the last 7 days | Menstrual phase for women | Pregnancy for women | |
| Lifestyle | Work | Diet | Physical activities |
Smoking (Y/N) |
Exercise |
In addition, it is essential to report relevant clinical data such as health status, medical history, medication usage, and lifestyle factors such as diet, exercise, and smoking habits Furthermore, documenting appropriate physiological parameters, like body mass index (BMI) and blood pressure, assists in understanding the sample relevance and ensures accurate interpretation of metabolomic findings
Healthy individuals often take dietary supplements, herbal medicines, or over-the-counter drugs, which can impact the metabolic composition of their blood. Therefore, a well-designed study should keep a record of the consumption of these substances. Although evidence of these compounds can be detected in the blood, often their identity is unknown without reference spectra. Lastly, it is worth mentioning that individuals can serve as their own controls, which should encourage the development of metabolomics baseline screening as a routine procedure for large populations.
Recommendations on reporting methodological information
As highlighted in our literature review of current practices (see Sect. 7) as well as a recent literature review published in (Cochran et al., 2024), most metabolomics researchers do not provide sufficient detail to allow the experiments or experimental measurements to be fully or properly reproduced. The paper by Powers et al. (Powers et al., 2024) provides many useful recommendations for reporting methodological data for all aspects of NMR-based metabolomics. However, we would also add a few more. We recommend that papers published on NMR-based serum/plasma/blood metabolomics should describe the methods and experimental design considerations to mitigate the most significant intrinsic or pre-analytical variables impacting metabolite concentrations in these biofluids. Descriptions of the methods (either pre-analytical or post hoc statistical) used to mitigate or control for subject specific confounders should be provided. In addition to the intrinsic or patient-specific data, additional information on how the blood samples were collected, how the serum/plasma samples were prepared, which tube types were used, the total time required for blood preparation, the blood processing temperatures, the centrifugation times and speeds, the thaw times, the thawing conditions and the delipidation or deproteinization protocols should all be provided. This information is most efficiently given in a table and should, ideally, be part of every reported study.
Conclusion
Standardization of sample selection, collection and preparation is essential to generate comparable, robust and reusable scientific data. Our goal in this review was to help metabolomics researchers in collecting reliable, standardized datasets for NMR-based blood/serum/plasma metabolomics. In doing so we first defined what is currently known about the NMR-detectable blood/serum/plasma metabolome. We then outlined the main factors that have been determined by ourselves and others that affect blood metabolite levels, including both intrinsic or patient-specific factors (such as age, sex, BMI, diet, health status, etc.) and external pre-analytical factors (such as sample preparation, handling and storage). We also assessed the current state of the field by conducting a literature survey of common practices and methods. Based on this information, we offered a series of practical recommendations for what to measure/expect, how to mitigate confounding factors, how to properly prepare, handle and store blood, plasma and serum and how to correctly report methodological or experimental design information in NMR-based metabolomics studies of blood, plasma and serum. We hope that this information and these recommendations will reduce the impact of confounding factors and enhance inter-laboratory comparability, enabling more meaningful outcome in NMR-based metabolomics studies.
Supplementary Information
Below is the link to the electronic supplementary material.
Acknowledgements
HUZ is supported by the German Federal Ministry of Education and Research (BMBF) within the framework of the e:Med research and funding concept (grant number 01ZX1912A).
Author contribution
All authors wrote and reviewed the manuscript. A-H. E. prepared Fig. 1 and M.L. prepared Figs. 2 and 3.
Funding
Open access publishing provided by King Abdullah University of Science and Technology (KAUST).
Data availability
Data is provided within the supplementary information file.
Declarations
Conflict of interest
Dr. Kaddurah-Daouk is an inventor on a series of patents on use of metabolomics for the diagnosis and treatment of CNS diseases and holds equity in Metabolon Inc., Chymia LLC and PsyProtix.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- Abdul Rahim, M. B. H., Chilloux, J., Martinez-Gili, L., Neves, A. L., Myridakis, A., Gooderham, N., & Dumas, M. E. (2019). Diet-induced metabolic changes of the human gut microbiome: Importance of short-chain fatty acids, methylamines and indoles. Acta Diabetologica,56(5), 493–500. 10.1007/s00592-019-01312-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aguilar, M. A., McGuigan, J., & Hall, M. A. (2021). Semi-automated NMR pipeline for environmental exposures: New insights on the metabolomics of smokers versus non-smokers. Pacific Symposium Biocomputer,26, 316–327. [PMC free article] [PubMed] [Google Scholar]
- Ala-Korpela, M., Hiltunen, Y., & Bell, J. D. (1995). Quantification of biomedical NMR data using artificial neural network analysis: Lipoprotein lipid profiles from 1H NMR data of human plasma. NMR in Biomedicine, 8(6), 235–244. 10.1002/nbm.1940080603 [DOI] [PubMed] [Google Scholar]
- Amathieu, R., Triba, M. N., Goossens, C., Bouchemal, N., Nahon, P., Savarin, P., & Moyec, L. L. (2016). Nuclear magnetic resonance based metabolomics and liver diseases: Recent advances and future clinical applications. World Journal of Gastroenterology,22(1), 417–426. 10.3748/wjg.v22.i1.417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amedei, A., & Morbidelli, L. (2019). Circulating metabolites originating from gut microbiota control endothelial cell function. Molecules (Basel, Switzerland),24(21), 3992. 10.3390/molecules24213992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ang, J. E., Revell, V., Mann, A., Mäntele, S., Otway, D. T., Johnston, J. D., et al. (2012). Identification of human plasma metabolites exhibiting time-of-day variation using an untargeted liquid chromatography-mass spectrometry metabolomic approach. Chronobiology International,29(7), 868–881. 10.3109/07420528.2012.699122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angioni, D., Delrieu, J., Hansson, O., Fillit, H., Aisen, P., Cummings, J., et al. (2022). Blood biomarkers from research use to clinical practice: What must be done? A report from the EU/US CTAD task force. The Journal of Prevention of Alzheimer’s Disease,9(4), 569–579. 10.14283/jpad.2022.85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anton, G., Wilson, R., Yu, Z. H., Prehn, C., Zukunft, S., Adamski, J., et al. (2015). Pre-analytical sample quality: Metabolite ratios as an intrinsic marker for prolonged room temperature exposure of serum samples. PLoS ONE,10(3), e0121495. 10.1371/JOURNAL.PONE.0121495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bahado-Singh, R. O., Syngelaki, A., Mandal, R., Han, B. S., Li, L., Bjorndahl, T. C., et al. (2018). First-trimester metabolomic prediction of stillbirth. The Journal of Maternal-Fetal and Neonatal Medicine,32(20), 3435–3441. 10.1080/14767058.2018.1465552 [DOI] [PubMed] [Google Scholar]
- Balling, M., Langsted, A., Afzal, S., Varbo, A., Davey Smith, G., & Nordestgaard, B. G. (2020). Reply to: “Methodological issues regarding: ‘A third of nonfasting plasma cholesterol is in remnant lipoproteins: Lipoprotein subclass profiling in 9293 individuals.’” Atherosclerosis,302, 57–58. 10.1016/j.atherosclerosis.2020.03.027 [DOI] [PubMed] [Google Scholar]
- Bando, K., Kawahara, R., Kunimatsu, T., Sakai, J., Kimura, J., Funabashi, H., et al. (2010). Influences of biofluid sample collection and handling procedures on GC–MS based metabolomic studies. Journal of Bioscience and Bioengineering,110(4), 491–499. 10.1016/j.jbiosc.2010.04.010 [DOI] [PubMed] [Google Scholar]
