Abstract
A multiplex PCR method was developed to identify simultaneously multiple fungal pathogens in a single reaction. Five sets of species-specific primers were designed from the internal transcribed spacer (ITS) regions, ITS1 and ITS2, of the rRNA gene to identify Candida albicans, Candida glabrata, Candida parapsilosis, Candida tropicalis, and Aspergillus fumigatus. Another set of previously published ITS primers, CN4 and CN5, were used to identify Cryptococcus neoformans. Three sets of primers were used in one multiplex PCR to identify three different species. Six different species of pathogenic fungi can be identified with two multiplex PCRs. Furthermore, instead of using templates of purified genomic DNA, we performed the PCR directly from yeast colonies or cultures, which simplified the procedure and precluded contamination during the extraction of DNA. A total of 242 fungal isolates were tested, representing 13 species of yeasts, four species of Aspergillus, and three zygomycetes. The multiplex PCR was tested on isolated DNA or fungal colonies, and both provided 100% sensitivity and specificity. However, DNA from only about half the molds could be amplified directly from mycelial fragments, while DNA from every yeast colony was amplified. This multiplex PCR method provides a rapid, simple, and reliable alternative to conventional methods to identify common clinical fungal isolates.
Invasive mycoses have become a major cause of infectious morbidity and mortality in patients receiving immunosuppressive chemotherapy for cancer or organ transplantation or in immunodeficient patients, such as individuals with AIDS (2, 6, 7, 9, 20, 26). Since opportunistic mycoses are often grave, the early, rapid, and accurate identification of the pathogenic fungus is critical for timely, appropriate management. The conventional identification of pathogenic fungi in the clinical microbiology laboratory is based on morphological and physiological tests, often requires 3 or more days, and may be inaccurate (12, 14).
In recent years, numerous DNA-based methods have been developed to improve the diagnosis of mycotic infections and the identification of pathogenic fungi (13, 27, 34, 37, 45). PCR methods are particularly promising because of their simplicity, specificity, and sensitivity. For example, PCR methods targeting different genes have been described elsewhere for identification of Cryptococcus neoformans (39), Aspergillus fumigatus (19, 36), and species of Candida (4, 5, 11, 15, 28-30, 33, 37, 42, 48). A number of studies have described probes, restriction fragment length polymorphism, or other methods to identify unique ribosomal DNA (rDNA) sequences (10, 16-18, 22, 23, 32, 40, 41, 43). The most common approaches have targeted portions of the rDNA of species of Candida (3, 8, 10, 31, 38, 44). Although these published PCR methods have been useful for the identification of fungal species, they either identify only one species at a time or require a probe hybridization procedure that incurs time and expense.
We describe here a sensitive and specific method to rapidly and simultaneously identify the most common pathogenic fungi in tandem multiplex PCRs. The method combines three species-specific primers in a single PCR tube. To obtain genomic DNA template, we demonstrate that a colony can be sampled directly from a pure culture. Primers CGL1-CGL2, CTR1-CTR2, and CPA1-CPA2 were combined in one multiplex PCR to identify Candida glabrata, Candida tropicalis, and Candida parapsilosis, respectively (multiplex G-T-P); primers AFUM1-AFUM2, CALB1-CALB2, and CN5-CN4 were combined in another multiplex PCR to identify A. fumigatus, Candida albicans, and C. neoformans, respectively (multiplex F-A-N). In a separate PCR, the ITS1-ITS4 primer pair provided a positive control to monitor the amplification of all fungal samples (47).
MATERIALS AND METHODS
Fungal isolates.
Most of the yeast strains and samples of A. fumigatus were isolated from clinical specimens at Duke University Medical Center and maintained in the collection of the Duke Medical Mycology Research Laboratory. The identification of all isolates was confirmed by conventional morphological and physiological methods (1, 35, 46). The samples of C. albicans included one isolate that was identified as Candida stellatoidea, which we deemed a variant of C. albicans (24). To provide a spectrum of medical fungi related to many of the target species, a total of 242 isolates were analyzed, including 10 species of Candida (181 isolates), 4 species of Aspergillus (38 isolates), 3 zygomycetous species (9 isolates), and 3 other yeast species: C. neoformans (10 isolates), Saccharomyces cerevisiae (3 isolates), and Kluyveromyces marianum (1 isolate). A rather large sampling of C. parapsilosis was available because of a concurrent study in which we obtained isolates from the oral cavities and fingernails of healthy undergraduate and medical students of Duke University and the University of North Carolina at Chapel Hill. Control isolates of C. parapsilosis groups I, II, and III were obtained from Paul F. Lehmann (21). A pure culture of each isolate was obtained by streaking the liquid culture on yeast extract-peptone-dextrose agar plates and grown overnight at 30°C.
DNA isolation.
