Abstract
Repetitive sequence-based PCR (rep-PCR) and amplified fragment length polymorphism (AFLP) were used to characterize a sample of 43 field isolates and 4 attenuated vaccine strains of Pasteurella multocida recovered from multiple avian species. Both rep-PCR and AFLP assays were rapid and reproducible, with high indices of discrimination. Concordance analyses of rep-PCR and AFLP with somatic serotyping indicate that, in general, somatic serotyping is a poor indicator of genetic relatedness among isolates of P. multocida. In addition, the data provide evidence of host specificity of P. multocida clones. Overall, the results of our study indicate that the rep-PCR and AFLP techniques enable rapid fingerprinting of P. multocida isolates from multiple avian species and enhance the investigation of fowl cholera outbreaks.
Pasteurella multocida is an aerobic, gram-negative, nonmotile bacterium that causes fowl cholera in domestic poultry and avian pasteurellosis in other avian species. The disease usually occurs in two forms, an acute septicemia with high morbidity and mortality rates and chronic localized infection of joints and sinuses (17). Five capsular serogroups, designated A, B, D, E, and F, and 16 somatic serotypes (designated 1 through 16) of P. multocida have been described (17, 18). Although serotyping of both capsular and somatic antigens has proved to be very useful for detection and identification of this bacterium, it is limited in that it provides insufficient information for epidemiological studies to distinguish among different strains of the same serotype.
While a variety of molecular subtyping techniques, including restriction endonuclease analysis (4, 31, 32, 33), plasmid profiling (4, 16), and ribotyping (2, 21), have been used for subtyping of P. multocida, these techniques in general have several limitations. Restriction endonuclease analysis and ribotyping are limited in that they are time-consuming and labor intensive. Plasmid profiling is relatively easy and inexpensive but has limited applications in epidemiological investigations because all isolates may not carry plasmids or they might be lost during growth in laboratory media (23).
The objective of this study was to determine the utility of two rapid PCR-based approaches, repetitive sequence-based PCR (rep-PCR) and amplified fragment length polymorphism (AFLP), for differentiation of P. multocida isolates from multiple avian species and fowl cholera outbreak investigations to the subspecies level.
Bacterial isolates and DNA isolation.
The geographic origin and source of the P. multocida isolates are presented in Table 1. Thirty-eight isolates of P. multocida were obtained from the National Animal Disease Center, Ames, Iowa, and the University of Minnesota, St. Paul, Minn. An additional nine clinical isolates (96-174, 96-226, 96-235, 96-240, 96-246, 96-248, 96-260, 96-265, and 96-269) were obtained from outbreaks in turkey farms in Minnesota. The methods used to prepare genomic DNA from bacterial isolates have been described previously (1).
TABLE 1.
Properties of 47 P. multocida isolates
| Straina | Host | Sourceb | Somatic typec | rep-PCR type | AFLP type |
|---|---|---|---|---|---|
| 96-265* | Turkey | Minnesota | NA | 1 | 1 |
| 96-248* | Turkey | Minnesota | 3 | 1 | 1 |
| 96-246* | Turkey | Minnesota | 3 | 1 | 1 |
| 96-174** | Turkey, lung | Minnesota | 3 | 1 | 1 |
| 2220 | Turkey | Iowa | 3 | 1 | 2 |
| 4038 | Turkey | Oregon | 3 | 1 | 2 |
| CU | Turkey | UMN | 3,4 | 1 | 3 |
| CU | Turkey | UMN | 3,4 | 1 | 3 |
| 4020 | Turkey | Texas | 3 | 1 | 3 |
| 4021 | Turkey | Texas | 3 | 1 | 3 |
| 96-235*** | Turkey | Minnesota | 3 | 2 | 3 |
| 96-226*** | Turkey, lung | Minnesota | 3 | 2 | 3 |
| 96-260**** | Turkey | Minnesota | NA | 3 | 4 |
| 3704 | Turkey | South Dakota | 3,4,12 | 3 | 4 |
| 3707 | Turkey | South Dakota | 3,4,12 | 3 | 4 |
| 3758 | Turkey | Kansas | 3,4,12 | 3 | 4 |
| 4031 | Turkey | Iowa | 3 | 3 | 4 |
