Abstract
Hepatocellular carcinoma (HCC) is a highly aggressive malignancy with poor prognosis, driven by complex molecular mechanisms that remain inadequately understood. Among these, the ubiquitin–proteasome system plays a crucial role in regulating protein stability and function, with E3 ubiquitin ligases emerging as key players in cancer progression. Here, we identify Tripartite Motif-containing 6 (TRIM6), an E3 ubiquitin ligase, as a critical regulator of HCC metastasis. We demonstrate that TRIM6 is significantly upregulated in HCC tissues and correlates with poor overall survival. Mechanistically, we uncover that STAT3 directly regulates TRIM6 by binding to its promoter and enhancing its transcription. Functionally, TRIM6 promotes epithelial-mesenchymal transition (EMT) and cell invasion by upregulating the key EMT transcription factor Snail1. Importantly, we reveal that TRIM6 interacts with and ubiquitinates DDX58 (RIG-I), leading to its proteasomal degradation. The degradation of DDX58 by TRIM6 alleviates its inhibitory effects on Snail1, thereby facilitating EMT and enhancing the invasive potential of HCC cells. These findings establish the STAT3-TRIM6-DDX58-Snail1 axis as a pivotal pathway in HCC progression, offering novel insights into the molecular underpinnings of HCC metastasis and highlighting TRIM6 as a potential therapeutic target and prognostic biomarker in HCC.
Keywords: Hepatocellular carcinoma, TRIM6, E3 ubiquitin ligase, STAT3, DDX58, Epithelial-mesenchymal transition (EMT), Metastasis
Subject terms: Oncogenes, Cell migration, Cell signalling, Proteolysis, Bioinformatics
Introduction
Hepatocellular carcinoma (HCC) is one of the most prevalent and deadly forms of cancer worldwide, accounting for the majority of primary liver cancers1. Despite advances in early detection and therapeutic strategies, HCC remains a major global health challenge due to its aggressive nature, high recurrence rates, and poor prognosis. The molecular mechanisms underlying HCC progression and metastasis are complex and multifaceted, involving dysregulated signaling pathways, genomic instability, and a hostile tumor microenvironment2. Identifying the key molecular drivers of HCC is crucial for developing targeted therapies that can improve patient outcomes.
Among the various molecular alterations in HCC, the dysregulation of the ubiquitin–proteasome system (UPS) has emerged as a significant contributor to tumorigenesis and cancer progression3. The UPS is responsible for the selective degradation of proteins, thereby regulating a wide array of cellular processes, including cell cycle progression, apoptosis, and signal transduction4. E3 ubiquitin ligases, which confer specificity to the ubiquitination process, are particularly important in maintaining cellular homeostasis. Dysregulation of E3 ligases has been implicated in the development of various cancers, including HCC, by promoting the degradation of tumor suppressors and stabilizing oncogenic proteins5.
Tripartite motif-containing protein 6 (TRIM6) is a member of the TRIM family of E3 ubiquitin ligases, characterized by a conserved tripartite motif comprising a RING finger domain, one or two B-box domains, and a coiled-coil region6. TRIM proteins are involved in numerous biological processes, including antiviral responses, immune regulation, and cell proliferation7,8. However, the role of TRIM6 in cancer, particularly in HCC, remains largely unexplored. Emerging evidence suggests that TRIM6 may play a pivotal role in cancer progression by modulating key signaling pathways involved in cell survival, proliferation, and metastasis9–11.
Epithelial-mesenchymal transition (EMT) is a well-recognized process by which epithelial cells acquire mesenchymal characteristics, leading to enhanced migratory and invasive capabilities12,13. EMT is critical for the progression of cancers, including HCC, as it facilitates the dissemination of cancer cells from the primary tumor to distant organs14. Central to the regulation of EMT are transcription factors such as Snail1, which repress epithelial markers like E-cadherin and induce the expression of mesenchymal markers such as N-cadherin and vimentin15,16. The molecular mechanisms that regulate Snail1 expression and activity are complex and involve multiple signaling pathways, including those mediated by E3 ubiquitin ligases17.
In addition to its well-established role in antiviral immunity, DDX58 (RIG-I) has recently been implicated in the regulation of EMT and cancer cell migration. DDX58 is an RNA helicase that plays a key role in the innate immune response by detecting viral RNA and initiating downstream signaling cascades18. Interestingly, DDX58 has been shown to inhibit EMT and metastasis in various cancers19,20. However, the regulatory mechanisms governing DDX58 stability and function in cancer cells remain poorly understood.
Given the potential interplay between TRIM6 and DDX58 in HCC, we hypothesized that TRIM6 may promote HCC progression by regulating DDX58 through ubiquitination, thereby influencing EMT and metastatic potential. In this study, we aimed to elucidate the role of TRIM6 in HCC and explore its potential as a therapeutic target. We employed a combination of bioinformatic analyses, in vitro experiments, and molecular biology techniques to investigate the expression and function of TRIM6 in HCC. Our findings reveal that TRIM6 is significantly upregulated in HCC and plays a critical role in promoting EMT and cell migration through the degradation of DDX58 and the subsequent regulation of Snail1. These insights contribute to our understanding of the molecular mechanisms driving HCC progression and highlight the TRIM6-DDX58-Snail1 axis as a novel target for therapeutic intervention.
Materials and methods
Identification of differentially expressed genes (DEGs)
To identify differentially expressed genes (DEGs) associated with HCC, we utilized two publicly available microarray datasets: GSE50579 and GSE112791. Raw microarray data were downloaded from the Gene Expression Omnibus (GEO) database and preprocessed using the R programming language with the Bioconductor packages. Background correction and normalization were performed using the Robust Multi-array Average (RMA) method. For differential expression analysis, the limma package was employed to compare gene expression profiles between HCC tissues and normal liver tissues in both datasets. The criteria for identifying DEGs were set as a p value < 0.05 and |log2 fold change (log2FC)|> 1.5. Volcano plots were generated using the ggplot2 package in R to visualize the distribution of upregulated and downregulated genes. Venn diagrams were constructed to identify common DEGs across the datasets using the VennDiagram package.
Enrichment analysis
To gain insights into the biological processes and pathways associated with TRIM6, we performed Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analyses21. The clusterProfiler package in R was used to analyze the enrichment of GO terms and KEGG pathways among the DEGs identified in TRIM6_Knockout versus wild-type (WT) cells (GSE138841 dataset). DEGs with a p value < 0.05 and a false discovery rate (FDR) < 0.25 were considered significantly enriched. The results were visualized using bar plots and dot plots generated by the enrichplot package in R.
Transcription factor binding site prediction
To predict potential transcription factors regulating TRIM6 expression, we used the TF_Target_Finder online tool (https://jingle.shinyapps.io/TF_Target_Finder/). This tool integrates data from several databases, including KnockTF, CHEA, GTRD, ChIP_Atlas, and the cor_TCGA database. STAT3 was identified as a potential transcription factor regulating TRIM6.
Molecular docking simulation
Molecular docking simulations were performed to predict the binding interface between TRIM6 and DDX58. The three-dimensional structures of TRIM6 and DDX58 were obtained from the Protein Data Bank (PDB) or predicted using the Alphafold3 model if the crystal structures were unavailable. The docking simulations were conducted using AutoDock Vina, a widely used docking software for predicting protein–ligand interactions. The grid box for docking was centered around the predicted interaction interface based on the PPI network analysis, with grid dimensions large enough to cover all potential binding sites. Docking parameters were set to default values, and the docking results were analyzed to identify the most favorable binding conformation based on the binding energy scores. The binding interactions between TRIM6 and DDX58 were visualized using PyMOL software, highlighting key residues involved in the interaction, such as ASN-409, PRO-429, and ARG-431 on TRIM6. These residues were predicted to form stable hydrogen bonds and hydrophobic interactions with DDX58, supporting the hypothesis that TRIM6 may regulate DDX58 through direct binding and subsequent ubiquitination.
Kaplan–Meier survival analysis
To assess the prognostic significance of TRIM6 expression in HCC, Kaplan–Meier survival analysis was performed using the Kaplan–Meier Plotter database (http://kmplot.com/). HCC patients were stratified into high and low TRIM6 expression groups based on the median expression value. Overall survival (OS) was compared between the two groups using the log-rank test, and a p value < 0.05 was considered statistically significant.
Cell lines and culture conditions
HCC cell lines HCCLM3 and Huh-7 were obtained from the Cell Bank of the Chinese Academy of Sciences (Shanghai, China). Cells were cultured in Dulbecco’s Modified Eagle Medium (DMEM, Gibco) supplemented with 10% fetal bovine serum (FBS, Gibco), 100 U/mL penicillin, and 100 µg/mL streptomycin (Gibco) at 37 °C in a humidified atmosphere containing 5% CO₂. Cells were passaged every 2–3 days and maintained at 70–80% confluency for all experiments.
