Abstract
Multiplex PCR assays for the detection and identification of various Streptococcus suis strains in tonsillar specimens from pigs were developed and evaluated. In two separate reactions, five distinct DNA targets were amplified. Three targets, based on the S. suis capsular polysaccharide (cps) genes specific for serotypes 1 (and 14), 7, and 9, were amplified in multiplex PCR I. Two other targets, based on the serotype 2- (and 1/2-) specific cps gene and the epf gene, encoding the EF proteins of virulent serotype 2 and highly virulent serotype 1 strains, were amplified in multiplex PCR II. To identify false-negative results, firefly luciferase (luc) DNA and primers based on the luc gene were included in the assay. The multiplex PCR assays were evaluated with tonsillar specimens from pigs infected with S. suis strains. The results obtained with the PCR assays were compared with the results obtained with a bacteriological examination. Most (94%) of the results obtained with multiplex PCR assays were confirmed by the bacteriological examination. The PCR method seems to be more sensitive compared to the bacteriological method, since the remaining 6% of the samples were positive by PCR and negative by bacteriological examination. These results indicate that the PCR method is highly specific for the detection of S. suis strains most frequently involved in clinical disease in infected pig herds. The serotypes found by PCR in tonsillar specimens from diseased pigs were compared with the serotypes of the strains isolated from the affected tissues of the same pigs. The results showed that there is significant association between carriership and clinical illness for S. suis serotype 9 and EF-positive serotype 2 strains and not for serotype 7 and EF-negative serotype 2 (or 1/2) strains.
Streptococcus suis is an important agent of meningitis, arthritis, pericarditis, peritonitis, pneumonia, and sudden death in young piglets. Most infections occur when pigs are at the age of 3 to 12 weeks. Especially after weaning, the pigs are susceptible to infection (11). The disease has a worldwide distribution and causes considerable losses to pig production (3). Attempts to control the disease are still hampered by the lack of effective vaccines and sensitive diagnostic tools.
So far, 35 serotypes of S. suis based on capsular antigens have been described (5, 6, 8, 14), with serotypes 1/2, 1, 2, 7, 9, and 14 being among the most prevalent serotypes recovered from diseased animals (29). S. suis colonizes the palatine tonsils of both healthy and diseased pigs (2, 12, 25). Subclinical carrier pigs are known to be the source of infection for young sensitive pigs (9). Detection of these carriers may lead to a better understanding of the epidemiology of S. suis infections and may help in the development of effective control measures. At present, no sensitive and specific methods are available for the detection of pigs carrying S. suis strains. The bacteria can be cultured from tonsillar specimens by traditional microbiological techniques. However, tonsils are also colonized by nonvirulent S. suis strains and other streptococcal species, which are difficult to distinguish on the basis of colony morphology. To meet this shortage, serotype-specific isolation techniques with selective-elective media (25) and an immunomagnetic separation technique (7) have been developed. However, so far their use has been limited to serotype 2 and 1/2 strains. Moreover, these methods are very laborious and time-consuming and have a low sensitivity.
For more convenient detection of specific serotypes and virulence-associated phenotypes of S. suis, PCR procedures could be used. Recently, a PCR assay was described and successfully applied to detect virulent S. suis serotype 2 and highly virulent serotype 1 strains and weakly virulent S. suis serotype 2 strains in tonsillar specimens from pigs (28). The PCR primers in this assay are based on the sequence of the extracellular protein factor (epf) gene encoding the EF protein, a marker of virulence in serotype 2 strains and highly virulent serotype 1 strains (23, 27). The PCR primers also amplified fragments of the related epf* gene, encoding the EF* protein of weakly virulent serotype 2 strains. (The epf and epf* genes are highly homologous; however, the epf* genes contain several repeated inserts [20] and thus encode proteins with higher molecular weights [asterisks are used herein to distinguish both the variant genes and the proteins they encode]. Therefore, EF*-producing strains gave rise to larger PCR products than the virulent EF-positive strains of S. suis serotype 1 and 2 [28]).
In addition, we recently developed PCR assays for detection of S. suis serotype 1 (and 14), 2 (and 1/2), 7, and 9 strains in tonsillar specimens from pigs (21, 22). These assays are based on the capsular polysaccharide (cps) biosynthesis loci of S. suis serotypes 1, 2, 7, and 9 (19, 21, 22). However, these PCR assays were not optimized for maximum sensitivity or evaluated for specificity. Moreover, a large number of individual PCR assays were required if single primer sets were used on large numbers of clinical specimens. To reduce the number of tests, we combined the single PCR assays with a set of two multiplex PCR assays. The tests were carried out in a 96-well microplate format, allowing large-scale application, and were evaluated with tonsillar specimens from pigs infected with S. suis strains.
The results showed that the multiplex PCR assays are specific and sensitive diagnostic tools suitable for the detection of pigs carrying S. suis serotype 1 (and 14), 2 (and 1/2), 7, and 9 and virulent S. suis serotype 2 and highly virulent S. suis serotype 1 strains.