| 4032 | Turkey | Iowa | 3 | 3 | 4 |
| 5314 | Turkey | Iowa | 3,4 | 3 | 4 |
| CU | Turkey | UMN | 3,4 | 3 | 5 |
| 5167 | Turkey | Germany | 3,4 | 4 | 2 |
| 1996 | Bald eagle | California | 1 | 5 | 6 |
| 2148 | Duck | Utah | 1 | 5 | 6 |
| 2410 | Duck | Utah | 1 | 5 | 6 |
| 3200 | NA | UMN | 3 | 6 | 16 |
| 4601 | Quail | Virginia | 1 | 7 | 7 |
| 4602 | Quail | Virginia | 1 | 7 | 7 |
| 2846 | Chicken | Singapore | 1 | 7 | 10 |
| 2847 | Duck | Singapore | 1 | 7 | 10 |
| 2855 | Chicken | Singapore | 1 | 7 | 10 |
| 4247 | Chicken | Michigan | 1 | 7 | 10 |
| 4251 | Duck | Michigan | 1 | 7 | 10 |
| 5439 | Chicken | Egypt | 1 | 7 | 10 |
| 5440 | Turkey | Egypt | 1 | 7 | 10 |
| 4910 | Chicken | Bangladesh | 1 | 8 | 8 |
| 2852 | Chicken | Singapore | 1 | 8 | 10 |
| 2848 | Duck | Singapore | 1 | 8 | 11 |
| V2283 | NA | UMN | 3,4 | 9 | 19 |
| 2853 | Duck | Singapore | 1 | 10 | 15 |
| 5288 | Turkey | California | 10 | 11 | 20 |
| 96-240***** | Turkey | Minnesota | 3 | 12 | 17 |
| 96-269****** | Turkey | Minnesota | NA | 13 | 18 |
| 5162 | Turkey | Germany | 3 | 14 | 12 |
| 1896 | Turkey | Canada | 4 | 15 | 13 |
| 4052 | Chicken | Arkansas | 10 | 16 | 9 |
| 2879 | Duck | Maryland | 3,12,15 | 17 | 14 |
| 2887 | Duck | Maryland | 3,4,12 | 17 | 14 |
*, isolates recovered from farm A; **, isolates recovered from farm B; ***, isolates recovered from farm C; ****, isolates recovered from farm D; *****, isolates recovered from farm E; ******, isolates recovered from farm F; CU, Clemson University.
UMN, University of Minnesota.
NA, not available.
rep-PCR.
rep-PCR was performed as previously described with some modifications (1, 28, 34). Oligonucleotide primers ERIC1R (5′-ATGTAAGCTCCTGGGGATTCAC) and ERIC2 (5′-AAGTAAGTGACTGGGGTGAGCG) were used to generate DNA fingerprints in this study. To evaluate the reproducibility of the rep-PCR fingerprints, three separate reactions were performed for each isolate.
AFLP.
AFLP was performed as described in the AFLP microbial fingerprinting kit protocol (Perkin Elmer-Applied Biosystems Division), Foster City, Calif.).
Selective amplification was performed with the EcoRI selective primer containing a fluorescent label on the 5′ end with additional base A (carboxyfluorescein dye [FAM]-EcoRI-A: FAM-5′-GACTGCGTACCAATTCA) and the MseI selective primer with no additional base (MseI-O: 5′-GATGAGTCCTGAGTAA). The AFLP fingerprint profiles were automatically analyzed with GeneScan (Perkin-Elmer) with GeneScan-500 Rhodamine X (ROX) size standard as an internal size control. Sizes of amplified products were then tabulated and exported for cluster analysis with Molecular Analyst software (Bio-Rad, Hercules, Calif.). To analyze AFLP fingerprints, DNA fragments from 100 to 500 bp in size were included for cluster analysis.
Computer-assisted analysis of rep-PCR and AFLP fingerprints.
Molecular Analyst software (Bio-Rad) was used to compare the rep-PCR and AFLP fingerprint profiles among P. multocida isolates. The program automatically computed the similarity for each pair of fingerprints on the basis of band positions with the Dice coefficient (SD). In this investigation, the cutoff value of SD was 0.90. This cutoff value is assigned based on experience with the gel analysis software to ensure that the same bacterial strain run on different PCRs and different gel electrophoreses will be identified as the same type. Pairs of isolates with a similarity coefficient (SD) of 0.90 were considered similar and not distinguishable. Cluster analysis among isolates was performed by the unweighted pair group average (UPGMA) linkage method (20).
Calculation of index of discrimination.
The index of discrimination (D) indicates the probability that the fingerprinting method will identify two unrelated bacterial strains as different fingerprint types. Statistical analysis to determine the discriminating power of rep-PCR and AFLP was performed with the discriminatory index (8).
Concordance analysis of fingerprinting methods.