Gene knockdown and overexpression in HCC cells
TRIM6 and DDX58 knockdown were achieved using lentivirus-delivered short hairpin RNAs (shRNAs) specifically targeting human TRIM6 or DDX58 mRNA. Two independent shRNA sequences for each gene were cloned into the pLKO.1-puro vector (Addgene). Lentiviral particles were produced in HEK293T cells by co-transfection with standard packaging plasmids using Lipofectamine 3000 (Invitrogen). Target HCC cell lines (HCCLM3 and Huh-7) were infected with viral supernatants in the presence of 8 μg/mL polybrene, followed by selection with 2 μg/mL puromycin for 48–72 h. Knockdown efficiency was confirmed by qRT-PCR and Western blot analysis. TRIM6 and DDX58 overexpression were performed by subcloning the full-length human TRIM6 or DDX58 coding sequences into the pcDNA3.1(+) mammalian expression vector (Invitrogen). Transient transfection of expression plasmids was carried out using Lipofectamine 3000 following the manufacturer’s instructions. Cells were harvested 48 h post-transfection for subsequent assays. Overexpression efficiency was validated by immunoblotting.
RNA extraction and quantitative real-time PCR (qRT-PCR)
Total RNA was extracted from HCCLM3 and Huh-7 cells using TRIzol reagent (Invitrogen) according to the manufacturer’s instructions. The concentration and purity of RNA were measured using a NanoDrop 2000 spectrophotometer (Thermo Fisher Scientific). cDNA was synthesized from 1 µg of total RNA using the PrimeScript RT reagent Kit with gDNA Eraser (Takara) following the manufacturer’s protocol. Quantitative real-time PCR (qRT-PCR) was performed using TB Green Premix Ex Taq II (Takara) on a StepOnePlus Real-Time PCR System (Applied Biosystems). The PCR conditions were as follows: initial denaturation at 95 °C for 30 s, followed by 40 cycles of denaturation at 95 °C for 5 s, annealing at 60 °C for 30 s, and extension at 72 °C for 30 s. The relative expression levels of target genes were normalized to GAPDH as the internal control, and the fold change in gene expression was calculated using the 2−ΔΔCt method.
Western blotting
Cells were lysed in RIPA buffer (Beyotime) containing protease and phosphatase inhibitors (Roche). Protein concentrations were determined using the BCA Protein Assay Kit (Thermo Fisher Scientific). Equal amounts of protein (20–40 µg) were separated by SDS-PAGE on 10% or 12% polyacrylamide gels and transferred onto polyvinylidene fluoride (PVDF) membranes (Millipore). Membranes were blocked with 5% non-fat milk in TBS-T (Tris-buffered saline with 0.1% Tween-20) for 1 h at room temperature and incubated overnight at 4 °C with the following primary antibodies: Anti-TRIM6 (1:1000, Abcam, ab141413), Anti-DDX58 (1:1000, Cell Signaling Technology, #4200), Anti-Snail1 (1:1000, Cell Signaling Technology, #3879), Anti-GAPDH (1:2000, Proteintech, 60004-1-Ig), Anti-β-Actin (1:2000, Proteintech, 66009-1-Ig). After washing with TBS-T, membranes were incubated with horseradish peroxidase-conjugated secondary antibodies (1:5000, Jackson ImmunoResearch) for 1 h at room temperature. Protein bands were visualized using the ECL Plus Western Blotting Detection System (GE Healthcare) and detected with the ChemiDoc MP Imaging System (Bio-Rad). Densitometric analysis was performed using ImageJ software.
Luciferase reporter assay
The TRIM6 promoter region, including putative STAT3 binding sites, was amplified by PCR from human genomic DNA and cloned into the pGL3-Basic vector (Promega) to generate the wild-type TRIM6 promoter luciferase reporter construct (TRIM6-WT). Site-directed mutagenesis was performed to create mutant constructs with disrupted STAT3 binding sites (TRIM6-Mut1, TRIM6-Mut2, and TRIM6-Mut3) using the QuikChange II XL Site-Directed Mutagenesis Kit (Agilent Technologies). HCCLM3 and Huh-7 cells were co-transfected with 500 ng of the luciferase reporter construct (WT or mutant), 100 ng of Renilla luciferase plasmid (pRL-TK, Promega) as an internal control, and 500 ng of STAT3 overexpression plasmid or control vector using Lipofectamine 3000 (Invitrogen) according to the manufacturer’s instructions. After 48 h, luciferase activity was measured using the Dual-Luciferase Reporter Assay System (Promega) on a GloMax 96 Microplate Luminometer (Promega). Firefly luciferase activity was normalized to Renilla luciferase activity, and the results were expressed as relative luciferase activity.
Chromatin immunoprecipitation (ChIP) assay
Chromatin immunoprecipitation (ChIP) was performed using the EZ-Magna ChIP A/G Kit (Millipore) following the manufacturer’s protocol. Briefly, HCCLM3 and Huh-7 cells were cross-linked with 1% formaldehyde for 10 min at room temperature, followed by quenching with 125 mM glycine. Cells were then lysed, and the chromatin was sonicated to obtain DNA fragments of approximately 200–1000 bp. The sonicated chromatin was incubated overnight at 4 °C with 5 µg of anti-STAT3 antibody (Cell Signaling Technology, #4904) or normal rabbit IgG as a negative control. Protein-DNA complexes were precipitated using protein A/G magnetic beads and washed extensively. The cross-links were reversed by incubation at 65 °C for 4 h, and the DNA was purified using the provided spin columns. The presence of STAT3 binding sites in the TRIM6 promoter region was analyzed by qPCR using specific primers: The qPCR conditions were identical to those described for qRT-PCR. The data were analyzed using the 2−ΔΔCt method, and the results were presented as the percentage of input chromatin.
Co-immunoprecipitation (Co-IP)
HCCLM3 and Huh-7 cells were lysed in IP lysis buffer (Thermo Fisher Scientific) containing protease and phosphatase inhibitors. The lysates were pre-cleared with protein A/G magnetic beads (Pierce) for 1 h at 4 °C, followed by incubation with 2 µg of anti-TRIM6 antibody (Abcam, ab141413) or anti-DDX58 antibody (Cell Signaling Technology, #4200) overnight at 4 °C. Normal rabbit IgG (Millipore) was used as a negative control. Immune complexes were captured by incubation with protein A/G magnetic beads for 2 h at 4 °C, and the beads were washed extensively with lysis buffer. Bound proteins were eluted by boiling in SDS loading buffer and analyzed by Western blotting as described above.
Transwell inviasion assay
The invasive capacity of HCCLM3 and Huh-7 cells was evaluated using 24-well transwell invasion chambers with 8 µm pore size polycarbonate membranes (Corning), pre-coated with Matrigel (BD Biosciences) to simulate the extracellular matrix barrier. A total of 5 × 104 cells suspended in serum-free DMEM were seeded into the upper chamber, while the lower chamber was filled with DMEM containing 10% FBS to act as a chemoattractant. Following 24 h of incubation at 37 °C, non-invaded cells on the upper surface of the membrane were carefully removed using a cotton swab. The invaded cells on the lower surface of the membrane were fixed with 4% paraformaldehyde, stained with 0.1% crystal violet, and subsequently quantified under a light microscope. The number of invaded cells was counted in five randomly selected fields to assess the invasive potential of the cells.
Statistical analysis
All experiments were performed in triplicate and independently repeated at least three times. Data are presented as the mean ± standard deviation (SD). Statistical comparisons were made using Student’s t-test for two-group comparisons or one-way ANOVA followed by Tukey’s post hoc test for multiple comparisons. Kaplan–Meier survival analysis was conducted using the log-rank test. A p value of < 0.05 was considered statistically significant. Statistical analyses were performed using GraphPad Prism 8.0 software.