- Banoei, M. M., Vogel, H. J., Weljie, A. M., Kumar, A., Yende, S., Angus, D. C., et al. (2017). Plasma metabolomics for the diagnosis and prognosis of H1N1 influenza pneumonia. Critical Care,21(1), 97. 10.1186/s13054-017-1672-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bar, N., Korem, T., Weissbrod, O., Zeevi, D., Rothschild, D., Leviatan, S., et al. (2020). A reference map of potential determinants for the human serum metabolome. Nature,588(7836), 135–140. 10.1038/S41586-020-2896-2 [DOI] [PubMed] [Google Scholar]
- Barba, I., Andrés, M., Picón, I., Aguade-Bruix, S., & Garcia-Dorado, D. (2019). Sex differences in the 1H NMR metabolic profile of serum in cardiovascular risk patients. Science and Reports,9(1), 2380. 10.1038/s41598-019-38881-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barba, I., Sanz, C., Barbera, A., Tapia, G., Mate, J. L., Garcia-Dorado, D., et al. (2009). Metabolic fingerprinting of fresh lymphoma samples used to discriminate between follicular and diffuse large B-cell lymphomas. Experimental Hematology,37(11), 1259–1265. 10.1016/j.exphem.2009.08.006 [DOI] [PubMed] [Google Scholar]
- Barrilero, R., Ramírez, N., Vallvé, J. C., Taverner, D., Fuertes, R., Amigó, N., & Correig, X. (2017). Unravelling and quantifying the “nMR-Invisible” metabolites interacting with human serum albumin by binding competition and T2 relaxation-based decomposition analysis. Journal of Proteome Research,16(5), 1847–1856. 10.1021/acs.jproteome.6b00814 [DOI] [PubMed] [Google Scholar]
- Barton, R. H., Waterman, D., Bonner, F. W., Holmes, E., Clarke, R., Nicholson, J. K., & Lindon, J. C. (2009). The influence of EDTA and citrate anticoagulant addition to human plasma on information recovery from NMR-based metabolic profiling studies. Molecular BioSystems,6(1), 215–224. 10.1039/b907021d [DOI] [PubMed] [Google Scholar]
- Bathen, T. F., Krane, J., Engan, T., Bjerve, K. S., & Axelson, D. (2000). Quantification of plasma lipids and apolipoproteins by use of proton NMR spectroscopy, multivariate and neural network analysis. NMR in Biomedicine,13(5), 271–288. [DOI] [PubMed] [Google Scholar]
- Beckonert, O., Keun, H. C., Ebbels, T. M. D., Bundy, J., Holmes, E., Lindon, J. C., & Nicholson, J. K. (2007). Metabolic profiling, metabolomic and metabonomic procedures for NMR spectroscopy of urine, plasma, serum and tissue extracts. Nature Protocols,2(11), 2692–2703. 10.1038/nprot.2007.376 [DOI] [PubMed] [Google Scholar]
- Bell, J. A., Santos Ferreira, D. L., Fraser, A., Soares, A. L. G., Howe, L. D., Lawlor, D. A., et al. (2021). Sex differences in systemic metabolites at four life stages: Cohort study with repeated metabolomics. BMC Medicine,19(1), 58. 10.1186/s12916-021-01929-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bermingham, K. M., Mazidi, M., Franks, P. W., Maher, T., Valdes, A. M., Linenberg, I., et al. (2023). Characterisation of fasting and postprandial NMR metabolites: Insights from the ZOE PREDICT 1 study. Nutrients,15(11), 2638. 10.3390/nu15112638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernini, P., Bertini, I., Luchinat, C., Nincheri, P., Staderini, S., & Turano, P. (2011a). Standard operating procedures for pre-analytical handling of blood and urine for metabolomic studies and biobanks. Journal of Biomolecular NMR,49(3), 231–243. 10.1007/S10858-011-9489-1 [DOI] [PubMed] [Google Scholar]
- Bernini, P., Bertini, I., Luchinat, C., Tenori, L., & Tognaccini, A. (2011b). The cardiovascular risk of healthy individuals studied by NMR metabonomics of plasma samples. Journal of Proteome Research,10(11), 4983–4992. 10.1021/pr200452j [DOI] [PubMed] [Google Scholar]
- Bervoets, L., Louis, E., Reekmans, G., Mesotten, L., Thomeer, M., Adriaensens, P., & Linsen, L. (2015). Influence of preanalytical sampling conditions on the 1H NMR metabolic profile of human blood plasma and introduction of the Standard PREanalytical Code used in biobanking. Metabolomics,11(5), 1197–1207. 10.1007/s11306-015-0774-y [Google Scholar]
- Bester, R., Stander, Z., Mason, S., Keane, K. M., Howatson, G., Clifford, T., et al. (2021). Characterizing marathon-induced metabolic changes using 1H-NMR metabolomics. Metabolites,11(10), 656. 10.3390/metabo11100656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bjerrum, J. T., Wang, Y. L., Seidelin, J. B., & Nielsen, O. H. (2021). IBD metabonomics predicts phenotype, disease course, and treatment response. eBioMedicine,71, 103551. 10.1016/j.ebiom.2021.103551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blaise, B. J., Correia, G., Tin, A., Young, J. H., Vergnaud, A. C., Lewis, M., et al. (2016). Power analysis and sample size determination in metabolic phenotyping. Analytical Chemistry,88(10), 5179–5188. 10.1021/acs.analchem.6b00188 [DOI] [PubMed] [Google Scholar]
- Bliziotis, N. G., Engelke, U. F. H., Aspers, R. L. E. G., Engel, J., Deinum, J., Timmers, H. J. L. M., et al. (2020). A comparison of high-throughput plasma NMR protocols for comparative untargeted metabolomics. Metabolomics,16(5), 64. 10.1007/s11306-020-01686-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brugnara, L., Vinaixa, M., Murillo, S., Samino, S., Rodriguez, M. A., Beltran, A., et al. (2012). Metabolomics approach for analyzing the effects of exercise in subjects with type 1 diabetes mellitus. PLoS ONE,7(7), e40600. 10.1371/journal.pone.0040600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunius, C., Pedersen, A., Malmodin, D., Karlsson, B. G., Andersson, L. I., Tybring, G., & Landberg, R. (2017). Prediction and modeling of pre-analytical sampling errors as a strategy to improve plasma NMR metabolomics data. Bioinformatics,33(22), 3567–3574. 10.1093/bioinformatics/btx442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruzzone, C., Bizkarguenaga, M., Gil-Redondo, R., Diercks, T., Arana, E., García de Vicuña, A., et al. (2020). SARS-CoV-2 infection dysregulates the metabolomic and lipidomic profiles of serum. iScience,23(10), 101645. 10.1016/j.isci.2020.101645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruzzone, C., Conde, R., Embade, N., Mato, J. M., & Millet, O. (2023). Metabolomics as a powerful tool for diagnostic, pronostic and drug intervention analysis in COVID-19. Frontiers in Molecular Biosciences,10, 1111482. 10.3389/fmolb.2023.1111482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buergel, T., Steinfeldt, J., Ruyoga, G., Pietzner, M., Bizzarri, D., Vojinovic, D., et al. (2022). Metabolomic profiles predict individual multidisease outcomes. Nature Medicine,28(11), 2309–2320. 10.1038/s41591-022-01980-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burger, B., Vaudel, M., & Barsnes, H. (2021). Importance of block randomization when designing proteomics experiments. Journal of Proteome Research,20(1), 122–128. 10.1021/acs.jproteome.0c00536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cacciatore, S., Wium, M., Licari, C., Ajayi-Smith, A., Masieri, L., Anderson, C., et al. (2021). Inflammatory metabolic profile of South African patients with prostate cancer. Cancer & Metabolism,9(1), 29. 10.1186/s40170-021-00265-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cannet, C., Pilotto, A., Rocha, J. C., Schäfer, H., Spraul, M., Berg, D., et al. (2020). Lower plasma cholesterol, LDL-cholesterol and LDL-lipoprotein subclasses in adult phenylketonuria (PKU) patients compared to healthy controls: Results of NMR metabolomics investigation. Orphanet Journal of Rare Diseases,15(1), 61. 10.1186/s13023-020-1329-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carraro, P., Zago, T., & Plebani, M. (2012). Exploring the initial steps of the testing process: Frequency and nature of pre-preanalytic errors. Clinical Chemistry,58(3), 638–642. 10.1373/clinchem.2011.175711 [DOI] [PubMed] [Google Scholar]