Concordance between subtyping techniques was calculated by comparing all possible pairs of 44 isolates (1,081 pairwise comparisons) and classifying each pair of isolates for whether both isolates were in the same or different subtype and whether they matched or mismatched in another subtype. The sum of the same somatic type match rep-PCR or AFLP types and of different somatic types mismatch rep-PCR or AFLP types represents the percent agreement of the pairwise comparisons of isolates. The statistical analysis of concordance was performed with the G test of independence (22).
DNA fingerprinting of avian isolates of P. multocida with rep-PCR.
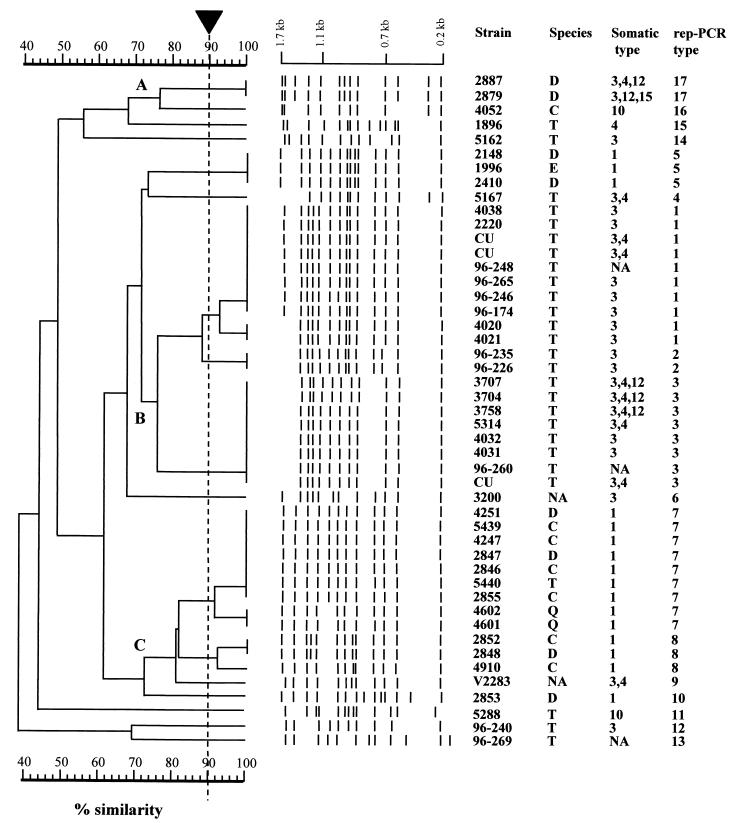
Cluster analysis assigned the 47 avian isolates of P. multocida into 17 distinct fingerprint patterns (Table 1 and Fig. 1). The average number of bands per rep-PCR fingerprint of P. multocida was 12.4 ± 1.3 (range, 10 to 14). Repeated PCRs for each isolate revealed identical rep-PCR fingerprint patterns. The index of discrimination of rep-PCR for differentiation of the P. multocida isolates was 0.89. At an arbitrary 75% similarity cutoff level, the 47 isolates could be further grouped into three major clusters, designated A through C (Fig. 1). It is noteworthy that all of the isolates (20 of 20) in cluster B (rep-PCR patterns 1, 2, and 3) were recovered from turkeys. On the other hand, only 7.69% (1 of 13) of the isolates recovered from turkeys were found in rep-PCR patterns 7, 8, and 9 (cluster C) (Fig. 1).
FIG. 1.
UPGMA dendrogram of genetic relatedness of 17 rep-PCR types (1 to 17) of 47 P. multocida isolates recovered from avian sources. The scale represents percent similarity generated by computer-assisted comparison of rep-PCR fingerprint profiles. The arrow indicates the cutoff value (90%) for cluster analysis. Species: D, duck; C, chicken; T, turkey; E, bald eagle; Q, quail. NA, not available; CU, Clemson University.
DNA fingerprinting of avian isolates of P. multocida with AFLP. (i) AFLP fingerprints of P. multocida.
The fingerprints obtained by AFLP with primer FAM-EcoRI-A:MseI-O were highly polymorphic. The average number of bands per isolate was 20.6 ± 2.9, ranging from 16 to 25 bands. A dendrogram generated by computer-assisted analysis of 47 AFLP fingerprints is shown in Fig. 2. AFLP characterized the 47 P. multocida isolates into 20 distinct AFLP patterns (Table 1). The reproducibility of AFLP fingerprinting was evaluated by generating at least two separate AFLP reactions for each isolate, and the results showed that the same AFLP fingerprint patterns were generated for each isolate at a cutoff value of 90% similarity. The index of discrimination of AFLP was 0.93, which was slightly higher than that obtained by rep-PCR (0.89).