Results
Upregulation of TRIM6 is associated with poor prognosis in hepatocellular carcinoma
To identify key regulatory genes in hepatocellular carcinoma (HCC), we conducted differential expression analysis on two microarray datasets, GSE50579 and GSE112791, which profile gene expression in HCC versus normal liver tissues. The GSE50579 dataset includes 67 tumor samples and 10 normal samples, while GSE112791 comprises 183 tumor samples and 15 normal samples. Applying a stringent cutoff of p < 0.05 and |log2FC|> 1.5, we identified 667 upregulated and 587 downregulated genes in HCC tissues compared to normal tissues in the GSE50579 dataset (Supplementary Table 1). These differentially expressed genes (DEGs) were visualized using volcano plots (Fig. 1A, B). Similarly, analysis of the GSE112791 dataset revealed 245 upregulated and 362 downregulated genes (Supplementary Table 2), also depicted in volcano plots (Fig. 1C, D). To identify potential key drivers of HCC tumorigenesis, we intersected the upregulated genes from both datasets with a curated list of E3 ubiquitin ligases. This approach identified TRIM6 as the only gene consistently upregulated in both datasets (Fig. 1E). Validation in the TCGA-LIHC cohort confirmed that TRIM6 expression is significantly elevated in HCC tissues compared to normal liver tissues (Fig. 1F). To explore the clinical relevance of TRIM6 expression, we used the UALCAN platform, which demonstrated a strong correlation between TRIM6 mRNA levels and both tumor grade and stage in HCC samples (Fig. 1G, H). These findings were further validated across three independent HCC cohorts (GSE62232, GSE84402, and GSE45267), where TRIM6 expression was consistently higher in tumor tissues compared to normal liver tissues (Fig. 1I, K). Kaplan–Meier survival analysis using the Kaplan–Meier Plotter database revealed that elevated TRIM6 expression is significantly associated with poorer overall survival in HCC patients (Fig. 1L). Together, these results indicate that TRIM6 is not only upregulated in HCC but also closely linked to adverse clinical outcomes, positioning TRIM6 as a potential prognostic biomarker and therapeutic target in HCC.
Fig. 1.
Upregulation of TRIM6 is associated with poor prognosis in hepatocellular carcinoma. (a) Volcano plot showing differentially expressed genes in the GSE50579 dataset. (B) Bar chart depicting the number of upregulated (667) and downregulated (587) genes identified in the GSE50579 dataset based on differential expression analysis. (C) Volcano plot showing differentially expressed genes in the GSE112791 dataset. (D) Bar chart depicting the number of upregulated (245) and downregulated (362) genes identified in the GSE112791 dataset based on differential expression analysis. (E) Venn diagram showing the intersection of upregulated genes from GSE50579 and GSE112791 with a curated list of E3 ubiquitin ligase genes, identifying TRIM6 as the only common gene. (F) Box plot showing TRIM6 expression in HCC tissues compared to normal liver tissues in the TCGA-LIHC cohort. (G) Box plot showing the correlation between TRIM6 expression and tumor grade in HCC samples. (H) Box plot showing the correlation between TRIM6 expression and tumor stage in HCC samples. (I–K) Box plots showing TRIM6 expression in three independent HCC cohorts (GSE62232, GSE84402, GSE45267) compared to normal liver tissues. (L) Kaplan–Meier survival curve showing the association between high TRIM6 expression and poor overall survival in HCC patients.
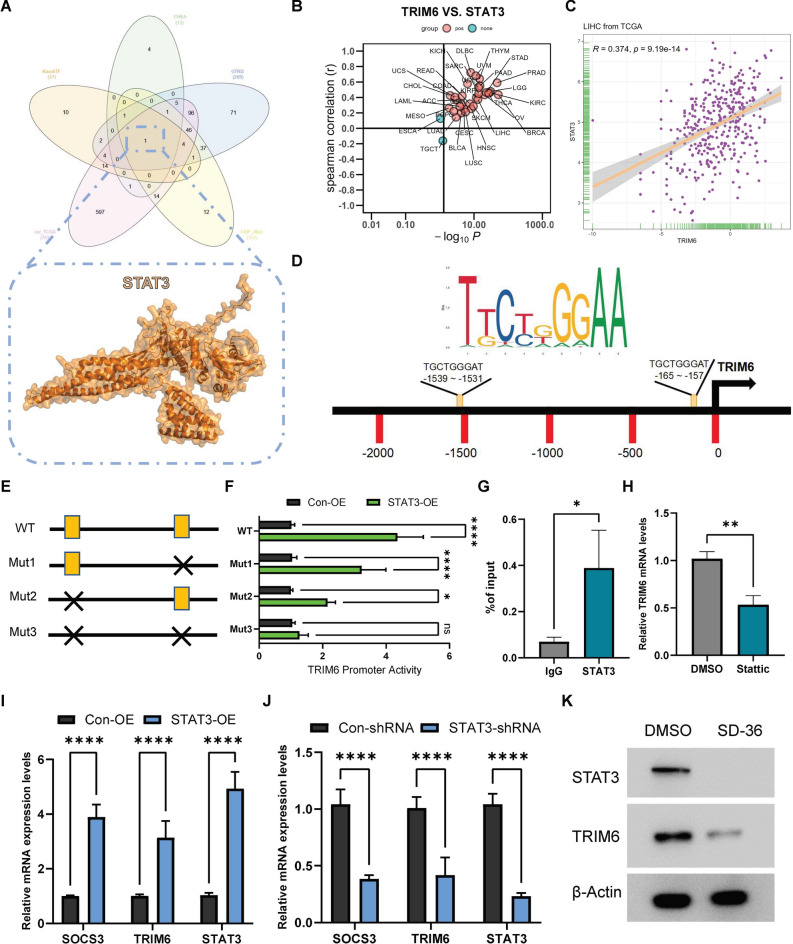
STAT3 directly regulates TRIM6 expression in hepatocellular carcinoma
To further explore the mechanisms underlying TRIM6 upregulation in hepatocellular carcinoma (HCC), we sought to identify its potential upstream regulatory factors. Using the TF_Target_Finder online tool (https://jingle.shinyapps.io/TF_Target_Finder/), we conducted a comprehensive transcription factor prediction analysis. This analysis integrated data from multiple sources, including KnockTF, CHEA, GTRD, ChIP_Atlas, and the cor_TCGA database. Through this integrative approach, we identified STAT3 as a potential transcription factor that may regulate TRIM6 expression (Fig. 2A). To confirm the regulatory relationship between STAT3 and TRIM6, we first analyzed the correlation between TRIM6 and STAT3 expression across multiple cancer types using TCGA data. Specifically, a scatter plot analysis of TRIM6 versus STAT3 expression levels in LIHC revealed a significant positive correlation, underscoring a strong association between STAT3 and TRIM6 expression (Fig. 2B, C). To determine whether STAT3 directly regulates TRIM6 transcription, we conducted an in silico analysis of the TRIM6 promoter region, identifying two putative STAT3 binding sites (Fig. 2D). To experimentally validate this regulatory relationship, we constructed luciferase reporter plasmids containing either the wild-type (WT) TRIM6 promoter or mutant promoters with disrupted STAT3 binding sites (Mut1, Mut2, and Mut3). The WT TRIM6 promoter construct included the entire promoter region with both intact STAT3 binding sites, while the mutant constructs contained site-specific mutations designed to disrupt STAT3 binding. HCCLM3 cells were co-transfected with these luciferase reporter constructs along with a STAT3 expression plasmid. Luciferase activity assays showed a significant increase in promoter activity with the WT TRIM6 construct upon STAT3 overexpression, indicating that STAT3 enhances TRIM6 promoter activity through direct binding (Fig. 2F). In contrast, mutation of one (Mut1 or Mut2) or both (Mut3) STAT3 binding sites resulted in a marked reduction in luciferase activity, particularly in the Mut3 construct, where both binding sites were disrupted (Fig. 2E, F). These findings suggest that STAT3’s transcriptional regulation of TRIM6 is directly dependent on its ability to bind specific sites within the TRIM6 promoter. To further confirm STAT3 binding to the TRIM6 promoter in vivo, we performed chromatin immunoprecipitation (ChIP) assays in HCCLM3 cells. The ChIP results confirmed that STAT3 binds directly to the TRIM6 promoter region (Fig. 2G). Additionally, treatment of HCC cells with Stattic, a specific STAT3 inhibitor22, led to a significant reduction in TRIM6 mRNA levels, underscoring the necessity of STAT3 for TRIM6 expression (Fig. 2H). Overexpression of STAT3 significantly elevated the mRNA levels of TRIM6, as well as SOCS3, a known STAT3 target gene23, confirming that STAT3 positively regulates TRIM6 transcription (Fig. 2I). Conversely, STAT3 knockdown via shRNA resulted in a marked reduction in the mRNA levels of both TRIM6 and SOCS3, indicating that STAT3 is crucial for sustaining TRIM6 expression (Fig. 2J). Furthermore, treatment with SD-36, a specific STAT3-targeted degrader24,25, resulted in substantial reduction of STAT3 protein levels, which in turn led to a significant decrease in TRIM6 protein levels (Fig. 2K). These findings underscore STAT3 as a key regulator of TRIM6 expression at both the mRNA and protein levels in HCC cells.
Fig. 2.