MATERIALS AND METHODS
Bacteria and growth conditions.
Reference strains of S. suis serotypes 1/2 and 1 to 34 (5, 6, 8, 14), EF-positive strains of S. suis serotypes 1 (strain 6388) and 2 (strain 3), serotype 2 strains belonging to the five different EF* classes (20, 23, 27), 18 other streptococcal strains belonging to Lancefield groups A to E, G, L, P, and Q, and 24 bacterial strains belonging to the genera Staphylococcus, Micrococcus, Aerococcus, Actinobacillus, Bordetella, Escherichia, Pasteurella, Proteus, Salmonella, and Serratia, and a yeast, a Cryptococcus sp. (collection ID-Lelystad), were used in this study.
Strains were plated on Columbia blood agar plates (code CM 331; Oxoid Ltd. Inc. Columbia, Md.) supplemented with 6% horse blood and grown overnight at 37°C in air with 5% CO2. Colonies were inoculated in Todd-Hewitt broth (code CM 189; Oxoid) and grown overnight at 37°C.
Tonsillar specimens.
Tonsils from 38 pigs obtained from 28 farms were collected at the Animal Health Service, Boxtel, The Netherlands, and stored at −20°C. All pigs showed clinical signs of disease such as meningitis, sepsis, pneumonia, or arthritis. S. suis strains were isolated from tissues typically affected by S. suis such as brains, organs, lungs, or joints.
Tonsillar specimens were prepared for PCR by the multiscreen method as described previously (28).
To evaluate the results obtained with the multiplex PCR assays, S. suis serotype 1 (and 14), 2 (and 1/2), 7, and 9 and EF-positive S. suis strains were directly isolated from the tonsillar specimens with a bacteriological examination (28). To do this, tonsillar specimens were plated, and colonies were lifted onto sterilized GeneScreen Plus membranes (New England Nuclear Corp., Boston, Mass.) and hybridized with serotype-specific cps probes or an epf-specific probe (see below). Hybridizing colonies were subcultured, characterized and serotyped by standard procedures (4, 26).
PCR conditions.
Oligonucleotide primers used in the multiplex PCR assays for the detection of S. suis serotype 1 (and 14), 2 (and 1/2), and 9 and EF-positive S. suis strains were as described before (22, 28), except for the reverse primer in the serotype 2-specific PCR. The new primer has the sequence 5′-CATTTCCTAAGTCTCGCACC-3′ and corresponds to positions 14027 to 14008 in the cps2J gene (22). The primers used for the detection of serotype 7 strains correspond to positions 3185 to 3206 and 3726 to 3705 in the serotype 7-specific cps gene (21). The sequences were 5′-GAATCAATCCAGTCAGTGTTGG-3′ and 5′-CTAATTCGATACGAAGCTAAAC-3′.
In multiplex PCR I, primers specific for serotype 1 (and 14), 7, and 9 strains were combined. Amplified fragments are 441, 541, and 388 bp in length, respectively. In multiplex PCR II, primers specific for serotype 2 and EF-positive strains were combined. In this PCR, amplified fragments are 236 and 626 bp in length, respectively.
To control for failure of DNA amplification and to confirm the reliability of the PCR assays, each sample was spiked with a positive control template. For this, the pGL2-Basic vector (Promega, Madison, Wis.) encoding the firefly luciferase gene (luc) was used. Oligonucleotide primers amplifying a part of the luc gene, corresponding to positions 723 to 744 and 1672 to 1651 on the pGL2-Basic vector, had the sequences 5′-CGTCAGATTCTCGCATGCCAGA-3′ and 5′-TTGCGTCGAGTTTTCCGGTAAG-3′. This resulted in a PCR product of 949 bp.
As the template for PCR, we used 1 ng of purified chromosomal DNA (16) from bacterial strains or 25 μl of clinical sample prepared for PCR as described above. The reaction mixtures (50 μl) contained 10 mM Tris-HCl (pH 8.3), 2 mM MgCl2, 50 mM KCl, 0.2 mM each of the four deoxynucleotide triphosphates, 0.4 μM each of the primers, and 1.5 U of AmpliTaq Gold DNA polymerase (Perkin Elmer Applied Biosystems, Roche Molecular Systems, Branchburg, N.J.).
PCR mixtures were overlaid with two drops of mineral oil. DNA amplification was carried out in microplates (Thermowell H; Corning Costar, Cambridge, Mass.) in a DNA thermal cycler (Omnigene; Hybaid, Teddington, Middlesex, United Kingdom). The program used for multiplex PCR I consisted of incubation for 10 min at 95°C and 40 cycles of 0.45 min at 94.8°C, 1.10 min at 60°C, and 3 min at 72°C, followed by incubation for 10 min at 72°C. The program used for multiplex PCR II consisted of incubation for 10 min at 95°C and 40 cycles of 0.45 min at 94.8°C, 1.10 min at 63°C, and 1.20 min at 72°C, followed by incubation for 10 min at 72°C. Then 20 μl of the PCR products was separated by electrophoresis on 2% agarose gels, stained with ethidium bromide (0.25 μg/ml), and photographed under UV light.