For DNA extraction, a single colony was transferred to a yeast extract-peptone-dextrose plate and grown overnight at 30°C, followed by DNA isolation as described previously (49). For direct yeast cell amplification, a single colony approximately 1 mm in diameter was picked with a micropipette tip, suspended in 5 μl of sterile, distilled water in a microcentrifuge tube, and vortexed; then, 0.5 μl of this suspension was used in the PCRs. Molds were cultured for at least 5 days to produce a visible colony, and a tiny portion of the colony was transferred directly to the PCR tube. We also tested molds that had been stored for more than 1 year.
Primer design.
Species-specific primer pairs—CGL1-CGL2, CTR1-CTR2, CPA1-CPA2, CALB1-CALB2, and AFUM1-AFUM2—were designed based on the sequence data for the internal transcribed spacer (ITS) region (Table 1) in the GenBank database to specifically amplify C. glabrata, C. tropicalis, C. parapsilosis, C. albicans, and A. fumigatus, respectively. The forward primers (primer 1 of each pair) were designed within the ITS1 region, and the reverse primers (primer 2) were designed from the ITS2 region. The C. neoformans-specific primers CN5 and CN4 were previously described (25). The universal fungal primers ITS1 and ITS4 provided a positive PCR control (47).
TABLE 1.
Primer pairs designed to amplify DNA specifically from the listed species of pathogenic fungi
| Species | Primer namea | Sequence (5′ → 3′) | GenBank accession nos.b | Amplicon size (bp) |
|---|---|---|---|---|
| All fungic | ITS1 | TCC GTA GGT GAA CCT GCG G | M27607, D89886 | Variabled |
| ITS4 | TCC TCC GCT TAT TGA TAT GC | |||
| Aspergillus fumigatus | AFUM1 | CGC CGA AGA CCC CAA CAT GAA CGC | AF176662, AF078889 | ≈385 |
| AFUM2 | TAA AGT TGG GTG TCG GCT GGC | |||
| Candida albicans | CALB1 | TTT ATC AAC TTG TCA CAC CAG A | L47111, L28817 | ≈273 |
| CALB2 | ATC CCG CCT TAC CAC TAC CG | |||
| Candida glabrata | CGL1 | TTA TCA CAC GAC TCG ACA CT | AB032177, AF167993 | ≈423 |
| CGL2 | CCC ACA TAC TGA TAT GGC CTA CAA | |||
| Candida parapsilosis | CPA1e | TTG GTA GGC CTT CTA TAT GGG | AF287909, L47109 | ≈320 |
| CPA3f | GCC AGA GAT TAA ACT CAA CCA A | ≈300 | ||
| CPA2 | CCT ATC CAT TAG TTT ATA CTC CGC | |||
| Candida tropicalis | CTR1 | CAA TCC TAC CGC CAG AGG TTA T | AF287910, AF268095 | ≈357 |
| CTR2 | TGG CCA CTA GCA AAA TAA GCG T | |||
| Cryptococcus neoformansg | CN5 | GAA GGG CAT GCC TGT TTG AGA G | M94516, M94517 | ≈136 |
| CN4 | ATC ACC TTC CCA CTA ACA CAT T |
Odd-numbered primers are forward primers, and even-numbered primers are reverse primers.
For the universal fungal primers ITS1 and ITS4, the GenBank accession number refers to primers in the 18S and ITS regions of S. cerevisiae. For all other primer pairs, the accession numbers are specific for the indicated taxa; strains evince slight variation in amplicon sizes.
Sequences are from reference 47.
The ITS1-ITS4 amplicon sizes vary with each organism.
Amplifies only group I strains of C. parapsilosis (21).
Amplifies all groups of C. parapsilosis.
Sequences are from reference 25.
PCR.
For PCR with individual primer pairs, each reaction mixture contained 2 μl (∼1 ng) of diluted genomic DNA template or 0.5 μl of yeast cell suspension, 20 mM Tris-HCl (pH 8.4), 50 mM KCl, 1.5 mM MgCl2, 0.2 mM (each of the four) deoxyribonucleotide triphosphates, 0.5 μM (each) primer, and 0.5 U of Taq DNA polymerase (Invitrogen Life Technologies, Carlsbad, Calif.) in a total volume of 20 μl. PCR amplification conditions were 5 min of denaturation at 96°C, followed by 40 cycles of 94°C for 30 s, 58°C for 30 s, and 72°C for 30 s and a final extension step of 72°C for 15 min. A sample of 10 μl of product from each PCR was electrophoresed in a 1.5% agarose gel with 0.5 μg of ethidium bromide/ml and 1× Tris-acetate-EDTA buffer for 1 to 2 h. DNA bands were visualized on a UV transilluminator and documented with an Alpha-Imager 2000 (Alpha Innotech, San Leandro, Calif.).
Multiplex PCR.