STAT3 directly regulates TRIM6 expression in hepatocellular carcinoma. (A) Identification of STAT3 as a potential upstream regulator of TRIM6 using the TF_Target_Finder tool, which integrates data from KnockTF, CHEA, GTRD, ChIP_Atlas, and cor_TCGA databases. (B) Scatter plot demonstrating the correlation between TRIM6 and STAT3 expression levels across different cancer types, highlighting the significant positive correlation in LIHC. (C) Correlation analysis of TRIM6 and STAT3 expression in LIHC, indicating a positive correlation with a linear regression line. (D) Schematic representation of the TRIM6 promoter region, showing the two predicted STAT3 binding sites. Red bars represent the positions of the putative binding sites. (E) Diagram of the wild-type (WT) and mutated (Mut1, Mut2, Mut3) TRIM6 promoter constructs used in luciferase reporter assays. The WT construct includes the full promoter region with both intact STAT3 binding sites, while the mutant constructs have specific mutations that disrupt one (Mut1, Mut2) or both (Mut3) STAT3 binding sites. (F) Luciferase reporter assay results showing TRIM6 promoter activity in STAT3-overexpressing (STAT3-OE) cells compared to control cells (Con-OE). Overexpression of STAT3 significantly increases luciferase activity in the WT construct, indicating enhanced TRIM6 promoter activity. In contrast, mutations in either of the STAT3 binding sites (Mut1, Mut2) lead to a partial reduction in luciferase activity, while mutation of both sites (Mut3) results in a substantial decrease in promoter activity, demonstrating that both STAT3 binding sites are crucial for full activation of the TRIM6 promoter. Statistical significance is indicated by *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001. (G) Chromatin immunoprecipitation (ChIP) assay confirming STAT3 binding to the TRIM6 promoter in HCC cells. *p < 0.05. (H) Relative TRIM6 mRNA levels in HCC cells treated with DMSO or the STAT3 inhibitor Stattic, showing that inhibition of STAT3 reduces TRIM6 expression. **p < 0.01. (I) Relative mRNA expression levels of SOCS3, TRIM6, and STAT3 in STAT3-overexpressing (STAT3-OE) and control (Con-OE) cells, as determined by qRT-PCR. Overexpression of STAT3 significantly increases the expression of TRIM6 and SOCS3. ****p < 0.0001. (J) Relative mRNA expression levels of SOCS3, TRIM6, and STAT3 in STAT3-knockdown (STAT3-shRNA) and control (Con-shRNA) cells. Knockdown of STAT3 leads to a significant reduction in TRIM6 and SOCS3 mRNA levels. ****p < 0.0001. (K) Western blot analysis of STAT3 and TRIM6 protein levels in HCC cells treated with DMSO (control) or the STAT3-targeted degrader SD-36. Overexpression of STAT3 increases TRIM6 protein levels, while STAT3 knockdown or targeted degradation by SD-36 significantly reduces TRIM6 protein levels. β-Actin serves as the loading control.
TRIM6 promotes epithelial-mesenchymal transition (EMT) and cell invasion in HCC
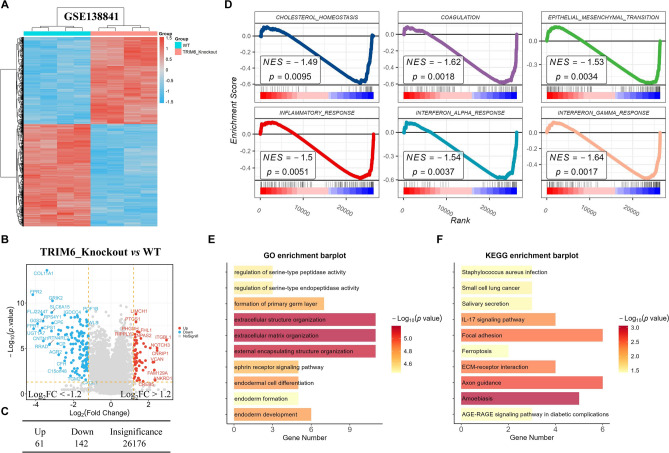
To investigate the potential mechanisms by which TRIM6 influences cancer progression, we utilized the GSE138841 dataset to analyze gene expression differences between the TRIM6_Knockout group and the WT (wild-type) group. Differentially expressed genes were identified using stringent criteria (p < 0.05, |log2FC|> 1.2), and the results were visualized using a heatmap (Fig. 3A) and a volcano plot (Fig. 3B). The analysis revealed that in the TRIM6_Knockout group, 61 genes were upregulated, and 142 genes were downregulated compared to the WT group (Fig. 3C and Supplementary Tables 3). Gene Set Enrichment Analysis (GSEA) showed that TRIM6 significantly activates several biological pathways, including CHOLESTEROL HOMEOSTASIS, COAGULATION, EPITHELIAL MESENCHYMAL TRANSITION (EMT), INFLAMMATORY RESPONSE, INTERFERON ALPHA RESPONSE, and INTERFERON GAMMA RESPONSE (Fig. 3D). The activation of the EMT pathway, in particular, is of interest due to its known role in enhancing the migratory and invasive capabilities of cancer cells. To gain further insights into the functional impact of TRIM6 knockout, we conducted Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analyses on the 142 downregulated genes in the TRIM6_Knockout group. GO enrichment analysis revealed significant involvement of these genes in biological processes such as extracellular structure organization, extracellular matrix organization, and external encapsulating structure organization (Fig. 3E). KEGG pathway analysis indicated that these downregulated genes were predominantly associated with ECM-receptor interaction, Focal adhesion, and Axon guidance pathways (Fig. 3F). These findings suggest that TRIM6 plays a critical role in the activation of the EMT signaling pathway, potentially contributing to the enhanced migration and invasion capabilities of cancer cells.
Fig. 3.
TRIM6 Activates the EMT Signaling Pathway. (A) Heatmap displaying the differential gene expression between TRIM6_Knockout and WT groups in the GSE138841 dataset. The analysis identified 61 upregulated and 142 downregulated genes in the TRIM6_Knockout group compared to the WT group. (B) Volcano plot illustrating the distribution of differentially expressed genes between TRIM6_Knockout and WT groups, highlighting 61 upregulated and 142 downregulated genes in the TRIM6_Knockout group. (C) Summary table showing the number of upregulated, downregulated, and insignificant genes in the TRIM6_Knockout group versus WT. (D) GSEA results showing the enrichment of several pathways, including CHOLESTEROL HOMEOSTASIS, COAGULATION, EPITHELIAL MESENCHYMAL TRANSITION (EMT), INFLAMMATORY RESPONSE, INTERFERON ALPHA RESPONSE, and INTERFERON GAMMA RESPONSE, in the TRIM6_Knockout group. (E) GO enrichment barplot showing the biological processes significantly enriched among the 142 downregulated genes in the TRIM6_Knockout group, focusing on extracellular structure organization, extracellular matrix organization, and external encapsulating structure organization. (F) KEGG enrichment barplot displaying the pathways significantly enriched among the 142 downregulated genes in the TRIM6_Knockout group, highlighting ECM-receptor interaction, Focal adhesion, and Axon guidance.
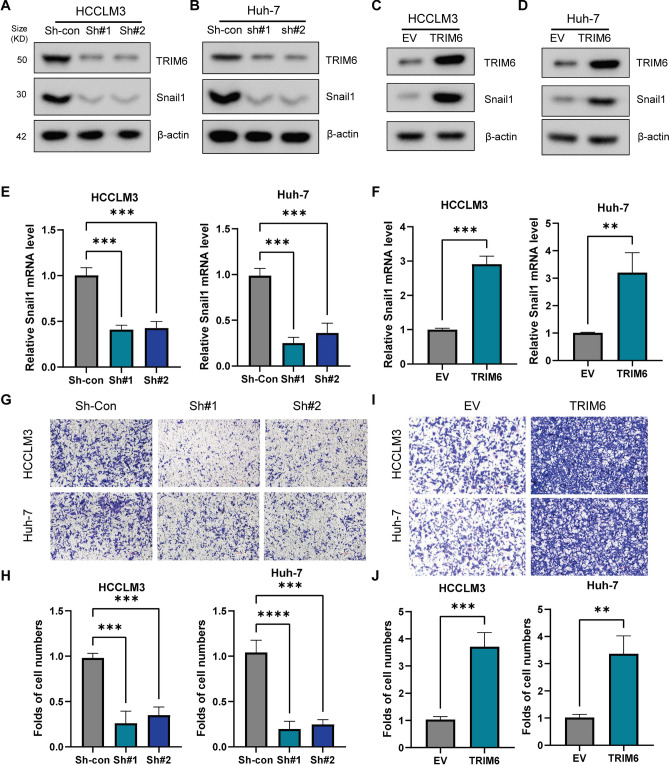
EMT is a critical process in cancer progression that enhances the migratory and invasive capabilities of cancer cells. Snail1 is a key transcription factor that drives EMT by repressing epithelial markers, such as E-cadherin, and upregulating mesenchymal markers, such as N-cadherin and vimentin15. Given the importance of Snail1 in EMT, we sought to determine whether TRIM6 influences EMT in HCC through the regulation of Snail1 expression. To explore this, we performed experiments in HCC cell lines HCCLM3 and Huh-7, where TRIM6 expression was either knocked down using shRNAs or overexpressed. Western blot analysis confirmed the effective knockdown of TRIM6 in both HCCLM3 and Huh-7 cells (Fig. 4A, B). Correspondingly, TRIM6 knockdown significantly reduced Snail1 mRNA and protein levels in both cell lines, suggesting that TRIM6 positively regulates Snail1 expression (Fig. 4A, B, E). This reduction in Snail1 expression upon TRIM6 knockdown indicates that TRIM6 may be an upstream regulator of EMT in HCC. Conversely, overexpression of TRIM6 in HCCLM3 and Huh-7 cells led to an increase in Snail1 mRNA and protein levels, further supporting the role of TRIM6 in promoting EMT (Fig. 4C, D, F). To assess the functional impact of TRIM6 on cell invasion, we conducted transwell invasion assays. In these assays, TRIM6 knockdown resulted in a marked decrease in the invasive capacity of both HCCLM3 and Huh-7 cells, as evidenced by the reduced number of invading cells (Fig. 4G, H). This reduction in invasion highlights the potential role of TRIM6 in facilitating EMT-related processes. Conversely, TRIM6 overexpression significantly enhanced cell invasion in both cell lines (Fig. 4G, J), consistent with its role in promoting Snail1 expression and EMT. These results suggest that TRIM6 plays a crucial role in promoting EMT and enhancing the invasive potential of HCC cells through the regulation of Snail1, potentially contributing to cancer metastasis.