Hybridization.
Reference strains of S. suis serotypes 1, 2, 7, and 9 and strain 3 (EF-positive) were used in individual PCR assays to amplify serotype 1-, 2-, 7-, and 9-specifc cps fragments and an epf-specific fragment, respectively. These fragments were purified from the amplification products with a High Pure PCR product purification kit (Roche) and labeled with [α-32P]dCTP (3,000 Ci mmol−1; Amersham Corp., Arlington Heights, Ill.) by use of a random primed labeling kit (Roche). The DNA on the blots was hybridized at 65°C with appropriate DNA probes, as recommended by the supplier of the GeneScreen Plus membranes. After hybridization, the membranes were washed twice with a solution of 40 mM sodium phosphate (pH 7.2)-1 mM EDTA-5% sodium dodecyl sulfate for 30 min at 65°C and twice with a solution of 40 mM sodium phosphate (pH 7.2)-1 mM EDTA-1% sodium dodecyl sulfate for 30 min at 65°C.
DNA sequence analysis.
DNA sequences were determined on an ABI Prism 3700 DNA analyzer system (Perkin Elmer Applied Biosystems). Samples were prepared by use of an ABI Prism BigDye terminator cycle sequencing ready reaction kit (Perkin Elmer Applied Biosystems).
Statistical analysis.
For each serotype, the association between clinical disease and tonsillar carriership was assessed by Fisher's exact test on independence in 2 by 2 tables. A P value smaller than 0.05 was considered significant. These tests were carried out with StatXact 4 for Windows (Cytel Software Corporation, Cambridge, Mass.).
RESULTS
Specificity of the multiplex PCR assays.
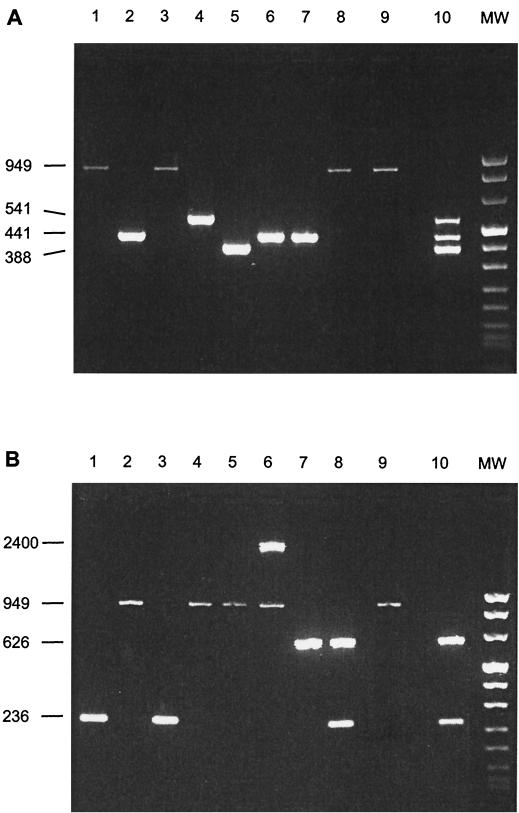
To test the specificity of the multiplex PCR assays, PCRs were performed on the various S. suis reference strains and on a panel of 18 streptococcal and 24 nonstreptococcal strains. As shown in Fig. 1A and B, PCR products of the predicted sizes were obtained on chromosomal DNAs (1 ng) of the S. suis serotype 1/2, 1, 2, 7, 9, and 14 and EF-expressing strains. Multiplex PCR II amplified a fragment of about 2,400 bp of chromosomal DNA of the S. suis serotype 14 strain (Fig. 1B, lane 6). This indicates the presence of an S. suis strain expressing an EF* protein. Multiple amplification products were observed in a mixture of chromosomal DNA of the above-mentioned eight strains (Fig. 1A and B, lane 10). No amplification products were obtained on any of the other S. suis serotypes or on any of the other strains examined (results not shown). This indicates that the multiplex PCR assays are highly specific for the detection of S. suis serotype 1, 2, 1/2, 7, 9, and 14 and EF-positive S. suis strains.
FIG. 1.
PCR products obtained in multiplex PCR I (A) or multiplex PCR II (B) with purified chromosomal DNA (1 ng) of eight S. suis strains spiked with 10 fg of pGL2-Basic vector. Samples were separated on a 2% agarose gel stained with ethidium bromide. Lane designations are indicated. S. suis serotype 1/2, strain 5209 (lane 1); S. suis serotype 1, strain 5210 (lane 2); S. suis serotype 2 (EF*-positive), strain 5211 (lane 3); S. suis serotype 7, strain 5216 (lane 4); S. suis serotype 9, strain 5218 (lane 5); S. suis serotype 14, strain 5223 (EF*-positive) (lane 6); S. suis type 1 (EF-positive), strain 6388 (lane 7); S. suis type 2 (EF-positive), strain 4005 (lane 8); negative control (without S. suis DNA) (lane 9); mixture of chromosomal DNA (1 ng) of strains in lanes 1 to 8 (lane 10). Lane MW contains DNA molecular size markers (0.019 to 1.11 kbp; Roche); the sizes (in base pairs) of the PCR products are indicated on the left.