- Castro, A., Catai, A. M., Rehder-Santos, P., Signini, É. F., de Abreu, R. M., Da Silva, C. D., et al. (2023). Insights into the serum metabolic adaptations in response to inspiratory muscle training: A metabolomic approach based on 1H NMR and UHPLC-HRMS/MS. International Journal of Molecular Sciences,24(23), 16764. 10.3390/ijms242316764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castro, A., Signini, É. F., De Oliveira, J. M., Di Medeiros Leal, M. C. B., Rehder-Santos, P., Millan-Mattos, J. C., et al. (2022). The aging process: A metabolomics perspective. Molecules,27(24), 8656. 10.3390/molecules27248656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cochran, D., Noureldein, M., Bezdeková, D., Schram, A., Howard, R., & Powers, R. (2024). A reproducibility crisis for clinical metabolomics studies. TrAC Trends in Analytical Chemistry,180, 117918. 10.1016/j.trac.2024.117918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coelho, W. S., Viveiros de Castro, L., Deane, E., Magno-França, A., Bassini, A., & Cameron, L.-C. (2016). Investigating the cellular and metabolic responses of world-class canoeists training: A sportomics approach. Nutrients,8(11), 719. 10.3390/nu8110719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohn, J. S., McNamara, J. R., & Schaefer, E. J. (1988). Lipoprotein cholesterol concentrations in the plasma of human subjects as measured in the fed and fasted states. Clinical Chemistry,34(12), 2456–2459. 10.1093/clinchem/34.12.2456 [PubMed] [Google Scholar]
- Cukkemane, A., Kumar, P., & Sathyamoorthy, B. (2020). A metabolomics footprint approach to understanding the benefits of synbiotics in functional foods and dietary therapeutics for health, communicable and non-communicable diseases. Food Research International,128, 108679. 10.1016/j.foodres.2019.108679 [DOI] [PubMed] [Google Scholar]
- Dasgupta, S., Ghosh, N., Bhattacharyya, P., Roy Chowdhury, S., & Chaudhury, K. (2023). Metabolomics of asthma, COPD, and asthma-COPD overlap: An overview. Critical Reviews in Clinical Laboratory Sciences,60(2), 153–170. 10.1080/10408363.2022.2140329 [DOI] [PubMed] [Google Scholar]
- De Backer, G., Ambrosioni, E., Borch-Johnsen, K., Brotons, C., Cifkova, R., Dallongeville, J., et al. (2003). European guidelines on cardiovascular disease prevention in clinical practice: Third joint task force of European and other societies on cardiovascular disease prevention in clinical practice. European Heart Journal,24(17), 1601–1610. 10.1016/S0195-668X(03)00347-6 [DOI] [PubMed] [Google Scholar]
- Debik, J., Isaksen, S. H., Strømmen, M., Spraul, M., Schäfer, H., Bathen, T. F., & Giskeødegård, G. F. (2022a). Effect of delayed centrifugation on the levels of NMR-measured lipoproteins and metabolites in plasma and serum samples. Analytical Chemistry,94(49), 17003–17010. 10.1021/acs.analchem.2c02167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Debik, J., Sangermani, M., Wang, F., Madssen, T. S., & Giskeødegård, G. F. (2022b). Multivariate analysis of NMR-based metabolomic data. NMR in Biomedicine,35(2), e4638. 10.1002/nbm.4638 [DOI] [PubMed] [Google Scholar]
- Deprez, S., Sweatman, B. C., Connor, S. C., Haselden, J. N., & Waterfield, C. J. (2002). Optimisation of collection, storage and preparation of rat plasma for 1H NMR spectroscopic analysis in toxicology studies to determine inherent variation in biochemical profiles. Journal of Pharmaceutical and Biomedical Analysis,30(4), 1297–1310. 10.1016/s0731-7085(02)00455-7 [DOI] [PubMed] [Google Scholar]
- Derveaux, E., Thomeer, M., Mesotten, L., Reekmans, G., & Adriaensens, P. (2021). Detection of lung cancer via blood plasma and1h-nmr metabolomics: Validation by a semi-targeted and quantitative approach using a protein-binding competitor. Metabolites. 10.3390/metabo11080537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeSilva, M. A., Shanaiah, N., Nagana Gowda, G. A., Rosa-Pérez, K., Hanson, B. A., & Raftery, D. (2009). Application of 31P NMR spectroscopy and chemical derivatization for metabolite profiling of lipophilic compounds in human serum. Magnetic Resonance in Chemistry. 10.1002/mrc.2480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dona, A. C., Jiménez, B., Schafer, H., Humpfer, E., Spraul, M., Lewis, M. R., et al. (2014). Precision high-throughput proton NMR spectroscopy of human urine, serum, and plasma for large-scale metabolic phenotyping. Analytical Chemistry,86(19), 9887–9894. 10.1021/ac5025039 [DOI] [PubMed] [Google Scholar]
- Du, D., Bruno, R., Blizzard, L., Venn, A., Dwyer, T., Smith, K. J., et al. (2020). The metabolomic signatures of alcohol consumption in young adults. European Journal of Preventive Cardiology,27(8), 840–849. 10.1177/2047487319834767 [DOI] [PubMed] [Google Scholar]
- Dunn, W. B., Broadhurst, D., Begley, P., Zelena, E., Francis-Mcintyre, S., Anderson, N., et al. (2011). Procedures for large-scale metabolic profiling of serum and plasma using gas chromatography and liquid chromatography coupled to mass spectrometry. Nature Protocols,6(7), 1060–1083. 10.1038/nprot.2011.335 [DOI] [PubMed] [Google Scholar]
- Edison, A. S., Colonna, M., Gouveia, G. J., Holderman, N. R., Judge, M. T., Shen, X., & Zhang, S. (2021). NMR: Unique strengths that enhance modern metabolomics research. Analytical Chemistry,93(1), 478–499. 10.1021/acs.analchem.0c04414 [DOI] [PubMed] [Google Scholar]
- Ellervik, C., & Vaught, J. (2015). Preanalytical variables affecting the integrity of human biospecimens in biobanking. Clinical Chemistry,61(7), 914–934. 10.1373/clinchem.2014.228783 [DOI] [PubMed] [Google Scholar]
- Ellul, S., Ponsonby, A.-L., Carlin, J. B., Collier, F., Mansell, T., Vuillermin, P., et al. (2020). Sex differences in infant blood metabolite profile in association with weight and adiposity measures. Pediatric Research,88(3), 473–483. 10.1038/s41390-020-0762-4 [DOI] [PubMed] [Google Scholar]
- Emwas, A.-H., Luchinat, C., Turano, P., Tenori, L., Roy, R., Salek, R., et al. (2015). Standardizing the experimental conditions for using urine in NMR-based metabolomic studies with a particular focus on diagnostic studies: A review. Metabolomics,11(4), 872–894. 10.1007/S11306-014-0746-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emwas, A.-H.M., Salek, R. M., Griffin, J. L., & Merzaban, J. (2013). NMR-based metabolomics in human disease diagnosis: Applications, limitations, and recommendations. Metabolomics,9(5), 1048–1072. 10.1007/s11306-013-0524-y [Google Scholar]
- Emwas, A.-H., Szczepski, K., McKay, R. T., Asfour, H., Chang, C., Lachowicz, J., & Jaremko, M. (2021). Pharmacometabolomics: a new horizon in personalized medicine. In X. Zhan (Ed.), Metabolomics Methodology and Applications in Medical Sciences and Life Sciences. IntechOpen: Rijeka. [Google Scholar]
- Fan, P., Li, L., Rezaei, A., Eslamfam, S., Che, D., & Ma, X. (2015). Metabolites of dietary protein and peptides by intestinal microbes and their impacts on gut. Current Protein and Peptide Science,16(7), 646–654. 10.2174/1389203716666150630133657 [DOI] [PubMed] [Google Scholar]
- Fang, L.-J., Lin, X.-C., Huang, D., Pan, T.-T., Yan, X.-M., Hu, W.-G., et al. (2020). 1H NMR-based metabolomics analyses in children with Helicobacter pylori infection and the alteration of serum metabolites after treatment. Microbial Pathogenesis,147(104292), 104292. 10.1016/j.micpath.2020.104292 [DOI] [PubMed] [Google Scholar]
- Farley, A., Hendry, C., & McLafferty, E. (2013). Blood components. Nursing Standard,27(13), 35–42. 10.7748/ns2012.11.27.13.35.c9449 [DOI] [PubMed] [Google Scholar]
- Ferreira, D. L. S., Maple, H. J., Goodwin, M., Brand, J. S., Yip, V., Min, J. L., et al. (2019). The effect of pre-analytical conditions on blood metabolomics in epidemiological studies. Metabolites,9(4), 64. 10.3390/metabo9040064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fomenko, M. V., Yanshole, L. V., & Tsentalovich, Y. P. (2022). Stability of metabolomic content during sample preparation: Blood and brain tissues. Metabolites,12(9), 811. 10.3390/metabo12090811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fotiou, M., Fotakis, C., Tsakoumaki, F., Athanasiadou, E., Kyrkou, C., Dimitropoulou, A., et al. (2018). 1H NMR-based metabolomics reveals the effect of maternal habitual dietary patterns on human amniotic fluid profile. Scientific Reports,8(1), 4076. 10.1038/s41598-018-22230-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Funderburg, N. T., Xu, D., Playford, M. P., Joshi, A. A., Andrade, A., Kuritzkes, D. R., et al. (2017). Treatment of HIV infection with a raltegravir-based regimen increases LDL levels, but improves HDL cholesterol efflux capacity. Antiviral Therapy,22(1), 71–75. 10.3851/IMP3091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghini, V., Abuja, P. M., Polasek, O., Kozera, L., Laiho, P., Anton, G., et al. (2022). Impact of the pre-examination phase on multicenter metabolomic studies. New Biotechnology. 10.1016/j.nbt.2022.01.006 [DOI] [PubMed] [Google Scholar]