A total of six species were tested in two multiplex PCR panels. Each multiplex panel contained three pairs of primers that were designed and comixed to produce amplicons sufficiently different in size and migration to identify three fungal species. The multiplex G-T-P panel contained primers that identified C. glabrata, C. tropicalis, and C. parapsilosis, and the F-A-N multiplex panel included primers that are specific for A. fumigatus, C. albicans, and C. neoformans. Primer sequences are presented in Table 1. The G-T-P multiplex PCR contained 0.7 μM primers CGL1 and CGL2, 0.4 μM primers CTR1 and CTR2, and 0.6 μM primers CPA1 and CPA2. The F-A-N multiplex PCR contained 0.5 μM (each of the following) primers AFUM1, AFUM2, CALB1, CALB2, CN5, and CN4 (Table 1). The 20-μl PCR mixtures contained 0.75 U of Taq DNA polymerase; all other reagents were the same as described above for single-primer-pair PCRs. The PCR tubes were kept on ice, and the PCRs were carried out in a Perkin-Elmer model 9700 thermal cycler preequilibrated at 96°C to provide a hot start. All PCRs were run with the same cycling program used for single-primer-pair PCR (described above). PCR products were electrophoresed in a 2% agarose gel with ethidium bromide for 2 to 3 h and evaluated as described above.
RESULTS
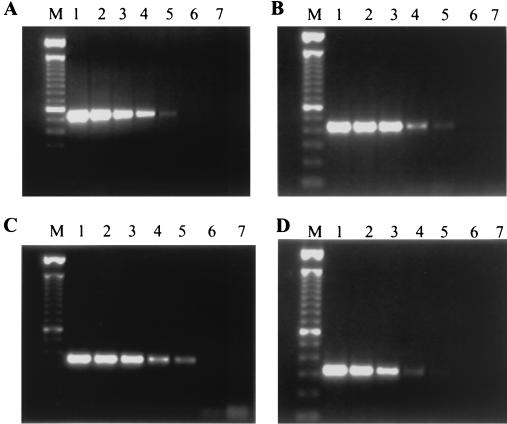
To determine the sensitivity and specificity of the PCR primers, each primer set was first tested with serial dilutions of PCR products from the corresponding species. Figure 1 shows a representative serial dilution of the PCR products (amplicons) with primers ITS1-ITS4, CGL1-CGL2, CTR1-CTR2, and CPA1-CPA2. The sensitivity of each set of primers ranged from 100 to 1,000 DNA molecules, which indicates the potential to amplify the appropriate amplicon from purified genomic DNA from 1 to 10 yeast cells.
FIG. 1.
Examples of the serial dilution of DNA amplicons. (A) Primers ITS1-ITS4; (B) primers CGL1-CGL2; (C) primers CTR1-CTR2; (D) primers CPA1-CPA2. Lanes 1 to 6, specific amplicons generated by using 106, 105, 104, 103, 102, and 101 molecules, respectively; lanes 7, negative control, lacking template DNA; lanes M, 100-bp DNA length ladder.
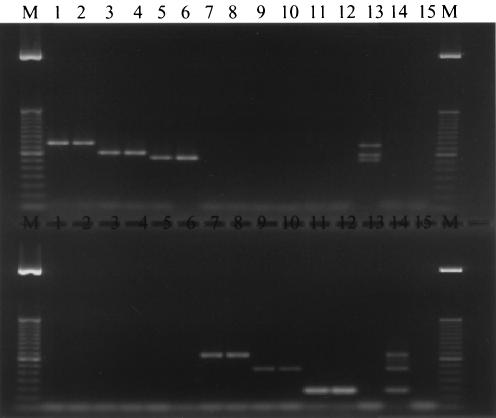
For multiplex PCR, three sets of primers were combined in a single tube to simultaneously identify three fungal pathogens. A total of six primer sets were used in two different multiplex PCR panels, as noted above and illustrated in Fig. 2. Multiplex G-T-P contained primer sets CGL1-CGL2, CTR1-CTR2, and CPA1-CPA2. Multiplex F-A-N consisted of primers AFUM1-AFUM2, CALB1-CALB2, and CN5-CN4. As shown in Fig. 2, both G-T-P and F-A-N multiplex PCRs generated specific amplicons of the correct sizes when templates from the corresponding species were present. When all three templates were included in each panel, three products of the signature size were correctly produced (Fig. 2, top, lane 13, and bottom, lane 14). The sensitivity of the multiplex PCR was similar to the sensitivity of single-primer-set PCR. These results indicate that up to three suspected pathogens can be identified in a single PCR.
FIG. 2.