FIG. 2.
UPGMA dendrogram of the genetic relatedness of 20 AFLP types from 47 P. multocida isolates from avian sources. The scale represents percent similarity generated by computer-assisted comparison of AFLP fingerprint profiles. The arrow indicates the cutoff value (90%) for cluster analysis. Species: D, duck; C, chicken; T, turkey; E, bald eagle; Q, quail. NA, not available; CU, Clemson University.
(ii) Concordance analysis of somatic types, rep-PCR types, and AFLP types.
The concordance between somatic types, rep-PCR types, and AFLP types is shown in Table 2. The overall percent agreement between somatic types and rep-PCR and AFLP types was 79.60% (kappa value [K] = 0.32) and 76.53% (K = 0.17), respectively, and these values were statistically significant (G test of independence, G = 104.8, df = 1, P < 0.001, and G = 37.9, df = 1, P < 0.001, respectively).
TABLE 2.
Concordance analysis of somatic type, rep-PCR, and AFLP fingerprintsa
| Somatic type | No. (%)
|
|||
|---|---|---|---|---|
| rep-PCR type
|
AFLP type
|
|||
| Match | Mismatch | Match | Mismatch | |
| Same | 70 (7.40) | 163 (17.23) | 40 (4.23) | 193 (20.40) |
| Different | 30 (3.17) | 683 (72.20) | 29 (3.07) | 684 (72.30) |
| Concordance | 753 (79.60) | 724 (76.53) | ||
The sum of the same somatic types matching rep-PCR types or AFLP types and different somatic types mismatching rep-PCR types or AFLP types represents the percent agreement for the total of 946 pairwise comparisons of isolates.
Forty-seven isolates were included for concordance analysis between rep-PCR types and AFLP types (Table 3). Isolates of the same AFLP patterns typically had identical rep-PCR patterns. From the total of 1,081 possible pairwise comparisons, 5.82% of the isolates of the same rep-PCR matched in AFLP patterns and 87.79% of the isolates with different rep-PCR had different AFLP patterns. On the other hand, only 1.39% of isolates with different rep-PCR patterns matched in AFLP patterns, and 5% of isolates with the same rep-PCR patterns differed in AFLP patterns. The overall concordance was 93.61% (K = 0.61) and was highly significant (G = 244.18, df = 1, P < 0.001), indicating a strong correlation between rep-PCR and AFLP fingerprints (Table 3).
TABLE 3.
Concordance analysis of rep-PCR and AFLP fingerprintsa
| Rep-PCR type | No. (%) of AFLP type
|
|
|---|---|---|
| Match | Mismatch | |
| Same | 63 (5.82) | 54 (5.00) |
| Different | 15 (1.39) | 949 (87.79) |
| Concordance | 1,012 (93.61) | |
The sum of the same rep-PCR types matching AFLP types and different rep-PCR types mismatching AFLP types represents the percent agreement for the total of 1,081 pairwise comparisons of isolates.
Investigation of fowl cholera outbreaks with rep-PCR and AFLP fingerprinting.
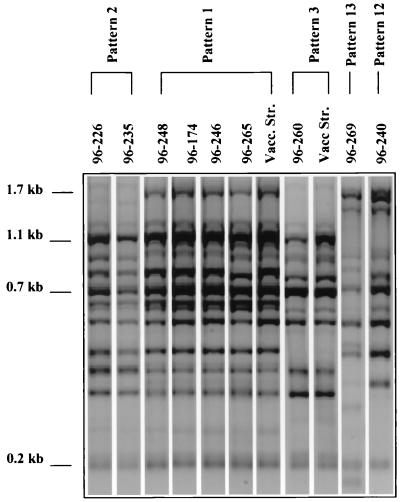
Nine field isolates of P. multocida recovered from recent fowl cholera outbreaks on turkey farms in Minnesota (Table 1) were characterized by rep-PCR and AFLP, and a total of five distinct fingerprint patterns were identified (Fig. 3 and 4). At a 90% cutoff value, rep-PCR results showed that four isolates from farm A (96-265, 96-248, and 96-246) and B (96-174) were classified into the common rep-PCR pattern 1 that included two vaccine strains. Two isolates from farm C (96-226 and 96-235) were classified into pattern 2, which differs from pattern 1 by two bands. One isolate (farm D), 96-260, was assigned to pattern 3, which differs from pattern 1 by three bands. However, two isolates, 96-240 and 96-269 (from farms E and F, respectively), displayed unique patterns, 12 and 13, respectively. These results show the ability of rep-PCR to identify epidemiologically related strains as the same pattern and suggest that the recent fowl cholera outbreaks in Minnesota resulted from infection with closely related (SD > 0.75) strains of patterns 1, 2, and 3 (cluster B).