Fig. 4.
TRIM6 Activates EMT and Promotes Snail1-Mediated Cell Migration in HCC. (A, B) Western blot analysis confirming the knockdown of TRIM6 in HCCLM3 (A) and Huh-7 (B) cells using two different shRNAs (sh#1 and sh#2). β-Actin was used as a loading control. (C, D) Western blot analysis showing the overexpression of TRIM6 in HCCLM3 (C) and Huh-7 (D) cells. β-Actin was used as a loading control. (E) Relative Snail1 mRNA levels in HCCLM3 and Huh-7 cells after TRIM6 knockdown (sh#1 and sh#2). TRIM6 knockdown significantly reduces Snail1 expression. ***p < 0.001. (F) Relative Snail1 mRNA levels in HCCLM3 and Huh-7 cells after TRIM6 overexpression. TRIM6 overexpression significantly increases Snail1 expression. **p < 0.01, ***p < 0.001. (G, H) Transwell migration assays showing the effect of TRIM6 knockdown on cell migration in HCCLM3 and Huh-7 cells. Knockdown of TRIM6 significantly reduces the number of migrating cells. ***p < 0.001, ****p < 0.0001. (I, J) Transwell migration assays showing the effect of TRIM6 overexpression on cell migration in HCCLM3 and Huh-7 cells. Overexpression of TRIM6 significantly increases the number of migrating cells. **p < 0.01, ***p < 0.001. These results demonstrate that TRIM6 enhances EMT and cell migration in HCC cells through the regulation of Snail1, highlighting its potential role in promoting cancer metastasis by facilitating the EMT process.
TRIM6 interacts with and promotes the degradation of DDX58 (RIG-I)
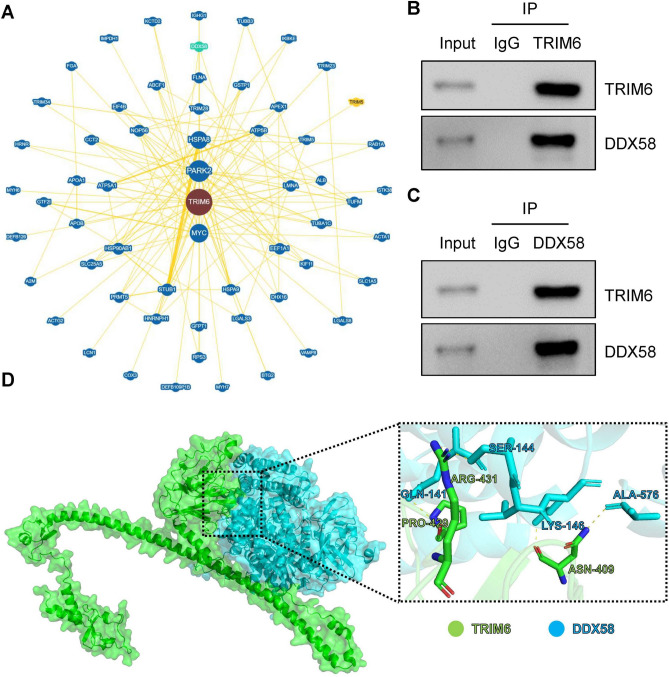
To identify potential protein interactions involving TRIM6 that could contribute to its role in HCC progression, we analyzed the TRIM6 protein interaction network using the BioGRID database. This analysis revealed DDX58 (also known as RIG-I) as a potential interacting partner of TRIM6 (Fig. 5A). DDX58 is primarily known for its role in the innate immune response as an RNA helicase that detects viral RNA18. However, literature research indicates that DDX58 also plays a role in regulating cancer cell migration and invasion26. To experimentally validate the interaction between TRIM6 and DDX58, we performed co-immunoprecipitation (Co-IP) assays in HCC cell lines. The results confirmed that TRIM6 physically interacts with DDX58 in both HCCLM3 (Fig. 5B) and Huh-7 (Fig. 5C) cells. This suggests that TRIM6 may influence DDX58-mediated signaling pathways that regulate migration and invasion in HCC cells. To further explore the molecular details of the TRIM6-DDX58 interaction, we utilized the Alphafold3 online platform to predict the interaction interface between TRIM6 and DDX58. The structural prediction revealed that TRIM6 recognizes and binds to DDX58 through specific residues, including ASN-409, PRO-429, and ARG-431 (Fig. 5D). These residues form a stable binding interface with DDX58, which may facilitate TRIM6’s regulatory effects on EMT and metastatic processes in HCC.
Fig. 5.
TRIM6 Interacts with DDX58. (A) Protein–protein interaction (PPI) network analysis using the BioGRID database, identifying DDX58 as a potential interacting partner of TRIM6. (B, C) Co-IP assays performed in HCCLM3 (B) and Huh-7 (C) cell lines demonstrate the physical nteraction between TRIM6 and DDX58. In (B), TRIM6 was immunoprecipitated, and the presence of DDX58 in the complex was confirmed by Western blotting. In (C), DDX58 was immunoprecipitated, and TRIM6 was detected in the complex. (D) Predicted interaction interface between TRIM6 and DDX58 based on structural modeling using Alphafold3. The zoomed-in view highlights key interacting residues on TRIM6 (ASN-409, PRO-429, and ARG-431) that recognize and bind to DDX58.