- Ghini, V., Quaglio, D., Luchinat, C., & Turano, P. (2019). NMR for sample quality assessment in metabolomics. New Biotechnology,52, 25–34. 10.1016/j.nbt.2019.04.004 [DOI] [PubMed] [Google Scholar]
- Ghini, V., Tenori, L., Pane, M., Amoruso, A., Marroncini, G., Squarzanti, D. F., et al. (2020). Effects of probiotics administration on human metabolic phenotype. Metabolites,10(10), 396. 10.3390/metabo10100396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- González-Domínguez, R., González-Domínguez, Á., Sayago, A., & Fernández-Recamales, Á. (2020). Recommendations and best practices for standardizing the pre-analytical processing of blood and urine samples in metabolomics. Metabolites,10(6), 229. 10.3390/METABO10060229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gralka, E., Luchinat, C., Tenori, L., Ernst, B., Thurnheer, M., & Schultes, B. (2015). Metabolomic fingerprint of severe obesity is dynamically affected by bariatric surgery in a procedure-dependent manner. American Journal of Clinical Nutrition,102(6), 1313–1322. 10.3945/ajcn.115.110536 [DOI] [PubMed] [Google Scholar]
- Gregory, J. F., Park, Y., Lamers, Y., Bandyopadhyay, N., Chi, Y. Y., Lee, K., et al. (2013). Metabolomic analysis reveals extended metabolic consequences of marginal vitamin B-6 deficiency in healthy human subjects. PLoS ONE,8(6), e63544. 10.1371/journal.pone.0063544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guder, W. G. (2014). History of the preanalytical phase: A personal view. Biochemia Medica,24(1), 25–30. 10.11613/BM.2014.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamaya, R., Mora, S., Lawler, P. R., Cook, N. R., Buring, J. E., Lee, I.-M., et al. (2022). Association of modifiable lifestyle factors with plasma branched-chain amino acid metabolites in women. Journal of Nutrition,152(6), 1515–1524. 10.1093/jn/nxac056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hefni, M. E., Witthöft, C. M., & Moazzami, A. A. (2018). Plasma metabolite profiles in healthy women differ after intervention with supplemental folic acid v. folate-rich foods. Journal of Nutritional Science. 10.1017/jns.2018.22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho, C. L., Quay, T. A. W., Devlin, A. M., & Lamers, Y. (2016). Prevalence and predictors of low vitamin B6 status in healthy young adult women in metro Vancouver. Nutrients,8(9), 538. 10.3390/nu8090538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ienaga, K., Nakamura, K., Fujisawa, T., Fukunaga, Y., Nihei, H., Narita, M., et al. (2007). Urinary excretion of creatol, an in vivo biomarker of hydroxyl radical, in patients with chronic renal failure. Renal Failure,29(3), 279. 10.1080/08860220701219863 [DOI] [PubMed] [Google Scholar]
- IOS. (2021). ISO 23118:2021(en) Molecular in vitro diagnostic examinations — Specifications for pre-examination processes in metabolomics in urine, venous blood serum and plasma. Geneva, Switzerland
- Jaguri, A., Al Thani, A. A., & Elrayess, M. A. (2023). Exercise metabolome: Insights for health and performance. Metabolites,13(6), 694. 10.3390/metabo13060694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- James, E. L., & Parkinson, E. K. (2015). Serum metabolomics in animal models and human disease. Current Opinion in Clinical Nutrition and Metabolic Care,18(5), 478–483. 10.1097/MCO.0000000000000200 [DOI] [PubMed] [Google Scholar]
- Jang, S. Y., Jung, Y., Lee, D.-H., & Hwang, G.-S. (2022). NMR-based metabolomic analysis of human plasma to examine the effect of exposure to persistent organic pollutants. Chemosphere,307(Pt 4), 135963. 10.1016/j.chemosphere.2022.135963 [DOI] [PubMed] [Google Scholar]
- Jendoubi, T. (2021). Approaches to integrating metabolomics and multi-omics data: A primer. Metabolites,11(3), 184. 10.3390/metabo11030184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeyarajah, E. J., Cromwell, W. C., & Otvos, J. D. (2006). Lipoprotein particle analysis by nuclear magnetic resonance spectroscopy. Clinics in Laboratory Medicine,26(4), 847–870. 10.1016/j.cll.2006.07.006 [DOI] [PubMed] [Google Scholar]
- Jiménez, B., Holmes, E., Heude, C., Tolson, R. F., Harvey, N., Lodge, S. L., et al. (2018). Quantitative lipoprotein subclass and low molecular weight metabolite analysis in human serum and plasma by 1H NMR spectroscopy in a multilaboratory trial. Analytical Chemistry,90(20), 11962–11971. 10.1021/acs.analchem.8b02412 [DOI] [PubMed] [Google Scholar]
- Julkunen, H., Cichońska, A., Tiainen, M., Koskela, H., Nybo, K., Mäkelä, V., et al. (2023). Atlas of plasma NMR biomarkers for health and disease in 118,461 individuals from the UK Biobank. Nature Communications,14(1), 604. 10.1038/s41467-023-36231-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung, J., Kim, S. H., Lee, H. S., Choi, G. S., Jung, Y. S., Ryu, D. H., et al. (2013). Serum metabolomics reveals pathways and biomarkers associated with asthma pathogenesis. Clinical and Experimental Allergy,43(4), 425–433. 10.1111/cea.12089 [DOI] [PubMed] [Google Scholar]
- Kalantari, S., & Nafar, M. (2019). An update of urine and blood metabolomics in chronic kidney disease. Biomarkers in Medicine,13(7), 577–597. 10.2217/bmm-2019-0008 [DOI] [PubMed] [Google Scholar]
- Kaluarachchi, M., Boulangé, C. L., Karaman, I., Lindon, J. C., Ebbels, T. M. D., Elliott, P., et al. (2018). A comparison of human serum and plasma metabolites using untargeted 1H NMR spectroscopy and UPLC-MS. Metabolomics,14(3), 32. 10.1007/s11306-018-1332-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato, T., Yamazaki, K., Nakajima, M., Date, Y., Kikuchi, J., Hase, K., et al. (2018). Oral administration of Porphyromonas gingivalis alters the gut microbiome and serum metabolome. mSphere,3(5), e0046018. 10.1128/mSphere.00460-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly, R. S., Kelly, M. P., & Kelly, P. (2020). Metabolomics, physical activity, exercise and health: A review of the current evidence. Biochimica et Biophysica Acta—Molecular Basis of Disease. 10.1016/j.bbadis.2020.165936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennedy, A. D., Ford, L., Wittmann, B., Conner, J., Wulff, J., Mitchell, M., et al. (2021). Global biochemical analysis of plasma, serum and whole blood collected using various anticoagulant additives. PLoS ONE,16(4), e0249797. 10.1371/journal.pone.0249797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khakimov, B., Hoefsloot, H. C. J., Mobaraki, N., Aru, V., Kristensen, M., Lind, M. V., et al. (2022). Human blood lipoprotein predictions from 1H NMR spectra: Protocol, model performances, and cage of covariance. Analytical Chemistry,94(2), 628–636. 10.1021/acs.analchem.1c01654 [DOI] [PubMed] [Google Scholar]
- Kim, H. S., Kim, S. W., Park, Y. S., Kwon, S. Y., Liu, J. R., Joung, H., & Jeon, J. H. (2009). Metabolic profiles of genetically modified potatoes using a combination of metabolite fingerprinting and multivariate analysis. Biotechnology and Bioprocess Engineering,14(6), 738–747. 10.1007/s12257-009-0168-y [Google Scholar]
- Kim, J. W., Ryu, S. H., Kim, S., Lee, H. W., Lim, M., Seong, S. J., et al. (2013). Pattern recognition analysis for hepatotoxicity induced by acetaminophen using plasma and urinary 1H NMR-based metabolomics in humans. Analytical Chemistry,85(23), 11326–11334. 10.1021/ac402390q [DOI] [PubMed] [Google Scholar]
- Kjeldby, I. K., Fosnes, G. S., Ligaarden, S. C., & Farup, P. G. (2013). Vitamin B6 deficiency and diseases in elderly people—A study in nursing homes. BMC Geriatrics,13(1), 13. 10.1186/1471-2318-13-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koeth, R. A., Wang, Z., Levison, B. S., Buffa, J. A., Org, E., Sheehy, B. T., et al. (2013). Intestinal microbiota metabolism of L-carnitine, a nutrient in red meat, promotes atherosclerosis. Nature Medicine,19(5), 576–585. 10.1038/nm.3145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondoh, H., Kameda, M., & Yanagida, M. (2020). Whole blood metabolomics in aging research. International Journal of Molecular Sciences,22(1), 175. 10.3390/IJMS22010175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krewski, D., Acosta, D., Andersen, M., Anderson, H., Bailar, J. C., Boekelheide, K., et al. (2010). Toxicity testing in the 21st century: A vision and a strategy. Journal of Toxicology and Environmental Health - Part b: Critical Reviews,13(2–4), 1–138. 10.1080/10937404.2010.483176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labaki, W. W., Gu, T., Murray, S., Curtis, J. L., Yeomans, L., Bowler, R. P., et al. (2019). Serum amino acid concentrations and clinical outcomes in smokers: SPIROMICS metabolomics study. Scientific Reports,9(1), 11367. 10.1038/s41598-019-47761-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau, C.-H.E., Manou, M., Markozannes, G., Ala-Korpela, M., Ben-Shlomo, Y., Chaturvedi, N., et al. (2023). NMR metabolomic modelling of age and lifespan: A multi-cohort analysis. medRxiv. 10.1101/2023.11.07.2329820037986826 [Google Scholar]