Multiplex PCR performed with genomic DNA from two clinical isolates of each species. (Top) Multiplex G-T-P PCR. (Bottom) Multiplex F-A-N PCR. Lanes 1 and 2, C. glabrata; lanes 3 and 4, C. tropicalis; lanes 5 and 6, C. parapsilosis; lanes 7 and 8, A. fumigatus; lanes 9 and 10, C. albicans; lanes 11 and 12, C. neoformans; lane 13, mixed DNA of C. glabrata, C. tropicalis, and C. parapsilosis; lane 14, mixed DNA of A. fumigatus, C. albicans, and C. neoformans; lane 15, negative control without template DNA; lanes M, 100-bp DNA length ladder.
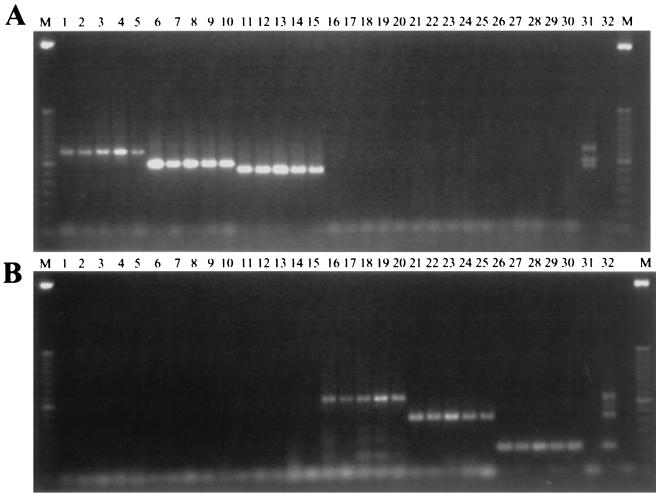
To further simplify the procedure and shorten the time required for identification, we explored the possibility of performing multiplex PCR directly from yeast colonies, bypassing the usual, time-consuming DNA isolation steps. All multiplex PCR conditions were the same as described above except that, instead of extracted DNA, the template consisted of 0.5 μl of a suspension of yeast cells or fragments of hyphae. As shown in Fig. 3, both multiplex panels amplified the specific bands from their corresponding isolates with similar sensitivity as that obtained with purified DNA templates.
FIG. 3.
Example of multiplex G-T-P (A) and multiplex F-A-N (B) PCRs directly from yeast colonies. The figure shows amplicons from each of five individual clinical isolates of C. glabrata (lanes 1 to 5), C. tropicalis (lanes 6 to 10), C. parapsilosis (lanes 11 to 15), A. fumigatus (lanes 16 to 20), C. albicans (lanes 21 to 25), and C. neoformans (lanes 26 to 30). Lanes 31, mixed DNA of C. glabrata, C. tropicalis, and C. parapsilosis. Lanes 32, mixed DNA of A. fumigatus, C. albicans, and C. neoformans. Lanes M, 50-bp DNA ladder.
Testing both individual primer pairs and multiplex PCR methods and using either extracted genomic DNA or a suspension of yeast cells as the template, we evaluated a total of 242 fungal strains (Table 2). As expected, the universal fungal primers, ITS1-ITS4, produced an amplicon of the appropriate size (Fig. 1A) from all the yeasts and about half the molds that were directly assayed. Older mold colonies were less likely to produce the universal amplicon than were fresh cultures. Since every isolate of C. albicans, C. glabrata, C. tropicalis, C. parapsilosis, and C. neoformans that was tested generated only species-specific products, their sensitivity was 100%. All isolates of A. fumigatus that could be amplified directly from hyphae (13 of 15 strains) also exhibited a sensitivity of 100%. The CALB1-CALB2 primer pair was positive for all 26 strains of C. albicans. Since all isolates of Candida dubliniensis, Candida guilliermondii, Candida kefyr, Candida krusei, Candida lusitaniae, Candida pichia, K. marianum, and S. cerevisiae tested negative with each of the six species-specific primer pairs, the specificity was 100%. There are at least three subgroups of C. parapsilosis (21). The first primer pair that we designed, CPA1-CPA2 (Table 1), amplified only isolates of group I, whereas the newly designed primer pair CPA3-CPA2 amplifies isolates from all groups of C. parapsilosis.
TABLE 2.