As expected, in the negative control (without chromosomal S. suis DNA), an amplification product of the internal positive control was detected (Fig. 1A and B, lane 9). No interference was observed between the internal positive control primer set with chromosomal DNA (1 ng) of the S. suis reference strains and vice versa between the S. suis primer sets and internal positive control DNA (1 ng) (results not shown).
We also used the multiplex PCR II on chromosomal DNA of EF*-producing serotype 2 strains. In addition to the serotype 2-specific cps PCR products, epf* PCR products of various sizes (1,278, 1,505, 2,313, 2,537, and 2,993 bp) were expected (28). However, a faint amplification product was obtained on chromosomal DNA of the strain expressing the smallest EF* protein, whereas no amplification products could be detected in the other EF*-producing strains (results not shown). Apparently, multiplex PCR II is not able to efficiently detect EF*-producing strains.
Sensitivity of the two multiplex PCR assays.
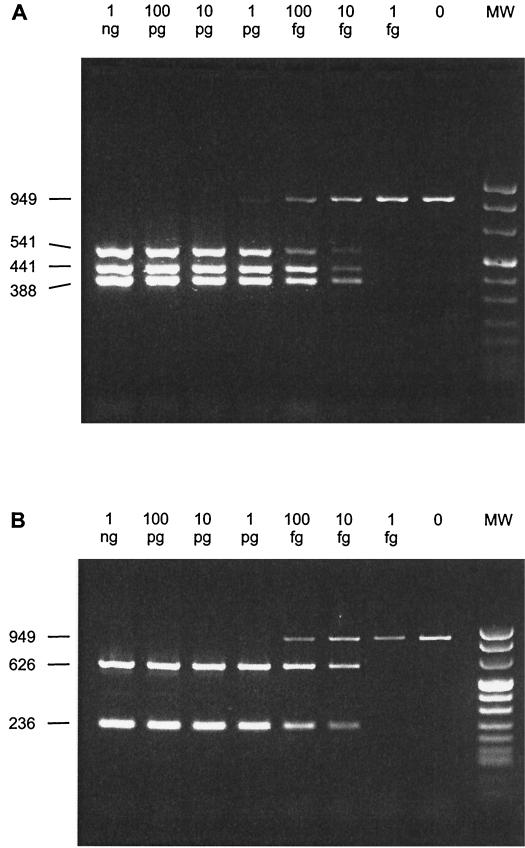
The sensitivity of the multiplex PCR assays was determined with chromosomal DNA of S. suis serotype 1, 2, 7, and 9 and an EF-positive serotype 2 strain. In both multiplex PCR assays, 10 fg of chromosomal DNA of the various serotypes and phenotypes was sufficient to amplify a fragment which could easily be detected by eye on an agarose gel (Fig. 2A and B).
FIG. 2.
Sensitivity of multiplex PCR I (A) and II (B) on a mixture of chromosomal DNAs of S. suis serotype 1, 7, and 9 (A) or on an EF-positive S. suis serotype 2 strain (B). Samples were separated on a 2% agarose gel stained with ethidium bromide. The amount of target DNA tested in the PCR assays is indicated above the lanes. All samples contained 10 fg of positive control DNA. Lane MW contains DNA molecular size markers; the sizes (in base pairs) of the PCR products are indicated on the left.
Evaluation of the multiplex PCR assays.
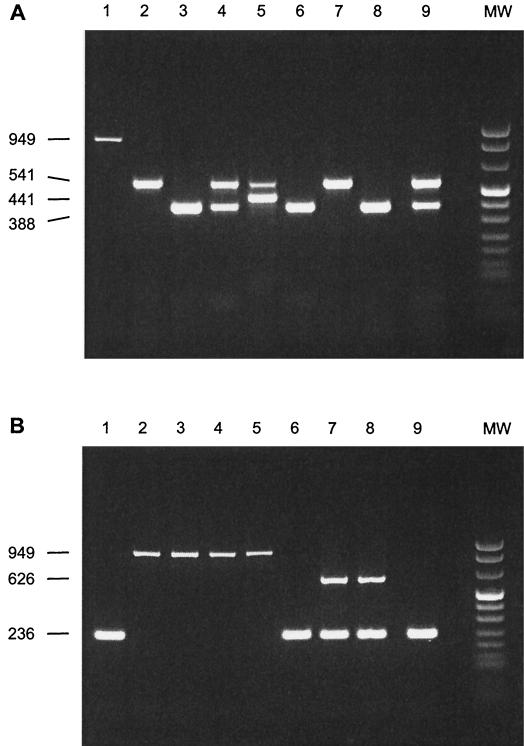
We subsequently analyzed tonsillar specimens by multiplex PCR assays and by bacteriological examination (28) to compare the two tests. The results are summarized in Table 1, and several examples are shown in Fig. 3A and B. All tonsillar specimens bacteriologically positive for a specific serotype or phenotype were also positive in the corresponding PCR. Moreover, tonsils negative in the multiplex PCR for a specific serotype or phenotype were also negative for this particular serotype or phenotype with the bacteriological method. Some samples positive in the multiplex PCR tested bacteriologically negative (3 of 17 for serotype 7, 4 of 28 for serotype 9, and 6 of 18 for serotype 2 [and 1/2]). This suggests that, especially for serotype 2 (and 1/2), the PCR method is more sensitive than the bacteriological examination. To corroborate this and to exclude false-positive PCR results, we repeatedly tested the samples both by PCR and by bacteriological examination. The same results were obtained. Moreover, the specificity of the serotype 2 PCR product of samples, which could not be confirmed bacteriologically, was further examined by sequence analysis and cross-hybridization experiments. All fragments showed the expected serotype 2-specific sequence and hybridized specifically with the serotype 2-specific probe (data not shown). From these data, we conclude that the multiplex PCR assays are more sensitive than the bacteriological examination.