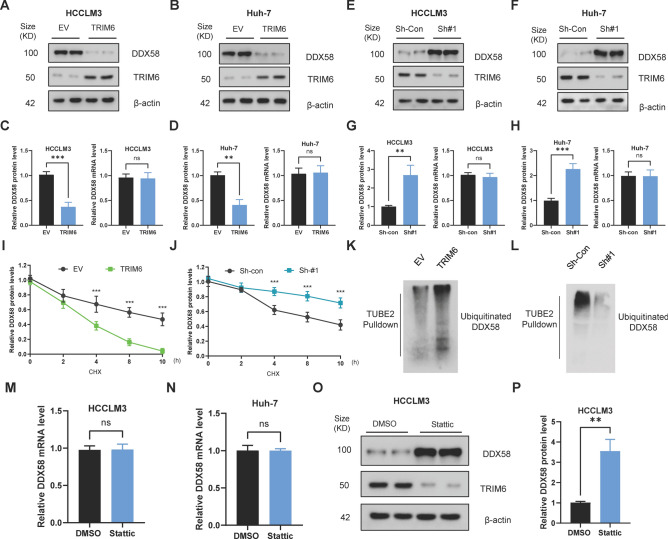
Given the known functions of TRIM family proteins as E3 ubiquitin ligases, we hypothesized that TRIM6 may regulate DDX58 through ubiquitination and subsequent proteasomal degradation. We first assessed the effect of TRIM6 overexpression on DDX58 protein levels in HCCLM3 and Huh-7 cell lines. Western blot analysis revealed that overexpression of TRIM6 resulted in a significant reduction in DDX58 protein levels in both cell lines (Fig. 6A, B). Quantification of these results showed a consistent decrease in DDX58 protein levels in TRIM6-overexpressing cells compared to empty vector controls. Interestingly, this reduction in DDX58 protein was not accompanied by a corresponding decrease in DDX58 mRNA levels, as determined by qRT-PCR, indicating that TRIM6 regulates DDX58 primarily at the post-transcriptional level (Fig. 6C, D). This suggests that TRIM6 may target DDX58 for degradation, rather than affecting its transcription. To further validate the role of TRIM6 in regulating DDX58 stability, we performed TRIM6 knockdown experiments using shRNA. Western blot analysis showed that TRIM6 knockdown led to a substantial increase in DDX58 protein levels in both HCCLM3 and Huh-7 cells (Fig. 6E, F). This increase was quantified and found to be statistically significant. Similar to the overexpression experiments, changes in DDX58 protein levels upon TRIM6 knockdown were not associated with changes in DDX58 mRNA levels, as determined by qRT-PCR (Fig. 6G, H). This reinforces the idea that TRIM6 modulates DDX58 stability at the protein level, likely through post-translational mechanisms. To determine whether TRIM6 affects the degradation rate of DDX58, we conducted a cycloheximide (CHX) chase assay to monitor the decay of DDX58 protein over time. In cells overexpressing TRIM6, the degradation of DDX58 was significantly accelerated, as indicated by a faster decline in DDX58 protein levels over a 10-h period compared to control cells (Fig. 6I). Conversely, in TRIM6 knockdown cells, the degradation of DDX58 was markedly slower, suggesting that TRIM6 is essential for the rapid turnover of DDX58 protein in HCC cells (Fig. 6J). These findings suggest that TRIM6 promotes the proteasomal degradation of DDX58. Finally, to investigate whether TRIM6 promotes the ubiquitination of DDX58, we performed a TUBE2 pulldown assay to isolate ubiquitinated proteins, followed by Western blotting to detect DDX58. The results showed that TRIM6 overexpression significantly increased the levels of ubiquitinated DDX58, while TRIM6 knockdown led to a noticeable decrease in DDX58 ubiquitination (Fig. 6K, L). Moreover, pharmacological inhibition of STAT3 using Stattic had no effect on DDX58 mRNA levels, but significantly increased DDX58 protein expression (Fig. 6M, P). This suggests that STAT3 regulates DDX58 at the post-transcriptional level. Given that TRIM6 is a transcriptional target of STAT3 and functions as an E3 ubiquitin ligase, the observed increase in DDX58 protein upon STAT3 inhibition is likely due to reduced TRIM6-mediated ubiquitination and proteasomal degradation of DDX58.
Fig. 6.
TRIM6 Promotes Ubiquitination and Degradation of DDX58 in Hepatocellular Carcinoma Cells. (A, B) Western blot analysis of DDX58 protein levels following TRIM6 overexpression in HCCLM3 (A) and Huh-7 (B) cells. TRIM6 overexpression significantly reduces DDX58 protein levels. β-Actin was used as a loading control. (C, D) Quantification of DDX58 protein and mRNA levels in HCCLM3 (C) and Huh-7 (D) cells after TRIM6 overexpression. The reduction in DDX58 protein levels is not accompanied by changes in DDX58 mRNA levels, suggesting post-transcriptional regulation. **p < 0.01, ***p < 0.001, ns = not significant. (E, F) Western blot analysis of DDX58 protein levels following TRIM6 knockdown in HCCLM3 (E) and Huh-7 (F) cells. TRIM6 knockdown increases DDX58 protein levels. β-Actin was used as a loading control. (G, H) Quantification of DDX58 protein and mRNA levels in HCCLM3 (G) and Huh-7 (H) cells after TRIM6 knockdown. The increase in DDX58 protein levels is not reflected in DDX58 mRNA levels. **p < 0.01, ***p < 0.001, ns = not significant. (I, J) Cycloheximide (CHX) chase assay showing the degradation rate of DDX58 in HCCLM3 cells. (I) TRIM6 overexpression accelerates DDX58 degradation, while (J) TRIM6 knockdown slows down DDX58 degradation. ***p < 0.001. (K, L) TUBE2 pulldown assay detecting ubiquitinated DDX58 in HCCLM3 cells. (K) TRIM6 overexpression increases DDX58 ubiquitination, whereas (L) TRIM6 knockdown reduces DDX58 ubiquitination. (M, N) Quantitative RT-PCR analysis of DDX58 mRNA levels in HCCLM3 (M) and Huh-7 (N) cells treated with the STAT3 inhibitor Stattic. ns = not significant. (O, P) Western blot analysis (O) and quantification (P) of DDX58 protein levels in HCCLM3 cells following Stattic treatment. Pharmacological inhibition of STAT3 with Stattic markedly elevates DDX58 protein abundance. **p < 0.01.
DDX58 mediates the effects of TRIM6 on Snail1 expression and cell invasion in HCC
Given that TRIM6 promotes the degradation of DDX58, we sought to determine whether DDX58 is a critical mediator of TRIM6’s effects on EMT and cell invasion in HCC. Specifically, we investigated whether manipulating DDX58 expression could rescue or reverse the changes in Snail1 expression and cell invasive capacity induced by TRIM6 modulation. We first examined the functional interaction between TRIM6 and DDX58 by performing simultaneous knockdown of both genes in HCCLM3 cells. TRIM6 was silenced using shRNA, and DDX58 was concurrently knocked down to determine whether loss of DDX58 could counteract the inhibitory effects of TRIM6 depletion on Snail1 expression and cell invasion. Conversely, to evaluate whether DDX58 overexpression could antagonize the pro-invasive effects of TRIM6 overexpression, we co-transfected HCCLM3 cells with TRIM6 and DDX58 expression constructs. This double overexpression approach allowed us to assess the functional hierarchy between TRIM6 and DDX58 in regulating EMT and metastatic potential. Simultaneous knockdown of DDX58 in TRIM6-deficient cells restored Snail1 expression, supporting the role of DDX58 as a suppressor of Snail1 downstream of TRIM6 (Fig. 7A). This observation was further supported by qRT-PCR analysis, which demonstrated a significant reduction in Snail1 mRNA levels upon TRIM6 knockdown, with partial recovery of Snail1 expression upon co-knockdown of DDX58 (Fig. 7B). These results suggest that DDX58 is involved in the regulation of Snail1 expression downstream of TRIM6. To assess the functional consequences of these molecular changes, we performed transwell invasion assays to measure the invasive capacity of HCCLM3 cells under the same experimental conditions. Consistent with the effects on Snail1 expression, TRIM6 knockdown led to a significant reduction in cell invasion, as evidenced by a marked decrease in the number of invading cells compared to control cells. However, the depletion of DDX58 in TRIM6-knockdown cells partially rescued the invasive ability, indicating that DDX58 plays a role in mediating the pro-invasive effects of TRIM6 (Fig. 7C, D). To further investigate the role of DDX58 in TRIM6-induced EMT and invasion, we reversed the experimental setup by overexpressing TRIM6 and then co-expression of DDX58 to observe if it could negate the TRIM6-induced effects. Western blot analysis showed that overexpression of TRIM6 resulted in a significant increase in Snail1 protein levels, consistent with the promotion of EMT. Co-overexpression of DDX58 in TRIM6-overexpressing cells counteracted the upregulation of Snail1, further confirming that TRIM6 regulates Snail1 via degradation of DDX58 (Fig. 7E). qRT-PCR analysis confirmed these findings at the mRNA level, where Snail1 expression increased with TRIM6 overexpression and was subsequently reduced by co-expression of DDX58 (Fig. 7F). The functional impact of these molecular changes was further evaluated using transwell invasion assays. TRIM6 overexpression significantly enhanced the invasive capacity of HCCLM3 cells, as indicated by a substantial increase in the number of invading cells. However, when DDX58 was overexpressed in the TRIM6-overexpressing cells, the enhanced invasive capacity was largely abrogated, leading to a significant reduction in invasion (Fig. 7G, H). This result highlights the importance of DDX58 as a mediator of TRIM6’s effects on cell invasion.
Fig. 7.
TRIM6 regulates Snail1 expression and HCC cell invasion via DDX58. (A) Immunoblot analysis of Snail1 protein levels in HCCLM3 cells following TRIM6 knockdown, with or without concomitant DDX58 knockdown. Silencing TRIM6 led to a reduction in Snail1 protein levels, which was partially restored upon co-knockdown of DDX58. β-Actin served as a loading control. (B) Quantitative RT-PCR analysis of Snail1 mRNA levels under the same conditions as in (A). TRIM6 knockdown reduced Snail1 transcript levels, which were partially rescued by simultaneous DDX58 knockdown. **p < 0.01, ***p < 0.001, ns = not significant. (C, D) Transwell invasion assay evaluating the invasive capacity of HCCLM3 cells following TRIM6 knockdown, with or without DDX58 knockdown. (C) Representative images of invaded cells stained with crystal violet. (D) Quantification of invasive cell numbers. TRIM6 silencing significantly impaired cell invasion, which was partially restored by DDX58 knockdown. **p < 0.01, ***p < 0.001. (E) Immunoblot analysis of Snail1 protein levels in HCCLM3 cells overexpressing TRIM6, with or without DDX58 overexpression. TRIM6 overexpression markedly elevated Snail1 protein expression, which was attenuated by co-expression of DDX58. β-Actin was used as a loading control. (F) Quantitative RT-PCR analysis of Snail1 mRNA levels in HCCLM3 cells upon TRIM6 overexpression, with or without DDX58 overexpression. The TRIM6-induced upregulation of Snail1 transcripts was reversed by enforced expression of DDX58. **p < 0.01, ***p < 0.001, ns = not significant. (G, H) Transwell invasion assay assessing the effects of TRIM6 and DDX58 overexpression on HCCLM3 cell invasiveness. (G) Representative images of invading cells stained with crystal violet. (H) Quantification of invasive cell numbers. TRIM6 overexpression significantly enhanced invasion, which was substantially suppressed by DDX58 overexpression. ***p < 0.001, ****p < 0.0001.