Sensitivity and specificity of multiplexed primer pairs for the identification of A. fumigatus, C. albicans, C. glabrata, C. parapsilosis, C. tropicalis, and C. neoformans, by using direct PCR from fungal colonies
| Group | Species | No. of isolates tested | No. of isolates amplified with each primer pair:
|
||||||
|---|---|---|---|---|---|---|---|---|---|
| ITS1-ITS2 | AFUM1-AFUM2 | CALB1-CALB2 | CGL1-CGL2 | CPA3-CPA2 | CTR1-CTR2 | CN5-CN4 | |||
| Yeasts | Candida albicans | 26 | 26 | 0 | 26 | 0 | 0 | 0 | 0 |
| Candida dubliniensis | 4 | 4 | 0 | 0 | 0 | 0 | 0 | 0 | |
| Candida glabrata | 17 | 17 | 0 | 0 | 17 | 0 | 0 | 0 | |
| Candida guilliermondii | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | |
| Candida kefyr | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | |
| Candida krusei | 7 | 7 | 0 | 0 | 0 | 0 | 0 | 0 | |
| Candida lusitaniae | 4 | 4 | 0 | 0 | 0 | 0 | 0 | 0 | |
| Candida parapsilosis | 85 | 85 | 0 | 0 | 0 | 85 | 0 | 0 | |
| Candida pichia | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | |
| Candida tropicalis | 35 | 35 | 0 | 0 | 0 | 0 | 35 | 0 | |
| Cryptococcus neoformans | 10 | 10 | 0 | 0 | 0 | 0 | 0 | 10 | |
| Kluyveromyces marianum | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | |
| Saccharomyces cerevisiae | 3 | 3 | 0 | 0 | 0 | 0 | 0 | 0 | |
| Hyphomycetes | Aspergillus flavus | 16 | 8 | 0 | 0 | 0 | 0 | 0 | 0 |
| Aspergillus fumigatus | 15 | 13 | 13 | 0 | 0 | 0 | 0 | 0 | |
| Aspergillus terreus | 5 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |
| Aspergillus niger | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |
| Zygomycetes | Absidia corymbifera | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| Rhizopus arrhizus | 6 | 3 | 0 | 0 | 0 | 0 | 0 | 0 | |
| Syncephalastrum spp. | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |
| Total isolates | 242 | 220a | 13 | 26 | 17 | 85 | 35 | 10 | |
All 195 yeast strains were amplified directly from samples of the colony, but only 53.2% (25 of 47) of the molds were amplified from mycelial fragments.
DISCUSSION
From the multicopy rRNA gene sequences, we designed six pairs of species-specific primers to amplify frequently encountered opportunistic pathogenic fungi (Table 1). We then developed a multiplex PCR protocol to rapidly and simultaneously identify these six species: A. fumigatus, C. albicans, C. glabrata, C. parapsilosis, C. tropicalis, and C. neoformans. All 173 isolates of the five yeast species and 13 of 15 isolates of A. fumigatus that were tested produced the signature amplicon, based on migration in agarose gels and comparison with positive controls (Table 2 and Fig. 2 and 3). (Two isolates of A. fumigatus could not be amplified directly from hyphae.) Among the 54 isolates of 14 related species that were tested, there were no false-positive PCR tests (Table 2).
In addition, we describe the use of whole yeast or hyphal cells as template for the PCRs. Omission of the DNA extraction procedure significantly decreases the time required to make an accurate identification by PCR. Although fungal cell breakage and the release of genomic DNA are undoubtedly less efficient without the preliminary extraction of DNA, adequate template was nevertheless available to yield positive PCR tests. Whole cells from all 195 yeast isolates, representing 13 species, yielded positive products with the universal fungal primers ITS1-ITS4 (Table 2). However, only 53.2% (25 of 47) of the molds produced amplicons directly from hyphal fragments. The intact yeast cells were consistently amplified probably because numerous cells were sampled and the rDNA genes being amplified are present in multiple copies (>100) per genome. Even if the DNA from most cells is not released, because of the many cells and copies of the target genes, sufficient template is available to yield positive PCR results. We have also been able to amplify fragments of many single-copy genes from several species of Candida (data not presented). For many purposes, this quick method has become routine for the amplification of DNA from cultures of yeast species. The molds were much less amenable to direct amplification, perhaps because of more intractable cell walls, abundant endogenous nucleases, inhibitors of the PCR, or other factors. There was a tendency for younger mold cultures to be more PCR positive than older cultures.
We are currently designing specific pairs of primers and multiplex PCR formats to identify additional medically relevant fungi. We are also applying these primers to the detection of pathogenic fungal DNA in clinical specimens.
Acknowledgments
This project was supported by grants from the North Carolina Biotechnology Center (9513-ARG-0018), Elan Pharmaceuticals, and the U.S. Public Health Service (AI 25783, AI 28836, and AI 44975).
REFERENCES
- 1.Ajello, L., and R. J. Hay (ed.). 1998. Medical mycology, p. 1-711. Edward Arnold, London, United Kingdom.