TABLE 1.
Comparison of multiplex PCR assays and bacteriological examination for 38 tonsillar specimens from diseased pigs
| Multiplex PCR resulta | No. (%) of tonsillar specimens
|
|||
|---|---|---|---|---|
| PCR positive, bacterial exam positive | PCR positive, bacterial exam negative | PCR negative, bacterial exam negative | PCR negative, bacterial exam positive | |
| Serotype 1 (or 14) | 1b (3) | 0 (0) | 37 (97) | 0 (0) |
| Serotype 2 (or 1/2) | 12c (31) | 6 (16) | 20 (53) | 0 (0) |
| EF positive | 5 (13) | 0 (0) | 33 (87) | 0 (0) |
| Serotype 7 | 14 (37) | 3 (8) | 21 (55) | 0 (0) |
| Serotype 9 | 28 (74) | 4 (10) | 6 (16) | 0 (0) |
| Total | 60 (31) | 13 (7) | 117 (62) | 0 (0) |
Similar PCR products were amplified for S. suis serotype 1 and 14 strains and for serotype 2 and 1/2 strains due to common capsular genes (18).
S. suis serotype 14 strain as detected by bacteriological examination.
S. suis serotype 1/2 (7 of 12) and EF-positive S. suis serotype 2 strains (5 of 12) as detected by bacteriological examination.
FIG. 3.
PCR products in multiplex PCR I (A) and multiplex PCR II (B) obtained directly on tonsillar specimens collected from diseased pigs carrying serotype 1 (and 14), 2 (and 1/2), 7, and 9 and EF-positive S. suis strains as detected by bacteriological examination. PCR products were obtained from tonsillar specimens collected from pigs carrying S. suis serotype 1/2 (lane 1); S. suis serotype 7 (lane 2); S. suis serotype 9 (lane 3); S. suis serotype 7 and 9 strains (lane 4); S. suis serotype 7 and 14 strains (lane 5); S. suis serotype 1/2 and 9 strains (lane 6); S. suis serotype 2 (EF-positive phenotype) and 7 strains (lane 7); S. suis serotype 2 (EF-positive phenotype) and 9 strains (lane 8); and S. suis serotype 1/2, 7, and 9 strains (lane 9). All samples contained 10 fg of positive control DNA. Lane MW contains DNA molecular size markers; the sizes (in base pairs) of the PCR products are indicated on the left.
Comparison of serotypes isolated from tonsils and affected tissues of diseased pigs.
We next compared the results of the serotypes found by PCR in tonsillar specimens of diseased pigs and the serotypes of the strains isolated from the affected tissues of the same pigs. Table 2 shows that in 89% (34 of 38) of the cases, an S. suis strain in the tonsils belonged to the same serotype as the disease-causing strain. For some serotypes, a significant (P < 0.05) association between clinical illness and carrier state could be found. Four of five pigs carrying EF-positive S. suis serotype 2 strains and most of the pigs (27 of 32) carrying a serotype 9 strain were also clinically ill due to such a strain. Such an association was not found in EF-negative serotype 2 (and 1/2) and 7 strains. One and two pigs suffered from serotype 1/2 and 7 infections, respectively, whereas 13 and 17 pigs, respectively, carried these bacteria on their tonsils. This could indicate that compared to the EF-positive serotype 2 and serotype 9 strains, EF-negative serotype 2 (and 1/2) and 7 strains are less virulent.
TABLE 2.
S. suis strains isolated from affected tissues compared with S. suis strains detected by multiplex PCR assays on tonsils from 38 diseased pigs
| S. suis strain | No. of pigs in which bacteria were:
|
||
|---|---|---|---|
| Isolated from affected tissues | Detected by PCR on tonsils | Isolated from affected tissues and detected by PCR on tonsils | |
| Serotype 1 (or 14) | 1 | 0 | 0 |
| Serotype 2 (or 1/2)a | 1 | 13 | 1 |
| EF positive | 4 | 5 | 4 |
| Serotype 7 | 2 | 17 | 2 |
| Serotype 9a | 29 | 32 | 27 |
| NTb | 1 | 0 | 0 |
| Total | 38 | 61 | 34 |
Significant association between clinical disease and tonsillar carriership.