Discussion
In this study, we identified TRIM6 as a novel player in HCC progression, demonstrating its significant upregulation in HCC tissues and its strong association with poor clinical outcomes. Our findings elucidate the mechanisms by which TRIM6 promotes HCC progression, specifically through the regulation of EMT and cell migration, mediated by its interaction with DDX58 and the downstream effects on Snail1 expression. These insights provide a deeper understanding of the molecular underpinnings of HCC metastasis and highlight TRIM6 as a potential therapeutic target in this aggressive cancer.
The upregulation of TRIM6 across multiple HCC datasets, as well as its correlation with tumor grade and stage, underscores its clinical relevance. These observations are consistent with prior studies indicating that the dysregulation of E3 ubiquitin ligases is a hallmark of various cancers, contributing to tumorigenesis through the modulation of protein stability and function27,28. Our analysis identified TRIM6 as a key E3 ligase consistently upregulated in HCC, suggesting its role in the oncogenic processes driving HCC progression. We further identified STAT3 as a direct upstream regulator of TRIM6, providing a mechanistic link between TRIM6 upregulation and STAT3 signaling, which is well-known for its role in inflammation and cancer29,30. The direct binding of STAT3 to the TRIM6 promoter, confirmed through chromatin immunoprecipitation (ChIP) assays, and the subsequent regulation of TRIM6 transcription, establish STAT3 as a critical driver of TRIM6 expression in HCC. This finding aligns with previous research showing that aberrant STAT3 activation promotes tumorigenesis by regulating a host of oncogenes31,32. The inhibition of TRIM6 expression following treatment with the STAT3 inhibitor or degrader further solidifies the role of STAT3 in controlling TRIM6 levels, offering a potential therapeutic avenue for targeting STAT3-TRIM6 signaling in HCC.
Beyond STAT3, TRIM6 expression also correlates with several other oncogenic transcription factors, including NF-κB (RELA), MYC, and HIF1A, in the TCGA-LIHC dataset. This raises the possibility that TRIM6 may be under complex transcriptional regulation involving multiple oncogenic pathways. While our experimental validation confirms direct transcriptional activation of TRIM6 by STAT3, further studies are warranted to investigate whether NF-κB, MYC, or HIF1A also participate in TRIM6 induction under specific tumor microenvironmental stimuli, such as inflammation or hypoxia. These interactions may contribute to the dynamic regulation of TRIM6 during hepatocellular carcinoma progression and metastasis.
Our study also reveals that TRIM6 exerts its oncogenic effects by promoting EMT. We demonstrated that TRIM6 promotes EMT in HCC by upregulating Snail1, a master regulator of EMT, and enhancing cell migration. The knockdown of TRIM6 resulted in a marked reduction of Snail1 expression and a corresponding decrease in cell migratory capacity, while TRIM6 overexpression had the opposite effect. These results are consistent with previous studies linking Snail1 to enhanced migratory and invasive behavior in various cancers33,34. The TRIM6-Snail1 axis thus appears to be a crucial pathway by which TRIM6 drives EMT and metastasis in HCC. In addition to its role in regulating Snail1, we discovered that TRIM6 interacts with and ubiquitinates DDX58, leading to its proteasomal degradation. DDX58, or RIG-I, is traditionally recognized for its role in antiviral responses, but recent studies have highlighted its involvement in cancer, where it functions as a tumor suppressor by inhibiting migration and invasion26,35.
Our findings suggest that TRIM6-mediated degradation of DDX58 alleviates its suppressive effects on Snail1 and EMT, thereby facilitating cancer cell migration and invasion. The ability of DDX58 overexpression suppressed Snail1 protein levels, further confirming its role as a negative regulator of Snail1 downstream of TRIM6. In addition to regulating Snail1, DDX58 may influence EMT and metastasis through broader signaling mechanisms. Emerging evidence suggests that activation of DDX58 can exert tumor-suppressive effects beyond antiviral immunity, including inhibition of key oncogenic pathways such as NF-κB and TGF-β, which are critically involved in EMT regulation26,35.
Specifically, DDX58 activation has been reported to suppress NF-κB signaling, thereby reducing transcription of EMT-promoting cytokines such as IL-6 and TNF-α. Furthermore, DDX58 can enhance type I IFN production, which in turn may antagonize TGF-β signaling and maintain epithelial differentiation. These findings raise the possibility that TRIM6-mediated degradation of DDX58 not only derepresses Snail1 but may also relieve inhibitory constraints on additional EMT-associated signaling cascades. Although our current study centers on the TRIM6–DDX58–Snail1 axis, future work will explore whether other EMT drivers or microenvironmental modulators are regulated downstream of DDX58. Such investigations may further elucidate the multifaceted tumor-suppressive role of DDX58 in hepatocellular carcinoma progression.
Furthermore, our data reveal that STAT3 indirectly regulates DDX58 protein stability via transcriptional activation of TRIM6, rather than by directly modulating DDX58 gene expression. Specifically, pharmacological inhibition of STAT3 with Stattic did not alter DDX58 mRNA levels, but led to a marked increase in DDX58 protein abundance. This effect coincided with a decrease in TRIM6 expression, suggesting that reduced TRIM6-mediated ubiquitination permits DDX58 stabilization. These findings support a post-translational regulatory model in which STAT3 promotes the degradation of DDX58 through upregulation of the E3 ligase TRIM6, rather than suppressing DDX58 transcription directly. This layered regulatory mechanism highlights how oncogenic signaling pathways can exert fine-tuned control over innate immune sensors like DDX58 to influence EMT. By driving TRIM6 expression, STAT3 indirectly facilitates the degradation of DDX58, thereby releasing its suppressive effect on Snail1. These results further reinforce the central role of the STAT3–TRIM6–DDX58–Snail1 axis in HCC metastasis and suggest that targeting STAT3 or TRIM6 may offer a means to restore DDX58-mediated tumor suppression.
This study has several strengths, including the integration of multiple datasets to validate the upregulation of TRIM6, the identification of STAT3 as a direct regulator of TRIM6, and the elucidation of the TRIM6-DDX58-Snail1 pathway as a key driver of EMT and migration in HCC. However, there are limitations that warrant further investigation. While we demonstrated the regulatory role of STAT3 in TRIM6 expression, the broader regulatory network involving other transcription factors and signaling pathways remains to be explored. Additionally, the functional role of TRIM6 in vivo and its impact on HCC metastasis in animal models need to be assessed to validate our findings in a physiological context. Future studies should also investigate the therapeutic potential of targeting the TRIM6-DDX58 axis in HCC. The use of small molecules or biologics to inhibit TRIM6 function, restore DDX58 activity, or disrupt the interaction between TRIM6 and STAT3 could offer novel treatment strategies for HCC patients. Moreover, given the role of STAT3 in regulating multiple oncogenic pathways, combination therapies that target both STAT3 and TRIM6 may enhance therapeutic efficacy and prevent resistance.
In conclusion, our study highlights TRIM6 as a critical regulator of EMT and migration in HCC, mediated by its interaction with DDX58 and regulation of Snail1. The STAT3-TRIM6-DDX58-Snail1 axis represents a novel pathway in HCC metastasis, offering potential targets for therapeutic intervention. These findings contribute to our understanding of the molecular mechanisms driving HCC progression and underscore the importance of TRIM6 as both a prognostic biomarker and a therapeutic target in this malignancy.
Supplementary Information
Acknowledgements
The project was supported by the Hainan Provincial Natural Science Foundation (Grant.821MS0833) and the Hainan Province Clinical Medical Center of China.