- 2.Barnes, R. A., D. W. Denning, E. G. V. Evans, R. J. Hay, C. C. Kibbler, A. G. Prentice, M. D. Richardson, M. M. Roberts, T. R. Rogers, D. C. Speller, D. W. Warnock, and R. E. Warren. 1996. Fungal infections: a survey of laboratory services for diagnosis and treatment. Commun. Dis. Rep. Rev. 6:R69-R75. [PubMed] [Google Scholar]
- 3.Bougnoux, M.-E., C. Dupont, J. Mateo, P. Saulnier, V. Faivre, D. Payen, and M.-H. Nicolas-Chanoine. 1999. Serum is more suitable than whole blood for diagnosis of systemic candidiasis by nested PCR. J. Clin. Microbiol. 37:925-930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Buchman, T. G., M. Rossier, W. G. Merz, and P. Charache. 1990. Detection of surgical pathogens by in vitro DNA amplification. Part I. Rapid identification of Candida albicans by in vitro amplification of a fungus-specific gene. Surgery 108:338-347. [PubMed] [Google Scholar]
- 5.Burgener-Kairuz, P., J.-P. Zuber, P. Jaunin, T. G. Buchman, J. L. Bille, and M. Rossier. 1994. Rapid detection and identification of Candida albicans and Torulopsis (Candida) glabrata in clinical specimens by species-specific nested PCR amplification of a cytochrome P-450 lanosterol-α-demethylase (L1A1) gene fragment. J. Clin. Microbiol. 32:1902-1907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Coleman, D. C., M. G. Rinaldi, K. A. Haynes, J. H. Rex, R. C. Summerbell, E. J. Anaissie, A. Li, and D. J. Sullivan. 1998. Importance of Candida species other than Candida albicans as opportunistic pathogens. Med. Mycol. 36(Suppl. 1):156-165. [PubMed] [Google Scholar]
- 7.Dasbach, E. J., G. M. Davies, and S. M. Teutsch. 2000. Burden of aspergillosis-related hospitalizations in the United States. Clin. Infect. Dis. 31:1524-1528. [DOI] [PubMed] [Google Scholar]
- 8.Elie, C. M., T. J. Lott, E. Reiss, and C. J. Morrison. 1998. Rapid identification of Candida species with species-specific DNA probes. J. Clin. Microbiol. 36:3260-3265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ellis, M. E., H. Al-Abdely, A. Sandridge, W. Greer, and W. Ventura. 2001. Fungal endocarditis: evidence in the world literature, 1965-1995. Clin. Infect. Dis. 32:50-62. [DOI] [PubMed] [Google Scholar]
- 10.Evertsson, U., H. J. Monstein, and A. G. Johansson. 2000. Detection and identification of fungi in blood using broad-range 28S rDNA PCR amplification and species-specific hybridisation. Acta Pathol. Microbiol. Immunol. Scand. 108:385-392. [DOI] [PubMed] [Google Scholar]
- 11.Flahaut, M., D. Sanglard, M. Monod, J. Bille, and M. Rossier. 1998. Rapid detection of Candida albicans in clinical samples by DNA amplification of common regions from C. albicans-secreted aspartic proteinase genes. J. Clin. Microbiol. 36:395-401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Goodwin, S. D., J. Fiedler-Kelly, T. H. Grasela, Jr., W. A. Schell, and J. R. Perfect. 1992. A nationwide survey of clinical laboratory methodologies for fungal infections. J. Med. Vet. Mycol. 30:153-160. [DOI] [PubMed] [Google Scholar]
- 13.Gottfredsson, M., G. M. Cox, and J. R. Perfect. 1998. Molecular methods for epidemiological and diagnostic studies of fungal infections. Pathology 30:405-418. [DOI] [PubMed] [Google Scholar]
- 14.Hazen, K. C. 1995. New and emerging yeast pathogens. Clin. Microbiol. Rev. 8:462-478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hidalgo, J. A., G. J. Alangaden, D. Eliott, R. A. Akins, J. Puklin, G. Abrams, and J. A. Vázquez. 2000. Fungal endophthalmitis diagnosis by detection of Candida albicans DNA in intraocular fluid by use of a species-specific polymerase chain reaction assay. J. Infect. Dis. 181:1198-1201. [DOI] [PubMed] [Google Scholar]
- 16.Hopfer, R. L., P. Walden, S. Setterquist, and W. E. Highsmith. 1993. Detection and differentiation of fungi in clinical specimens using polymerase chain reaction (PCR) amplification and restriction enzyme analysis. J. Med. Vet. Mycol. 31:65-75. [DOI] [PubMed] [Google Scholar]
- 17.Kappe, R., C. N. Okeke, C. Fauser, M. Maiwald, and H.-G. Sonntag. 1998. Molecular probes for the detection of pathogenic fungi in the presence of human tissue. J. Med. Microbiol. 47:811-820. [DOI] [PubMed] [Google Scholar]
- 18.Kauffman, C. A., J. A. Vázquez, J. D. Sobel, H. A. Gallis, D. S. McKinsey, A. W. Karchmer, A. M. Sugar, P. K. Sharkey, G. J. Wise, R. Mangi, A. Mosher, J. Y. Lee, W. E. Dismukes, et al. 2000. Prospective multicenter surveillance study of funguria in hospitalized patients. Clin. Infect. Dis. 30:14-18. [DOI] [PubMed] [Google Scholar]