- Lawton, K. A., Berger, A., Mitchell, M., Milgram, K. E., Evans, A. M., Guo, L., et al. (2008). Analysis of the adult human plasma metabolome. Pharmacogenomics. 10.2217/14622416.9.4.383 [DOI] [PubMed] [Google Scholar]
- Lehtovirta, M., Pahkala, K., Rovio, S. P., Magnussen, C. G., Laitinen, T. T., Niinikoski, H., et al. (2023). Association of tobacco smoke exposure with metabolic profile from childhood to early adulthood. The special turku coronary risk factor intervention project (STRIP). European Journal of Preventive Cardiology. 10.1093/eurjpc/zwad285 [DOI] [PubMed] [Google Scholar]
- Letertre, M. P. M., Dervilly, G., & Giraudeau, P. (2021). Combined nuclear magnetic resonance spectroscopy and mass spectrometry approaches for metabolomics. Analytical Chemistry,93(1), 500–518. 10.1021/acs.analchem.0c04371 [DOI] [PubMed] [Google Scholar]
- Li, R., Liang, N., Tao, Y., & Yin, H. (2023). Metabolomics in hyperuricemia and gout. Gout, Urate, and Crystal Deposition Disease,1(1), 49–61. 10.3390/gucdd1010006 [Google Scholar]
- Likhitweerawong, N., Thonusin, C., Boonchooduang, N., Louthrenoo, O., Nookaew, I., Chattipakorn, N., & Chattipakorn, S. C. (2021). Profiles of urine and blood metabolomics in autism spectrum disorders. Metabolic Brain Disease,36(7), 1641–1671. 10.1007/s11011-021-00788-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin, H.-T., Cheng, M.-L., Lo, C.-J., Lin, G., Lin, S.-F., Yeh, J.-T., et al. (2019). 1H Nuclear magnetic resonance (NMR)-based cerebrospinal fluid and plasma metabolomic analysis in type 2 diabetic patients and risk prediction for diabetic microangiopathy. Journal of Clinical Medicine,8(6), 874. 10.3390/jcm8060874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindqvist, H. M., Rådjursöga, M., Malmodin, D., Winkvist, A., & Ellegård, L. (2019). Serum metabolite profiles of habitual diet: Evaluation by 1H-nuclear magnetic resonance analysis. American Journal of Clinical Nutrition,110(1), 53–62. 10.1093/ajcn/nqz032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipfert, M., Rout, M. K., Berjanskii, M., & Wishart, D. S. (2019). Automated tools for the analysis of 1D-NMR and 2D-NMR spectra. Methods in Molecular Biology,2037, 429–449. 10.1007/978-1-4939-9690-2_24 [DOI] [PubMed] [Google Scholar]
- Lippi, G., Becan-McBride, K., Behúlová, D., Bowen, R. A., Church, S., Delanghe, J., et al. (2012). Preanalytical quality improvement: In quality we trust. Clinical Chemistry and Laboratory Medicine (CCLM),51(1), 229–241. 10.1515/cclm-2012-0597 [DOI] [PubMed] [Google Scholar]
- Lippi, G., Luca Salvagno, G., Montagnana, M., Brocco, G., & Cesare Guidi, G. (2006). Influence of hemolysis on routine clinical chemistry testing. Clinical Chemistry and Laboratory Medicine (CCLM),44(3), 311–316. 10.1515/cclm.2006.054 [DOI] [PubMed] [Google Scholar]
- Liu, L., Aa, J., Wang, G., Yan, B., Zhang, Y., Wang, X., et al. (2010). Differences in metabolite profile between blood plasma and serum. Analytical Biochemistry,406(2), 105–112. 10.1016/j.ab.2010.07.015 [DOI] [PubMed] [Google Scholar]
- Liu, X., & Locasale, J. W. (2017). Metabolomics: A primer. Trends in Biochemical Sciences,42(4), 274–284. 10.1016/j.tibs.2017.01.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, Y. T., Xu, W. Q., Wang, G. H., Li, A. P., Li, K., & Qin, X. M. (2019). A comparison of serum and plasma specimens in NMR-based metabonomics and its application in chronic atrophic gastritis rats. Analytical Methods,11(15), 2018–2026. 10.1039/c9ay00156e [Google Scholar]
- Lodge, S., Nitschke, P., Loo, R. L., Kimhofer, T., Bong, S. H., Richards, T., et al. (2021). Low volume in vitro diagnostic proton NMR spectroscopy of human blood plasma for lipoprotein and metabolite analysis: Application to SARS-CoV-2 biomarkers. Journal of Proteome Research,20(2), 1415–1423. 10.1021/acs.jproteome.0c00815 [DOI] [PubMed] [Google Scholar]
- Loo, R. L., Lodge, S., Kimhofer, T., Bong, S. H., Begum, S., Whiley, L., et al. (2020). Quantitative in-vitro diagnostic NMR spectroscopy for lipoprotein and metabolite measurements in plasma and serum: Recommendations for analytical artifact minimization with special reference to COVID-19/SARS-CoV-2 samples. Journal of Proteome Research,19(11), 4428–4444. 10.1021/acs.jproteome.0c00537 [DOI] [PubMed] [Google Scholar]
- Macias, S., Kirma, J., Yilmaz, A., Moore, S. E., McKinley, M. C., McKeown, P. P., et al. (2019). Application of 1H-NMR metabolomics for the discovery of blood plasma biomarkers of a Mediterranean diet. Metabolites,9(10), 201. 10.3390/metabo9100201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madrid-Gambin, F., Oller, S., Marco, S., Pozo, Ó. J., Andres-Lacueva, C., & Llorach, R. (2023). Quantitative plasma profiling by 1H NMR-based metabolomics: Impact of sample treatment. Frontiers in Molecular Biosciences,10, 1125582. 10.3389/fmolb.2023.1125582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallol, R., Amigó, N., Rodríguez, M. A., Heras, M., Vinaixa, M., Plana, N., et al. (2015). Liposcale: A novel advanced lipoprotein test based on 2D diffusion-ordered 1H NMR spectroscopy. Journal of Lipid Research,56(3), 737–746. 10.1194/jlr.D050120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandal, R., Zheng, J., Zhang, L., Oler, E., LeVatte, M. A., Berjanskii, M., et al. (2025). Comprehensive, quantitative analysis of SRM 1950: The NIST human plasma reference material. Analytical Chemistry,97, 667–675. 10.1021/acs.analchem.4c05018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marzocco, S., Di Paola, R., Ribecco, M. T., Sorrentino, R., Domenico, B., Genesio, M., et al. (2004). Effect of methylguanidine in a model of septic shock induced by LPS. Free Radical Research,38(11), 1143–1153. 10.1080/10715760410001725517 [DOI] [PubMed] [Google Scholar]
- Mickiewicz, B., Thompson, G. C., Blackwood, J., Jenne, C. N., Winston, B. W., Vogel, H. J., & Joffe, A. R. (2018). Biomarker phenotype for early diagnosis and triage of sepsis to the pediatric intensive care unit. Scientific Reports,8(1), 16606. 10.1038/s41598-018-35000-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller, T. L., & Wolin, M. J. (1996). Pathways of acetate, propionate, and butyrate formation by the human fecal microbial flora. Applied and Environmental Microbiology,62(5), 1589–1592. 10.1128/aem.62.5.1589-1592.1996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moazzami, A. A., Zhang, J. X., Kamal-Eldin, A., Åman, P., Hallmans, G., Johansson, J. E., & Andersson, S. O. (2011). Nuclear magnetic resonance-based metabolomics enable detection of the effects of a whole grain rye and rye bran diet on the metabolic profile of plasma in prostate cancer patients. Journal of Nutrition,141(12), 2126–2132. 10.3945/jn.111.148239 [DOI] [PubMed] [Google Scholar]
- Mohr, A. E., Jasbi, P., Vander Wyst, K. B., van Woerden, I., Shi, X., Gu, H., et al. (2022). Association of food insecurity on gut microbiome and metabolome profiles in a diverse college-based sample. Scientific Reports,12(1), 14358. 10.1038/s41598-022-18515-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moitra, S., Bandyopadhyay, A., & Lacy, P. (2023). Metabolomics of respiratory diseases. Handbook of Experimental Pharmacology,277, 339–365. 10.1007/164_2022_614 [DOI] [PubMed] [Google Scholar]
- Moussallieh, F. M., Elbayed, K., Chanson, J. B., Rudolf, G., Piotto, M., De Seze, J., & Namer, I. J. (2014). Serum analysis by 1H nuclear magnetic resonance spectroscopy: A new tool for distinguishing neuromyelitis optica from multiple sclerosis. Multiple Sclerosis Journal,20(5), 558–565. 10.1177/1352458513504638 [DOI] [PubMed] [Google Scholar]