Author contributions
Conceptualization: Y.W., J.W. Methodology: Y.W., J.W., S.H. Validation: Y.W., J.W., S.H., X.L. Formal Analysis: Y.W., J.W., S.H., X.L., Y.C. Investigation: Y.W., J.W., S.H., X.L., Y.C., T.W. Resources: Y.W., J.W., S.H., X.L., Y.C., T.W., H.Z. Data Curation: Y.W., J.W., S.H., X.L., Y.C., T.W., H.Z., X.L. Writing—Original Draft: Y.W., J.W., S.H., X.L., Y.C., T.W., H.Z., X.L., X.W. Visualization: Y.W., J.W., S.H., X.L., Y.C., T.W., H.Z., X.L., X.W., P.L. Supervision: P.L. Funding Acquisition: P.L.
Data availability
All data generated or analysed during this study are included in this published article and its supplementary information files.
Declaration
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Yiqiao Wang and Jie Wang contributed equally.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-025-96548-9.
References
- 1.Bray, F. et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin.68, 394–424. 10.3322/caac.21492 (2018). [DOI] [PubMed] [Google Scholar]
- 2.Villanueva, A. Hepatocellular carcinoma. N. Engl. J. Med.380, 1450–1462. 10.1056/NEJMra1713263 (2019). [DOI] [PubMed] [Google Scholar]
- 3.Popovic, D., Vucic, D. & Dikic, I. Ubiquitination in disease pathogenesis and treatment. Nat. Med.20, 1242–1253. 10.1038/nm.3739 (2014). [DOI] [PubMed] [Google Scholar]
- 4.Hershko, A. & Ciechanover, A. The ubiquitin system. Annu. Rev. Biochem.67, 425–479. 10.1146/annurev.biochem.67.1.425 (1998). [DOI] [PubMed] [Google Scholar]
- 5.Wertz, I. E. & Dixit, V. M. Regulation of death receptor signaling by the ubiquitin system. Cell Death Differ.17, 14–24. 10.1038/cdd.2009.168 (2010). [DOI] [PubMed] [Google Scholar]
- 6.Hatakeyama, S. TRIM family proteins: Roles in autophagy, immunity, and carcinogenesis. Trends Biochem. Sci.42, 297–311. 10.1016/j.tibs.2017.01.002 (2017). [DOI] [PubMed] [Google Scholar]
- 7.Lu, K. et al. TRIM proteins in hepatocellular carcinoma. J. Biomed. Sci.29, 69. 10.1186/s12929-022-00854-7 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cai, C., Tang, Y. D., Zhai, J. & Zheng, C. The RING finger protein family in health and disease. Signal Transduct. Target Ther.7, 300. 10.1038/s41392-022-01152-2 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Guo, J. et al. TRIM6: An upregulated biomarker with prognostic significance and immune correlations in gliomas. Biomolecules10.3390/biom13091298 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhang, Y., Yuan, L., Cui, S. & Wu, S. Tripartite motif protein 6 promotes hepatocellular carcinoma progression via multiple pathways. Turk. J. Med. Sci.53, 1032–1044. 10.55730/1300-0144.5668 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang, Y. et al. TRIM6 reduces ferroptosis and chemosensitivity by targeting SLC1A5 in lung cancer. Oxid. Med. Cell. Longev.2023, 9808100. 10.1155/2023/9808100 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Seton-Rogers, S. Epithelial-mesenchymal transition: Untangling EMT’s functions. Nat. Rev. Cancer16, 1. 10.1038/nrc.2015.6 (2016). [DOI] [PubMed] [Google Scholar]
- 13.Wrighton, K. H. Cell migration: EMT promotes contact inhibition of locomotion. Nat. Rev. Mol. Cell. Biol.16, 518. 10.1038/nrm4045 (2015). [DOI] [PubMed] [Google Scholar]
- 14.Brabletz, T., Kalluri, R., Nieto, M. A. & Weinberg, R. A. EMT in cancer. Nat. Rev. Cancer18, 128–134. 10.1038/nrc.2017.118 (2018). [DOI] [PubMed] [Google Scholar]
- 15.Garcia de Herreros, A. Dual role of Snail1 as transcriptional repressor and activator. Biochim. Biophys. Acta Rev. Cancer1879, 189037. 10.1016/j.bbcan.2023.189037 (2024). [DOI] [PubMed] [Google Scholar]
- 16.Peinado, H., Olmeda, D. & Cano, A. Snail, Zeb and bHLH factors in tumour progression: An alliance against the epithelial phenotype?. Nat. Rev. Cancer7, 415–428. 10.1038/nrc2131 (2007). [DOI] [PubMed] [Google Scholar]
- 17.Zhou, B. P. et al. Dual regulation of Snail by GSK-3beta-mediated phosphorylation in control of epithelial-mesenchymal transition. Nat. Cell. Biol.6, 931–940. 10.1038/ncb1173 (2004). [DOI] [PubMed] [Google Scholar]
- 18.Goubau, D., Deddouche, S. & Reis e Sousa, C. Cytosolic sensing of viruses. Immunity38, 855–869. 10.1016/j.immuni.2013.05.007 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Deng, Y. et al. Activation of DDX58/RIG-I suppresses the growth of tumor cells by inhibiting STAT3/CSE signaling in colon cancer. Int. J. Oncol.10.3892/ijo.2022.5410 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chang, Y. C. et al. Glucose transporter 4 promotes head and neck squamous cell carcinoma metastasis through the TRIM24-DDX58 axis. J. Hematol. Oncol.10, 11. 10.1186/s13045-016-0372-0 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kanehisa, M., Furumichi, M., Sato, Y., Kawashima, M. & Ishiguro-Watanabe, M. KEGG for taxonomy-based analysis of pathways and genomes. Nucleic Acids Res.51, D587-d592. 10.1093/nar/gkac963 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Busker, S., Page, B. & Arner, E. S. J. To inhibit TrxR1 is to inactivate STAT3-Inhibition of TrxR1 enzymatic function by STAT3 small molecule inhibitors. Redox Biol.36, 101646. 10.1016/j.redox.2020.101646 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Auernhammer, C. J. & Melmed, S. The central role of SOCS-3 in integrating the neuro-immunoendocrine interface. J. Clin. Invest.108, 1735–1740. 10.1172/JCI14662 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhou, H. et al. Structure-based discovery of SD-36 as a potent, selective, and efficacious PROTAC degrader of STAT3 protein. J. Med. Chem.62, 11280–11300. 10.1021/acs.jmedchem.9b01530 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bai, L. et al. A potent and selective small-molecule degrader of STAT3 achieves complete tumor regression in vivo. Cancer Cell36, 498-511 e417. 10.1016/j.ccell.2019.10.002 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Song, J. et al. Friend or foe: RIG- I like receptors and diseases. Autoimmun. Rev.21, 103161. 10.1016/j.autrev.2022.103161 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sampson, C. et al. The roles of E3 ubiquitin ligases in cancer progression and targeted therapy. Clin. Transl. Med.13, e1204. 10.1002/ctm2.1204 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cruz-Walma, D. A., Chen, Z., Bullock, A. N. & Yamada, K. M. Ubiquitin ligases: Guardians of mammalian development. Nat. Rev. Mol. Cell. Biol.23, 350–367. 10.1038/s41580-021-00448-5 (2022). [DOI] [PubMed] [Google Scholar]
- 29.Li, Y. J., Zhang, C., Martincuks, A., Herrmann, A. & Yu, H. STAT proteins in cancer: Orchestration of metabolism. Nat. Rev. Cancer23, 115–134. 10.1038/s41568-022-00537-3 (2023). [DOI] [PubMed] [Google Scholar]
- 30.Johnson, D. E., O’Keefe, R. A. & Grandis, J. R. Targeting the IL-6/JAK/STAT3 signalling axis in cancer. Nat. Rev. Clin. Oncol.15, 234–248. 10.1038/nrclinonc.2018.8 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sun, L. et al. STAT3-specific nanocarrier for shRNA/drug dual delivery and tumor synergistic therapy. Bioact. Mater.41, 137–157. 10.1016/j.bioactmat.2024.07.010 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Duan, Y. et al. STAT3-mediated up-regulation of DAB2 via SRC-YAP1 signaling axis promotes Helicobacter pylori-driven gastric tumorigenesis. Biomark. Res.12, 33. 10.1186/s40364-024-00577-x (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Xie, B. et al. DDR2 facilitates hepatocellular carcinoma invasion and metastasis via activating ERK signaling and stabilizing SNAIL1. J. Exp. Clin. Cancer Res.34, 101. 10.1186/s13046-015-0218-6 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kaufhold, S. & Bonavida, B. Central role of Snail1 in the regulation of EMT and resistance in cancer: a target for therapeutic intervention. J. Exp. Clin. Cancer Res.33, 62. 10.1186/s13046-014-0062-0 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yoneyama, M., Kato, H. & Fujita, T. Physiological functions of RIG-I-like receptors. Immunity57, 731–751. 10.1016/j.immuni.2024.03.003 (2024). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data generated or analysed during this study are included in this published article and its supplementary information files.