- 19.Kobayashi, M., H. Sonobe, T. Ikezoe, E. Hakoda, Y. Ohtsuki, and H. Taguchi. 1999. In situ detection of Aspergillus 18S ribosomal RNA in invasive pulmonary aspergillosis. Intern. Med. 38:563-569. [DOI] [PubMed] [Google Scholar]
- 20.Krcméry, V., Jr., I. Krupova, and D. W. Denning. 1999. Invasive yeast infections other than Candida spp. in acute leukaemia. J. Hosp. Infect. 41:181-194. [DOI] [PubMed] [Google Scholar]
- 21.Lin, D., L.-C. Wu, M. G. Rinaldi, and P. F. Lehmann. 1995. Three distinct genotypes within Candida parapsilosis from clinical sources. J. Clin. Microbiol. 33:1815-1821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Loeffler, J., N. Henke, H. Hebart, D. Schmidt, L. Hagmeyer, U. Schumacher, and H. Einsele. 2000. Quantification of fungal DNA by using fluorescence resonance energy transfer and the light cycler system. J. Clin. Microbiol. 38:586-590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Martin, C., D. Roberts, M. van Der Weide, R. Rossau, G. Jannes, T. Smith, and M. Maher. 2000. Development of a PCR-based line probe assay for identification of fungal pathogens. J. Clin. Microbiol. 38:3735-3742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McCullough, M. J., K. V. Clemons, and D. A. Stevens. 1999. Molecular and phenotypic characterization of genotypic Candida albicans subgroups and comparison with Candida dubliniensis and Candida stellatoidea. J. Clin. Microbiol. 37:417-421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mitchell, T. G., E. Z. Freedman, T. J. White, and J. W. Taylor. 1994. Unique oligonucleotide primers in PCR for identification of Cryptococcus neoformans. J. Clin. Microbiol. 32:253-255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mitchell, T. G., and J. R. Perfect. 1995. Cryptococcosis in the era of AIDS—100 years after the discovery of Cryptococcus neoformans. Clin. Microbiol. Rev. 8:515-548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mitchell, T. G., R. L. Sandin, B. H. Bowman, W. Meyer, and W. G. Merz. 1994. Molecular mycology: DNA probes and applications of PCR technology. J. Med. Vet. Mycol. 32(Suppl. 1):351-366. [DOI] [PubMed] [Google Scholar]
- 28.Morace, G., L. Pagano, M. Sanguinetti, B. Posteraro, L. Mele, F. Equitani, G. D'Amore, G. Leone, and G. Fadda. 1999. PCR-restriction enzyme analysis for detection of Candida DNA in blood from febrile patients with hematological malignancies. J. Clin. Microbiol. 37:1871-1875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Morace, G., M. Sanguinetti, B. Posteraro, G. Lo Cascio, and G. Fadda. 1997. Identification of various medically important Candida species in clinical specimens by PCR-restriction enzyme analysis. J. Clin. Microbiol. 35:667-672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Okhravi, N., P. Adamson, R. Mant, M. M. Matheson, G. Midgley, H. M. Towler, and S. Lightman. 1998. Polymerase chain reaction and restriction fragment length polymorphism mediated detection and speciation of Candida spp causing intraocular infection. Investig. Ophthalmol. Vis. Sci. 39:859-866. [PubMed] [Google Scholar]
- 31.Polanco, A. M., E. Mellado, C. Castilla, and J. L. Rodríguez-Tudela. 1999. Detection of Candida albicans in blood by PCR in a rabbit animal model of disseminated candidiasis. Diagn. Microbiol. Infect. Dis. 34:177-183. [DOI] [PubMed] [Google Scholar]
- 32.Prariyachatigul, C., A. Chaiprasert, V. Meevootisom, and S. Pattanakitsakul. 1996. Assessment of a PCR technique for the detection and identification of Cryptococcus neoformans. J. Med. Vet. Mycol. 34:251-258. [DOI] [PubMed] [Google Scholar]
- 33.Reichard, U., S. Margraf, B. Hube, and R. Rüchel. 1997. A method for recovery of Candida albicans DNA from larger blood samples and its detection by polymerase chain reaction on proteinase genes. Mycoses 40:249-253. [DOI] [PubMed] [Google Scholar]
- 34.Reiss, E., K. Tanaka, G. Bruker, V. Chazalet, D. C. Coleman, J.-P. Debeaupuis, R. Hanazawa, J.-P. Latgé, J. Lortholary, K. Makimura, C. J. Morrison, S. Y. Murayama, S. Naoe, S. Paris, J. Sarfati, K. Shibuya, D. J. Sullivan, K. Uchida, and H. Yamaguchi. 1998. Molecular diagnosis and epidemiology of fungal infections. Med. Mycol. 36(Suppl. 1):249-257. [PubMed] [Google Scholar]
- 35.Sigler, L., and M. A. Kennedy. 1999. Aspergillus, Fusarium, and other opportunistic moniliaceous fungi, p. 1212-1241. In P. R. Murray, E. J. Baron, M. A. Pfaller, F. C. Tenover, and R. H. Yolken (ed.), Manual of clinical microbiology, 7th ed. ASM Press, Washington, D.C.