- Nagana Gowda, G. A., Gowda, Y. N., & Raftery, D. (2015). Expanding the limits of human blood metabolite quantitation using NMR spectroscopy. Analytical Chemistry,87(1), 706–715. 10.1021/ac503651e [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagana Gowda, G. A., Pascua, V., Hill, L., Djukovic, D., Wang, D., & Raftery, D. (2024). Discovery of hypoxanthine and inosine as robust biomarkers for predicting the preanalytical quality of human plasma and serum for metabolomics. Analytical Chemistry,96(39), 15754–15764. 10.1021/acs.analchem.4c03719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagana Gowda, G. A., Pascua, V., & Raftery, D. (2022). A new limit for blood metabolite analysis using 1H NMR spectroscopy. Journal of Magnetic Resonance Open,12–13, 100082. 10.1016/j.jmro.2022.100082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagana Gowda, G. A., Pascua, V., & Raftery, D. (2023). Anomalous dynamics of labile metabolites in cold human blood detected using 1H NMR spectroscopy. Analytical Chemistry,95(34), 12923–12930. 10.1021/acs.analchem.3c02478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagana Gowda, G. A., & Raftery, D. (2014). Quantitating metabolites in protein precipitated serum using NMR spectroscopy. Analytical Chemistry,86(11), 5433–5440. 10.1021/ac5005103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagana Gowda, G. A., & Raftery, D. (2017). Whole blood metabolomics by 1H NMR spectroscopy provides a new opportunity to evaluate coenzymes and antioxidants. Analytical Chemistry,89(8), 4620–4627. 10.1021/acs.analchem.7b00171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagana Gowda, G. A., & Raftery, D. (2019). Analysis of plasma, serum, and whole blood metabolites using 1H NMR spectroscopy. Methods in Molecular Biology,2037, 17–34. 10.1007/978-1-4939-9690-2_2 [DOI] [PubMed] [Google Scholar]
- Nannini, G., Meoni, G., Amedei, A., & Tenori, L. (2020). Metabolomics profile in gastrointestinal cancers: Update and future perspectives. World Journal of Gastroenterology,26(20), 2514–2532. 10.3748/WJG.V26.I20.2514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navarro, S. L., Nagana Gowda, G. A., Bettcher, L. F., Pepin, R., Nguyen, N., Ellenberger, M., et al. (2023). Demographic, health and lifestyle factors associated with the metabolome in older women. Metabolites,13(4), 514. 10.3390/metabo13040514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicholson, J. K., O’Flynn, M. P., Sadler, P. J., Macleod, A. F., Juul, S. M., & Sönksen, P. H. (1984). Proton-nuclear-magnetic-resonance studies of serum, plasma and urine from fasting normal and diabetic subjects. Biochemical Journal,217(2), 365–375. 10.1042/bj2170365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Org, E., Blum, Y., Kasela, S., Mehrabian, M., Kuusisto, J., Kangas, A. J., et al. (2017). Relationships between gut microbiota, plasma metabolites, and metabolic syndrome traits in the METSIM cohort. Genome Biology,18(1), 70. 10.1186/s13059-017-1194-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otvos, J. D., Shalaurova, I., Wolak-Dinsmore, J., Connelly, M. A., Mackey, R. H., Stein, J. H., & Tracy, R. P. (2015). GlycA: A composite nuclear magnetic resonance biomarker of systemic inflammation. Clinical Chemistry,61(5), 714–723. 10.1373/clinchem.2014.232918 [DOI] [PubMed] [Google Scholar]
- Partula, V., Deschasaux-Tanguy, M., Mondot, S., Victor-Bala, A., Bouchemal, N., Lécuyer, L., et al. (2021). Associations between untargeted plasma metabolomic signatures and gut microbiota composition in the Milieu Intérieur population of healthy adults. British Journal of Nutrition,126(7), 982–992. 10.1017/S0007114520004870 [DOI] [PubMed] [Google Scholar]
- Patt, A., Siddiqui, J., Zhang, B., & Mathé, E. (2019). Integration of metabolomics and transcriptomics to identify gene-metabolite relationships specific to phenotype. Methods in Molecular Biology,1928, 441–468. 10.1007/978-1-4939-9027-6_23 [DOI] [PubMed] [Google Scholar]
- Patti, G. J., Yanes, O., & Siuzdak, G. (2012). Innovation: Metabolomics: The apogee of the omics trilogy. Nature Reviews Molecular Cell Biology,13(4), 263–269. 10.1038/nrm3314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pechlivanis, A., Kostidis, S., Saraslanidis, P., Petridou, A., Tsalis, G., Veselkov, K., et al. (2012). 1H NMR study on the short- and long-term impact of two training programs of sprint running on the metabolic fingerprint of human serum. Journal of Proteome Research,12(1), 470–480. 10.1021/pr300846x [DOI] [PubMed] [Google Scholar]
- Pedrini, M., Cao, B., Nani, J. V. S., Cerqueira, R. O., Mansur, R. B., Tasic, L., et al. (2019). Advances and challenges in development of precision psychiatry through clinical metabolomics on mood and psychotic disorders. Progress in Neuro-Psychopharmacology and Biological Psychiatry,93, 182–188. 10.1016/j.pnpbp.2019.03.010 [DOI] [PubMed] [Google Scholar]
- Pertinhez, T. A., Casali, E., Lindner, L., Spisni, A., Baricchi, R., & Berni, P. (2014). Biochemical assessment of red blood cells during storage by 1H nuclear magnetic resonance spectroscopy. Identification of a biomarker of their level of protection against oxidative stress. Blood Transfusion,12(4), 548–556. 10.2450/2014.0305-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinto, J., et al. (2014). Human plasma stability during handling and storage: Impact on NMR metabolomics. Analyst,139(5), 1168–1177. 10.1039/c3an02188b [DOI] [PubMed] [Google Scholar]
- Porzel, A., Farag, M. A., Mülbradt, J., & Wessjohann, L. A. (2014). Metabolite profiling and fingerprinting of Hypericum species: A comparison of MS and NMR metabolomics. Metabolomics,10(4), 574–588. 10.1007/s11306-013-0609-7 [Google Scholar]
- Powers, R., Andersson, E. R., Bayless, A. L., Brua, R. B., Chang, M. C., Cheng, L. L., et al. (2024). Best practices in NMR metabolomics: Current state. TrAC—Trends in Analytical Chemistry,171, 117478. 10.1016/j.trac.2023.117478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preuss, H. G., & Burris, J. F. (1996). Adverse metabolic effects of antihypertensive drugs implications for treatment. Drug Safety,14(6), 355–364. 10.2165/00002018-199614060-00001 [DOI] [PubMed] [Google Scholar]
- Psychogios, N., Hau, D. D., Peng, J., Guo, A. C., Mandal, R., Bouatra, S., et al. (2011). The human serum metabolome. PLoS ONE,6(2), e16957. 10.1371/journal.pone.0016957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rådjursöga, M., Lindqvist, H. M., Pedersen, A., Karlsson, G. B., Malmodin, D., Brunius, C., et al. (2019). The 1 H NMR serum metabolomics response to a two meal challenge: A cross-over dietary intervention study in healthy human volunteers. Nutrition Journal,18(1), 25. 10.1186/s12937-019-0446-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Remaley, A. T., Rifai, N., & Warnick, G. R. (2006). Lipids, lipoproteins, apolipoproteins, and other cardiovascular risk factors. In Burtis CA, Ashwood ER, & Bruns DE (Eds.), Tietz Textbook of Clinical Chemistry and Molecular Diagnostics (4th ed., pp. 903–982). Philadelphia, Pa: Elsevier Saunders. 10.1016/b978-1-4160-6164-9.00027-5
- Ren, Z., Rajan, C., & Jia, W. (2021). The distinctive serum metabolomes of gastric, esophageal and colorectal cancers. Cancers,13(4), 720. 10.3390/cancers13040720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rief, M., Raggam, R., Rief, P., Metnitz, P., Stojakovic, T., Reinthaler, M., et al. (2022). Comparison of two nuclear magnetic resonance spectroscopy methods for the measurement of lipoprotein particle concentrations. Biomedicines,10(7), 1766. 10.3390/biomedicines10071766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rist, M. J., Roth, A., Frommherz, L., Weinert, C. H., Krüger, R., Merz, B., et al. (2017). Metabolite patterns predicting sex and age in participants of the Karlsruhe Metabolomics and Nutrition (KarMeN) study. PLoS ONE,12(8), e0183228. 10.1371/journal.pone.0183228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts, L. D., Koulman, A., & Griffin, J. L. (2014). Towards metabolic biomarkers of insulin resistance and type 2 diabetes: Progress from the metabolome. The Lancet Diabetes and Endocrinology,2(1), 65–75. 10.1016/S2213-8587(13)70143-8 [DOI] [PubMed] [Google Scholar]