NT, nontypeable.
Multiple serotypes in tonsillar specimens from pigs.
In 66% (25 of 38) of the tonsillar specimens examined, two or more S. suis strains were identified by the multiplex PCR assays on the same tonsillar specimen (Table 3). Most frequently, a combination of serotypes 9 and 2 (and 1/2) (18%) and serotypes 9 and 7 (16%) strains were detected by the multiplex PCR assays. In five (13%) of the tonsillar samples, the multiplex PCR assays identified S. suis strains belonging to three different serotypes, serotypes 2 (and 1/2), 7, and 9.
TABLE 3.
Multiple serotypes of S. suis strains in tonsillar specimens from diseased pigs as detected by multiplex PCR assays
| No. of S. suis strains on tonsillar specimens | No. (%) of specimens |
S. suis strain(s) detecteda
|
||||
|---|---|---|---|---|---|---|
| Serotype 1 (or 14) | Serotype 2 (or 1/2) | EF positive | Serotype 7 | Serotype 9 | ||
| 1 | 11 (29) | − | − | − | − | + |
| 1 (3) | − | − | − | + | − | |
| 1 (3) | − | + | − | − | − | |
| 2 | 7 (18) | − | + | − | − | + |
| 6 (16) | − | − | − | + | + | |
| 3 (8) | − | +* | +* | − | + | |
| 2 (5) | − | +* | +* | + | − | |
| 1 (3) | − | + | − | + | − | |
| 1 (3) | + | − | − | + | − | |
| 3 | 5 (13) | − | + | − | + | + |
S. suis serotype 1 (and 14), 7, and 9 strains were detected by multiplex PCR I, and S. suis serotype 2 (and 1/2) and EF-positive S. suis strains were detected by multiplex PCR II. −, negative result; +, positive result; *, tonsillar specimens which contained an EF-positive serotype 2 strain.
DISCUSSION
We previously showed that a PCR based on the epf gene specifically detects virulent strains of S. suis serotype 2 and highly virulent strains of S. suis serotype 1 (28). In addition, PCR assays on the serotype-specific cps genes of S. suis serotypes 1, 2, 7, and 9 specifically identified S. suis serotype 1 (and 14), 2 (and 1/2), 7, and 9 strains (21, 22). In the present study, we improved the diagnostic value of these PCR methods by a multiplex approach. In two separate reactions, we could easily identify serotype 1 (and 14), 7, and 9 (multiplex PCR I) as well as serotype 2 (and 1/2) and EF-positive S. suis strains (multiplex PCR II). The multiplex PCR assays are easy to perform and allow large-scale application; 96 samples can be processed simultaneously. Compared to standard bacteriological assays, the PCR assays are much more rapid to perform. Therefore, these assays may be important diagnostic tools to detect pigs carrying the most frequently isolated serotypes and virulence-associated phenotypes of S. suis. It may be applicable for epidemiological and transmission studies and can contribute to efforts to control or eradicate S. suis infections.
Evaluation of the multiplex PCR assays with tonsillar specimens from diseased pigs showed that both assays are highly specific and sensitive. The bacteriological examination confirmed most (94%) of the results obtained with the multiplex PCR assays. The PCR method seemed to be more sensitive than the bacteriological method, since 13 of 73 samples positive by PCR were negative by bacteriological examination. Low levels of bacterial cells (live or dead) in the tonsillar specimens may explain the differences in results obtained with the two methods. S. suis strains producing an EF* protein yielded in multiplex PCR II only the serotype 2 products, although larger epf* PCR products of various sizes were also expected (28). Apparently, in the multiplex PCR, the epf* gene was far less efficiently amplified than the 236-bp amplicon of the serotype 2-specific cps gene. Since the frequency of S. suis strains isolated from diseased pigs which produce the EF* protein is very low (29), detecting strains producing an EF* protein in the multiplex PCR is a not a necessity.
In the bacteriological examination, we isolated serotype 1/2 and 14 strains which hybridized specifically with the serotype 2 and 1 probes, respectively. In early studies it was shown that serotype 1/2 strains cross-react in agglutination tests with antiserum against serotype 1 and serotype 2 strains (13). For serotype 1 and 14 strains, a one-way capsular cross-reaction in agglutination tests was described (6).
A significant association between carriership and clinical illness was observed for S. suis serotype 9 and EF-positive strains and not for serotype 7 and EF-negative serotype 2 (or 1/2) strains. This could indicate that, compared to serotype 9 and EF-positive strains, serotype 7 and EF-negative serotype 2 (or 1/2) strains are less virulent. This idea corresponds well with the observation that in The Netherlands, serotype 9 and EF-positive strains were more frequently isolated from diseased pigs than serotype 7 and EF-negative serotype 2 (or 1/2) strains (29). Moreover, our data confirmed earlier studies indicating that EF-negative serotype 2 and 1/2 strains are possibly less or nonvirulent for pigs compared to EF-positive serotype 2 strains (27, 29). A role for EF in the virulence of S. suis serotype 7 strains is not very likely because it was found in recent studies that all serotype 7 strains isolated from diseased pigs were EF-negative (1, 29). Whether the serotype 7 strains detected by the multiplex PCR assays in this study are less virulent than serotype 9 or EF-positive serotype 2 strains or whether these strains are totally nonvirulent is unknown. Experimental infections in pigs will be necessary to examine the virulence of the serotype 7 strains.