- 36.Skladny, H., D. Buchheidt, C. Baust, F. Krieg-Schneider, W. Seifarth, C. Leib-Mosch, and R. Hehlmann. 1999. Specific detection of Aspergillus species in blood and bronchoalveolar lavage samples of immunocompromised patients by two-step PCR. J. Clin. Microbiol. 37:3865-3871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sullivan, D. J., M. C. Henman, G. P. Moran, L. C. O'Neill, D. E. Bennett, D. B. Shanley, and D. C. Coleman. 1996. Molecular genetic approaches to identification, epidemiology and taxonomy of non-albicans Candida species. J. Med. Microbiol. 44:399-408. [DOI] [PubMed] [Google Scholar]
- 38.Tamura, M., K. Watanabe, T. Imai, Y. Mikami, and K. Nishimura. 2000. New PCR primer pairs specific for Candida dubliniensis and detection of the fungi from the Candida albicans clinical isolates in Japan. Clin. Lab. 46:33-40. [PubMed] [Google Scholar]
- 39.Tanaka, K., T. Miyazaki, S. Maesaki, K. Mitsutake, H. Kakeya, Y. Yamamoto, K. Yanagihara, M. A. Hossain, T. Tashiro, and S. Kohno. 1996. Detection of Cryptococcus neoformans gene in patients with pulmonary cryptococcosis. J. Clin. Microbiol. 34:2826-2828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Turenne, C. Y., S. E. Sanche, D. J. Hoban, J. A. Karlowsky, and A. M. Kabani. 1999. Rapid identification of fungi by using the ITS2 genetic region and an automated fluorescent capillary electrophoresis system. J. Clin. Microbiol. 37:1846-1851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Turin, L., F. Riva, G. Galbiati, and T. Cainelli. 2000. Fast, simple and highly sensitive double-rounded polymerase chain reaction assay to detect medically relevant fungi in dermatological specimens. Eur. J. Clin. Investig. 30:511-518. [DOI] [PubMed] [Google Scholar]
- 42.van Deventer, A. J. M., W. H. F. Goessens, A. van Belkum, E. W. M. van Etten, H. J. A. van Vliet, and H. A. Verbrugh. 1996. PCR monitoring of response to liposomal amphotericin B treatment of systemic candidiasis in neutropenic mice. J. Clin. Microbiol. 34:25-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Velegraki, A., M. E. Kambouris, G. Skiniotis, M. Savala, A. Mitroussia-Ziouva, and N. J. Legakis. 1999. Identification of medically significant fungal genera by polymerase chain reaction followed by restriction enzyme analysis. FEMS Immunol. Med. Microbiol. 23:303-312. [DOI] [PubMed] [Google Scholar]
- 44.Wahyuningsih, R., H. J. Freisleben, H.-G. Sonntag, and P. Schnitzler. 2000. Simple and rapid detection of Candida albicans DNA in serum by PCR for diagnosis of invasive candidiasis. J. Clin. Microbiol. 38:3016-3021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Walsh, T. J., and S. J. Chanock. 1998. Diagnosis of invasive fungal infections: advances in nonculture systems. Curr. Clin. Top. Infect. Dis. 18:101-153. [PubMed] [Google Scholar]
- 46.Warren, N. G., and K. C. Hazen. 1999. Candida, Cryptococcus, and other yeasts of medical importance, p. 1184-1199. In P. R. Murray, E. J. Baron, M. A. Pfaller, F. C. Tenover, and R. H. Yolken (ed.), Manual of clinical microbiology, 7th ed. ASM Press, Washington, D.C.
- 47.White, T. J., T. D. Bruns, S. B. Lee, and J. W. Taylor. 1990. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics, p. 315-322. In M. A. Innis, D. H. Gelfand, J. J. Sninsky, and T. J. White (ed.), PCR protocols. A guide to methods and applications. Academic Press, San Diego, Calif.
- 48.Wildfeuer, A., R. Schlenk, and W. Friedrich. 1996. Detection of Candida albicans DNA with a yeast-specific primer system by polymerase chain reaction. Mycoses 39:341-346. [DOI] [PubMed] [Google Scholar]
- 49.Xu, J., A. R. Ramos, R. J. Vilgalys, and T. G. Mitchell. 2000. Clonal and spontaneous origins of fluconazole resistance in Candida albicans. J. Clin. Microbiol. 38:1214-1220. [DOI] [PMC free article] [PubMed] [Google Scholar]