- Roberts, L. D., Souza, A. L., Gerszten, R. E., & Clish, C. B. (2012). Targeted metabolomics. Current Protocols in Molecular Biology. 10.1002/0471142727.mb3002s98 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson, O., Chadeau Hyam, M., Karaman, I., Climaco Pinto, R., Ala-Korpela, M., Handakas, E., et al. (2020). Determinants of accelerated metabolomic and epigenetic aging in a UK cohort. Aging Cell,19(6), e13149. 10.1111/ACEL.13149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothman, D. L., Magnusson, I., Katz, L. D., Shulman, R. G., & Shulman, G. I. (1991). Quantitation of hepatic glycogenolysis and gluconeogenesis in fasting humans with 13C NMR. Science,254(5031), 573–576. 10.1126/science.1948033 [DOI] [PubMed] [Google Scholar]
- Rout, M., Lipfert, M., Lee, B. L., Berjanskii, M., Assempour, N., Fresno, R. V., et al. (2023). MagMet: A fully automated web server for targeted nuclear magnetic resonance metabolomics of plasma and serum. Magnetic Resonance in Chemistry,61(12), 681–704. 10.1002/mrc.5371 [DOI] [PubMed] [Google Scholar]
- Saito, K., Maekawa, K., Pappan, K. L., Urata, M., Ishikawa, M., Kumagai, Y., & Saito, Y. (2014). Differences in metabolite profiles between blood matrices, ages, and sexes among Caucasian individuals and their inter-individual variations. Metabolomics,10(3), 402–413. 10.1007/s11306-013-0591-0 [Google Scholar]
- Saner, C., Harcourt, B. E., Pandey, A., Ellul, S., McCallum, Z., Kao, K.-T., et al. (2019). Sex and puberty-related differences in metabolomic profiles associated with adiposity measures in youth with obesity. Metabolomics,15(5), 75. 10.1007/s11306-019-1537-y [DOI] [PubMed] [Google Scholar]
- Sardeli, A. V., Castro, A., Gadelha, V. B., Dos Santos, W. M., Lord, J. M., Cavaglieri, C. R., & Chacon-Mikahil, M. P. T. (2022). Metabolomic response throughout 16 weeks of combined aerobic and resistance exercise training in older women with Metabolic Syndrome. Metabolites,12(11), 1041. 10.3390/metabo12111041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schicho, R., Nazyrova, A., Shaykhutdinov, R., Duggan, G., Vogel, H. J., & Storr, M. (2010). Quantitative metabolomic profiling of serum and urine in DSS-induced ulcerative colitis of mice by (1)H NMR spectroscopy. Journal of Proteome Research,9(12), 6265–6273. 10.1021/pr100547y [DOI] [PubMed] [Google Scholar]
- Schicho, R., Shaykhutdinov, R., Ngo, J., Nazyrova, A., Schneider, C., Panaccione, R., et al. (2012). Quantitative metabolomic profiling of serum, plasma, and urine by (1)H NMR spectroscopy discriminates between patients with inflammatory bowel disease and healthy individuals. Journal of Proteome Research,11(6), 3344–3357. 10.1021/pr300139q [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schrimpe-Rutledge, A. C., Codreanu, S. G., Sherrod, S. D., & McLean, J. A. (2016). Untargeted metabolomics strategies—Challenges and emerging directions. Journal of the American Society for Mass Spectrometry,27(12), 1897–1905. 10.1007/s13361-016-1469-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultheiss, U. T., Kosch, R., Kotsis, F., Altenbuchinger, M., & Zacharias, H. U. (2021). Chronic kidney disease cohort studies: A guide to metabolome analyses. Metabolites,11(7), 460. 10.3390/metabo11070460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwedes, U., Siebolds, M., & Mertes, G. (2002). Meal-related structured self-monitoring of blood glucose: Effect on diabetes control in non-insulin-treated type 2 diabetic patients. Diabetes Care,25(11), 1928–1932. 10.2337/diacare.25.11.1928 [DOI] [PubMed] [Google Scholar]
- Serkova, N. J., Standiford, T. J., & Stringer, K. A. (2011). The emerging field of quantitative blood metabolomics for biomarker discovery in critical illnesses. American Journal of Respiratory and Critical Care Medicine,184(6), 647–655. 10.1164/rccm.201103-0474CI [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shrestha, A., Müllner, E., Poutanen, K., Mykkänen, H., & Moazzami, A. A. (2017). Metabolic changes in serum metabolome in response to a meal. European Journal of Nutrition,56(2), 671–681. 10.1007/s00394-015-1111-y [DOI] [PubMed] [Google Scholar]
- Silva, R. A., Pereira, T. C. S., Souza, A. R., & Ribeiro, P. R. (2020). 1H NMR-based metabolite profiling for biomarker identification. Clinica Chimica Acta,502, 269–279. 10.1016/j.cca.2019.11.015 [DOI] [PubMed] [Google Scholar]
- Sitole, L. J., Williams, A. A., & Meyer, D. (2013). Metabonomic analysis of HIV-infected biofluids. Molecular BioSystems,9(1), 18–28. 10.1039/c2mb25318f [DOI] [PubMed] [Google Scholar]
- Sliz, E., Kettunen, J., Holmes, M. V., Williams, C. O., Boachie, C., Wang, Q., et al. (2018). Metabolomic consequences of genetic inhibition of PCSK9 compared with statin treatment. Circulation,138(22), 2499–2512. 10.1161/CIRCULATIONAHA.118.034942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snytnikova, O. A., Khlichkina, A. A., Sagdeev, R. Z., & Tsentalovich, Y. P. (2019). Evaluation of sample preparation protocols for quantitative NMR-based metabolomics. Metabolomics,15(6), 84. 10.1007/s11306-019-1545-y [DOI] [PubMed] [Google Scholar]
- Soininen, P., Kangas, A. J., Würtz, P., Suna, T., & Ala-Korpela, M. (2015). Quantitative serum nuclear magnetic resonance metabolomics in cardiovascular epidemiology and genetics. Circulation: Cardiovascular Genetics,8(1), 192–206. 10.1161/CIRCGENETICS.114.000216 [DOI] [PubMed] [Google Scholar]
- Somashekar, B. S., Ijare, O. B., Nagana Gowda, G. A., Ramesh, V., Gupta, S., & Khetrapal, C. L. (2006). Simple pulse-acquire NMR methods for the quantitative analysis of calcium, magnesium and sodium in human serum. Spectrochimica Acta—Part A: Molecular and Biomolecular Spectroscopy,65(2), 254–260. 10.1016/j.saa.2005.10.039 [DOI] [PubMed] [Google Scholar]
- Teruya, T., Chaleckis, R., Takada, J., Yanagida, M., & Kondoh, H. (2019). Diverse metabolic reactions activated during 58-hr fasting are revealed by non-targeted metabolomic analysis of human blood. Scientific Reports,9(1), 854. 10.1038/s41598-018-36674-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thachil, A., Wang, L., Mandal, R., Wishart, D., & Blydt-Hansen, T. (2024). An overview of pre-analytical factors impacting metabolomics analyses of blood samples. Metabolites,14(9), 474. 10.3390/metabo14090474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas, C. E., Wang, R., Adams-Haduch, J., Murphy, S. E., Ueland, P. M., Midttun, Ø., et al. (2020). Urinary cotinine is as good a biomarker as serum cotinine for cigarette smoking exposure and lung cancer risk prediction. Cancer Epidemiology Biomarkers and Prevention,29, 127–132. 10.1158/1055-9965.EPI-19-0653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiziani, S., Emwas, A. H., Lodi, A., Ludwig, C., Bunce, C. M., Viant, M. R., & Günther, U. L. (2008). Optimized metabolite extraction from blood serum for 1H nuclear magnetic resonance spectroscopy. Analytical Biochemistry,377(1), 16–23. 10.1016/j.ab.2008.01.037 [DOI] [PubMed] [Google Scholar]
- Townsend, M. K., Bao, Y., Poole, E. M., Bertrand, K. A., Kraft, P., Wolpin, B. M., et al. (2016). Impact of pre-analytic blood sample collection factors on metabolomics. Cancer Epidemiology Biomarkers and Prevention,25(5), 823–829. 10.1158/1055-9965.EPI-15-1206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trabado, S., Al-Salameh, A., Croixmarie, V., Masson, P., Corruble, E., Fève, B., et al. (2017). The human plasma-metabolome: Reference values in 800 French healthy volunteers; Impact of cholesterol, gender and age. PLoS ONE,12(3), e0173615. 10.1371/journal.pone.0173615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trautwein, C. (2025). Quantitative blood serum IVDr NMR spectroscopy in clinical metabolomics of cancer, neurodegeneration, and internal medicine. Methods in Molecular Biology,2855, 427–443. 10.1007/978-1-0716-4116-3_24 [DOI] [PubMed] [Google Scholar]
- Trimigno, A., Münger, L., Picone, G., Freiburghaus, C., Pimentel, G., Vionnet, N., et al. (2018). GC-MS based metabolomics and NMR spectroscopy investigation of food intake biomarkers for milk and cheese in serum of healthy humans. Metabolites,8(2), 26. 10.3390/metabo8020026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ubaida-Mohien, C., Tanaka, T., Tian, Q., Moore, Z., Moaddel, R., Basisty, N., et al. (2023). Blood biomarkers for healthy aging. Gerontology,69(10), 1167–1174. 10.1159/000530795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ussher, J. R., Elmariah, S., Gerszten, R. E., & Dyck, J. R. B. (2016). The emerging role of metabolomics in the diagnosis and prognosis of cardiovascular disease. Journal of the American College of Cardiology,68(25), 2850–2870. 10.1016/j.jacc.2016.09.972 [DOI] [PubMed] [Google Scholar]