So far, the multiplex PCR assays have been carried out on specimens consisting of tonsillar specimens from pigs. However, for routine detection of carriers, tonsillar swab specimens from live animals would be preferred as a sampling method. Our experience is that the PCR assay is applicable to tonsillar swab specimens from live pigs, perhaps with a slightly lower sensitivity (24). Nevertheless, additional experiments in which carrier rates obtained on whole tonsils are compared with those on tonsillar swabs and tonsillar biopsies are necessary.
Multiple S. suis serotypes were found on tonsils of diseased pigs. Earlier, it was found that pigs can be infected with multiple serotypes of S. suis (15, 17). These findings may account in part for the difficulty in disease control by vaccines. At present, for control of the disease by vaccines, autogenous bacterins are used. It seemed that these vaccines confer protection only against challenge with strains of a homologous serotype (10). It can be hypothesized that a bacterin vaccine prepared from one serotype may suppress clinical disease caused by that particular serotype, but that new outbreaks caused by S. suis strains belonging to other serotypes may occur. Therefore, identification of specific strains, not only those involved in clinical disease in infected herds but also those involved in the carrier state, may be needed to provide adequate control measures. The current multiplex PCR assays can contribute to such an approach.
Acknowledgments
We gratefully acknowledge K. Peperkamp, Animal Health Service, Boxtel, The Netherlands, for kindly collecting and providing tonsillar specimens.
REFERENCES
- 1.Allgaier, A., R. Goethe, H. J. Wisselink, H. E. Smith, and P. Valentin Weigand. 2001. Relatedness of Streptococcus suis isolates of various serotypes and clinical backgrounds as evaluated by macrorestriction analysis and expression of potential virulence traits. J. Clin. Microbiol. 39:445-453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arends, J. P., N. Hartwig, M. Rudolphy, and H. C. Zanen. 1984. Carrier rate of Streptococcus suis capsular type 2 in palatine tonsils of slaughtered pigs. J. Clin. Microbiol. 20:945-947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chengappa, M. M., L. W. Pace, J. A. Williams, C. H. Herren, and S. E. Ascher. 1990. Efficacy of tiamulin against experimentally induced Streptococcus suis type-2 infection in swine. J. Am. Vet. Med. Assoc. 197:1467-1470. [PubMed] [Google Scholar]
- 4.Devriese, L. A., K. Ceyssens, J. Hommez, R. Kilpper Balz, and K. H. Schleifer. 1991. Characteristics of different Streptococcus suis ecovars and description of a simplified identification method. Vet. Microbiol. 26:141-150. [DOI] [PubMed] [Google Scholar]
- 5.Gottschalk, M., R. Higgins, M. Jacques, M. Beaudoin, and J. Henrichsen. 1991. Characterization of six new capsular types (23 through 28) of Streptococcus suis. J. Clin. Microbiol. 29:2590-2594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gottschalk, M., R. Higgins, M. Jacques, K. R. Mittal, and J. Henrichsen. 1989. Description of 14 new capsular types of Streptococcus suis. J. Clin. Microbiol. 27:2633-2636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gottschalk, M., S. Lacouture, and L. Odierno. 1999. Immunomagnetic isolation of Streptococcus suis serotypes 2 and 1/2 from swine tonsils. J. Clin. Microbiol. 37:2877-2881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Higgins, R., M. Gottschalk, M. Boudreau, A. Lebrun, and J. Henrichsen. 1995. Description of six new capsular types (29-34) of Streptococcus suis. J. Vet. Diagn. Investig. 7:405-406. [DOI] [PubMed] [Google Scholar]
- 9.Higgins, R., M. Gottschalk, K. R. Mittal, and M. Beaudoin. 1990. Streptococcus suis infection in swine. A sixteen month study. Can. J. Vet. Res. 54:170-173. [PMC free article] [PubMed] [Google Scholar]
- 10.Kebede, M., M. M. Chengappa, and J. G. Stuart. 1990. Isolation and characterization of temperature-sensitive mutants of Streptococcus suis: efficacy trial of the mutant vaccine in mice. Vet. Microbiol. 22:249-257. [DOI] [PubMed] [Google Scholar]
- 11.Lamont, M. H., P. T. Edwards, and R. S. Windsor. 1980. Streptococcal meningitis in pigs: results of a five-year survey. Vet. Rec. 107:467-469. [DOI] [PubMed] [Google Scholar]
- 12.Mwaniki, C. G., I. D. Robertson, and D. J. Hampson. 1994. The prevalence of Streptococcus suis type 2 in Western Australian piggeries. Aust. Vet. J. 71:385-386. [DOI] [PubMed] [Google Scholar]