- van den Akker, E. B., Trompet, S., Barkey Wolf, J. J. H., Beekman, M., Suchiman, H. E. D., Deelen, J., et al. (2020). Metabolic age based on the BBMRI-NL 1H-NMR metabolomics repository as biomarker of age-related disease. Circulation. Genomic and Precision Medicine,13(5), 541–547. 10.1161/CIRCGEN.119.002610 [DOI] [PubMed] [Google Scholar]
- Van Doorn, M., Vogels, J., Tas, A., Van Hoogdalem, E. J., Burggraaf, J., Cohen, A., & Van Der Greef, J. (2007). Evaluation of metabolite profiles as biomarkers for the pharmacological effects of thiazolidinediones in type 2 diabetes mellitus patients and healthy volunteers. British Journal of Clinical Pharmacology,63(5), 562–574. 10.1111/j.1365-2125.2006.02816.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanhove, K., Derveaux, E., Mesotten, L., Thomeer, M., Criel, M., Mariën, H., & Adriaensens, P. (2022). Unraveling the rewired metabolism in lung cancer using quantitative NMR metabolomics. International Journal of Molecular Sciences. 10.3390/ijms23105602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vázquez-Fresno, R., Llorach, R., Urpi-Sarda, M., Lupianez-Barbero, A., Estruch, R., Corella, D., et al. (2015). Metabolomic pattern analysis after Mediterranean diet intervention in a nondiabetic population: A 1- and 3-year follow-up in the PREDIMED study. Journal of Proteome Research,14(1), 531–540. 10.1021/pr5007894 [DOI] [PubMed] [Google Scholar]
- Vignoli, A., Ghini, V., Meoni, G., Licari, C., Takis, P. G., Tenori, L., et al. (2019). High-throughput metabolomics by 1D NMR. Angewandte Chemie - International Edition,58(4), 968–994. 10.1002/anie.201804736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vignoli, A., Risi, E., McCartney, A., Migliaccio, I., Moretti, E., Malorni, L., et al. (2021). Precision oncology via NMR-based metabolomics: A review on breast cancer. International Journal of Molecular Sciences,22(9), 4687. 10.3390/ijms22094687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallmeier, J., Samol, C., Ellmann, L., Zacharias, H. U., Vogl, F. C., Garcia, M., et al. (2017). Quantification of metabolites by NMR spectroscopy in the presence of protein. Journal of Proteome Research,16(4), 1784–1796. 10.1021/acs.jproteome.7b00057 [DOI] [PubMed] [Google Scholar]
- Wang, F., Debik, J., Andreassen, T., Euceda, L. R., Haukaas, T. H., Cannet, C., et al. (2019). Effect of repeated freeze-thaw cycles on NMR-measured lipoproteins and metabolites in biofluids. Journal of Proteome Research,18(10), 3681–3688. 10.1021/acs.jproteome.9b00343 [DOI] [PubMed] [Google Scholar]
- Wang, F., Tessier, A. J., Liang, L., Wittenbecher, C., Haslam, D. E., Fernández-Duval, G., et al. (2023). Plasma metabolomic profiles associated with mortality and longevity in a prospective analysis of 13,512 individuals. Nature Communications,14(1), 5744. 10.1038/s41467-023-41515-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, Z., Pisano, S., Ghini, V., Kadeřávek, P., Zachrdla, M., Pelupessy, P., et al. (2021). Detection of metabolite-protein interactions in complex biological samples by high-resolution relaxometry: Toward interactomics by NMR. Journal of the American Chemical Society,143(25), 9393–9404. 10.1021/jacs.1c01388 [DOI] [PubMed] [Google Scholar]
- Wevers, R. A., Engelke, U., & Heerschap, A. (1994). High-resolution 1H-NMR spectroscopy of blood plasma for metabolic studies. Clinical Chemistry. 10.1093/clinchem/40.7.1245 [PubMed] [Google Scholar]
- Wishart, D. S. (2016). Emerging applications of metabolomics in drug discovery and precision medicine. Nature Reviews Drug Discovery,15(7), 473–484. 10.1038/nrd.2016.32 [DOI] [PubMed] [Google Scholar]
- Wishart, D. S. (2022). Metabolomics and the multi-omics view of cancer. Metabolites,12(2), 154. 10.3390/METABO12020154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wishart, D. S., Cheng, L. L., Copié, V., Edison, A. S., Eghbalnia, H. R., Hoch, J. C., et al. (2022a). NMR and metabolomics—A roadmap for the future. Metabolites,12(8), 678. 10.3390/METABO12080678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wishart, D. S., Guo, A., Oler, E., Wang, F., Anjum, A., Peters, H., et al. (2022). HMDB 5.0: The human metabolome database for 2022. Nucleic Acids Research. 10.1093/nar/gkab1062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wishart, D. S., Rout, M., Lee, B. L., Berjanskii, M., LeVatte, M., & Lipfert, M. (2022c). Practical aspects of NMR-based metabolomics. Handbook of Experimental Pharmacology,277, 1–41. 10.1007/164_2022_613 [DOI] [PubMed] [Google Scholar]
- World Health Organization. (2012). Guidelines on assessing donor suitability for blood donation. Blood Donor Selection. [PubMed]
- Wu, Q., Huang, Q.-X., Zeng, H.-L., Ma, S., Lin, H.-D., Xia, M.-F., et al. (2021). Prediction of metabolic disorders using NMR-based metabolomics: The shanghai changfeng study. Phenomics,1(4), 186–198. 10.1007/s43657-021-00021-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wulaningsih, W., Proitsi, P., Wong, A., Kuh, D., & Hardy, R. (2019). Metabolomic correlates of central adiposity and earlier-life body mass index. Journal of Lipid Research,60(6), 1136–1143. 10.1194/jlr.P085944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Würtz, P., Cook, S., Wang, Q., Tiainen, M., Tynkkynen, T., Kangas, A. J., et al. (2016). Metabolic profiling of alcohol consumption in 9778 young adults. International Journal of Epidemiology,45(5), 1493–1506. 10.1093/ije/dyw175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Würtz, P., Kangas, A. J., Soininen, P., Lawlor, D. A., Davey Smith, G., & Ala-Korpela, M. (2017). Quantitative serum nuclear magnetic resonance metabolomics in large-scale epidemiology: A primer on—Omic technologies. American Journal of Epidemiology,186(9), 1084–1096. 10.1093/aje/kwx016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin, P., Peter, A., Franken, H., Zhao, X., Neukamm, S. S., Rosenbaum, L., et al. (2013). Preanalytical aspects and sample quality assessment in metabolomics studies of human blood. Clinical Chemistry,59(5), 833–845. 10.1373/clinchem.2012.199257 [DOI] [PubMed] [Google Scholar]
- Yu, Z., Kastenmüller, G., He, Y., Belcredi, P., Möller, G., Prehn, C., et al. (2011). Differences between human plasma and serum metabolite profiles. PLoS ONE,6(7), e21230. 10.1371/JOURNAL.PONE.0021230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zacharias, H. U., Hochrein, J., Vogl, F. C., Schley, G., Mayer, F., Jeleazcov, C., et al. (2015). Identification of plasma metabolites prognostic of acute kidney injury after cardiac surgery with cardiopulmonary bypass. Journal of Proteome Research,14(7), 2897–2905. 10.1021/acs.jproteome.5b00219 [DOI] [PubMed] [Google Scholar]
- Zacharias, H. U., Kaleta, C., Cossais, F., Schaeffer, E., Berndt, H., Best, L., et al. (2022). Microbiome and metabolome insights into the role of the gastrointestinal–brain axis in Parkinson’s and Alzheimer’s disease: Unveiling potential therapeutic targets. Metabolites,12(12), 1222. 10.3390/metabo12121222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeleznik, O. A., Kang, J. H., Lasky-Su, J., Eliassen, A. H., Frueh, L., Clish, C. B., et al. (2023). Plasma metabolite profile for primary open-angle glaucoma in three US cohorts and the UK Biobank. Nature Communications,14(1), 2860. 10.1038/s41467-023-38466-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, A., Sun, H., & Wang, X. (2012). Serum metabolomics as a novel diagnostic approach for disease: A systematic review. Analytical and Bioanalytical Chemistry,404(4), 1239–1245. 10.1007/s00216-012-6117-1 [DOI] [PubMed] [Google Scholar]
- Zhang, Y., Lin, L., Xu, Y., Lin, Y., Jin, Y., & Zheng, C. (2013). 1H NMR-based spectroscopy detects metabolic alterations in serum of patients with early-stage ulcerative colitis. Biochemical and Biophysical Research Communications,433(4), 547–551. 10.1016/j.bbrc.2013.03.012 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data is provided within the supplementary information file.