- 13.Perch, B., E. Kjems, P. Slot, and K. B. Pedersen. 1981. Biochemical and serological properties of R, S, and RS streptococci. Acta Pathol. Microbiol. Scand. B 89:167-171. [DOI] [PubMed] [Google Scholar]
- 14.Perch, B., K. B. Pedersen, and J. Henrichsen. 1983. Serology of capsulated streptococci pathogenic for pigs: six new serotypes of Streptococcus suis. J. Clin. Microbiol. 17:993-996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Reams, R. Y., D. D. Harrington, L. T. Glickman, H. L. Thacker, and T. L. Bowersock. 1996. Multiple serotypes and strains of Streptococcus suis in naturally infected swine herds. J. Vet. Diagn. Investig. 8:119-121. [DOI] [PubMed] [Google Scholar]
- 16.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 17.Sihvonen, L., D. N. Kurl, and P. Salmela. 1986. Infection with Streptococcus suis serotypes 1 and 2 in the same diseased pig. Acta Vet. Scand. 27:626-628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Smith, H. E., M. Damman, J. van der Velde, F. Wagenaar, H. J. Wisselink, N. Stockhofe-Zurwieden, and M. A. Smits. 1999. Identification and characterization of the cps locus of Streptococcus suis serotype 2: the capsule protects against phagocytosis and is an important virulence factor. Infect. Immun. 67:1750-1756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Smith, H. E., R. de Vries, R. van't Slot, and M. A. Smits. 2000. The cps locus of Streptococcus suis serotype 2: genetic determinant for the synthesis of sialic acid. Microb. Pathog. 29:127-134. [DOI] [PubMed] [Google Scholar]
- 20.Smith, H. E., F. H. Reek, U. Vecht, A. L. Gielkens, and M. A. Smits. 1993. Repeats in an extracellular protein of weakly pathogenic strains of Streptococcus suis type 2 are absent in pathogenic strains. Infect. Immun. 61:3318-3326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Smith, H. E., L. van Bruijnsvoort, H. Buijs, H. J. Wisselink, and M. A. Smits. 1999. Rapid PCR test for Streptococcus suis serotype 7. FEMS Microbiol. Lett. 178:265-270. [DOI] [PubMed] [Google Scholar]
- 22.Smith, H. E., V. Veenbergen, J. van der Velde, M. Damman, H. J. Wisselink, and M. A. Smits. 1999. The cps genes of Streptococcus suis serotypes 1, 2, and 9: development of rapid serotype-specific PCR assays. J. Clin. Microbiol. 37:3146-3152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Stockhofe-Zurwieden, N., U. Vecht, H. J. Wisselink, H. van Lieshout, and H. E. Smith. 1996. Comparative studies on the pathogenicity of different Streptococcus suis serotype 1 strains, p. 299. In P. G. Monetti and G. Vignola (ed.), Proceedings of the 14th International Pig Veterinary Society Congress. University of Bologna, Bologna, Italy.
- 24.Swildens, B., H. J. Wisselink, H. E. Smith, and J. H. M. Verheijden. 2000. Tonsillar swabs can be used in a PCR assay for the detection of virulent Streptococcus suis serotype 2 strains in weaned piglets, p. 535. In C. Cargill and S. McOrist (ed.), Proceedings of the 16th International Pig Veterinary Society Congress, Melbourne, Australia.
- 25.Van Leengoed, L. A., U. Vecht, and E. R. M. Verheyen. 1987. Streptococcus suis type 2 infections in pigs in the Netherlands (part two). Vet. Q. 9:111-117. [DOI] [PubMed] [Google Scholar]
- 26.Vecht, U., L. A. van Leengoed, and E. R. M. Verheijen. 1985. Streptococcus suis infections in pigs in the Netherlands (part one). Vet. Q. 7:315-321. [DOI] [PubMed] [Google Scholar]
- 27.Vecht, U., H. J. Wisselink, J. E. van Dijk, and H. E. Smith. 1992. Virulence of Streptococcus suis type 2 strains in newborn germfree pigs depends on phenotype. Infect. Immun. 60:550-556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wisselink, H. J., F. H. Reek, U. Vecht, N. Stockhofe-Zurwieden, M. A. Smits, and H. E. Smith. 1999. Detection of virulent strains of Streptococcus suis type 2 and highly virulent strains of Streptococcus suis type 1 in tonsillar specimens from pigs by PCR. Vet. Microbiol. 67:143-157. [DOI] [PubMed] [Google Scholar]
- 29.Wisselink, H. J., H. E. Smith, N. Stockhofe-Zurwieden, K. Peperkamp, and U. Vecht. 2000. Distribution of capsular types and production of muramidase-released protein (MRP) and extracellular factor (EF) of Streptococcus suis strains isolated from diseased pigs in seven European countries. Vet. Microbiol. 74:237-248. [DOI] [PubMed] [Google Scholar]