FIG. 3.
Identification of clinical P. multocida isolates recovered from fowl cholera outbreaks in Minnesota by comparing their rep-PCR patterns with fingerprint patterns obtained from reference vaccine strains.
FIG. 4.
(A) AFLP gel image of P. multocida isolates generated with primers FAM-EcoRI-A and MseI-O. Blue bands represent primer-generated DNA fragments. Red bands represent the internal size standard (GS-500 ROX) included in each lane to help correct interlane variation and provide accurate band sizing. (B) Electrophoretogram of clinical isolates and vaccine strains of P. multocida.
It is interesting that three vaccine strains that have been described previously also belong to these common clones. AFLP analyses of the outbreak strains showed similar results, indicating that both rep-PCR and AFLP fingerprinting techniques will be useful for epidemiological investigations of fowl cholera outbreaks.
rep-PCR and AFLP fingerprinting of P. multocida isolates.
Traditional epidemiologic investigations of fowl cholera out-breaks have been based primarily on a thorough examination of case histories and detailed characterization of bacterial isolates by tests such as biochemical profiling, antimicrobial sensitivity profiling, and serotyping. However, not all isolates are typeable by these methods. For example, although serotyping has proved to be a useful tool for classifying isolates of P. multocida, it has been difficult to conduct epidemiologic investigations and assess the relationships of P. multocida isolates involved in fowl cholera outbreaks by serotyping because of the existence of considerable genetic variation within serotypes (3, 31, 32).
Therefore, the application of direct genetic methods of testing, especially PCR-based techniques, has gained widespread acceptance, and they are used in the investigation of putative disease outbreaks. An important advantage of PCR-based methods is that, in many instances, they enable the sensitive detection and typing of pathogens, as the tests require only a small amount of target DNA. Moreover, since these methods do not depend on the expression of bacterial gene products, all isolates are typeable.
DNA fingerprinting of bacterial isolates involves the generation of distinct “signature” profiles that enable bacterial strain identification. These fingerprints are usually represented by band patterns that result from the migration of different sizes of bands in a gel matrix. PCR-based typing methods include randomly amplified polymorphic DNA (RAPD) (30) and rep-PCR (28). rep-PCR has an advantage over RAPD analysis in that it involves specific rather than arbitrary amplification of target DNA, since oligonucleotide primers used in the amplification reactions are based on conserved repetitive elements that are dispersed throughout the bacterial genome. Thus, in our hands, and as described previously (1), this technique yields reliable and easily reproducible fingerprint patterns.
For instance, our analyses have shown that both AFLP and rep-PCR fingerprints were stable in testing an isolate of Mycobacterium avium over time (10 passages) and in testing 10 different colonies from the same isolate, indicating high reproducibility of the two techniques (A. Amonsin and V. Kapur, unpublished data). DNA fingerprinting of P. multocida recovered from poultry, pigs, and cattle with rep-PCR has also been reported previously (7, 9, 23, 24, 25, 26).
AFLP is a relatively recently described PCR-based DNA fingerprinting method that can be used for the characterization and comparison of DNA samples regardless of their origin or complexity (29). This method of genotyping has been used for a number of different bacterial species (5, 6, 11, 12, 13, 15, 27). For instance, the use of AFLP to trace the source of infection for a nosocomial outbreak of gentamicin-resistant Klebsiella pneumoniae has recently been documented and resulted in controlling of the disease outbreak (27). AFLP has been shown to have several advantages compared to various other typing methods, including increased discriminatory power, reproducibility, and easy compatibility with computerized analysis of the banding patterns.
Hence, the present investigation was conducted to assess the utility of two rapid PCR-based approaches, rep-PCR and AFLP, for the differentiation of P. multocida isolates recovered from multiple avian species and fowl cholera outbreak investigations in turkey farms. The results of our investigations clearly show that rep-PCR has a high discriminatory ability (D = 0.89) when used for the differentiation of P. multocida isolates recovered from avian sources. In addition, the method is rapid, reproducible, and easy to perform. Together, the data suggest that rep-PCR has great utility for investigating the epidemiology of fowl cholera and conducting outbreak investigations.
We have standardized AFLP to characterize P. multocida isolates from multiple avian sources, and the results show that the FAM-EcoRI-A and MseI-O primers yield highly polymorphic fingerprints for this bacterium. Overall, AFLP was found to be a powerful tool for epidemiological investigations of fowl cholera outbreaks in our study, and the index of discrimination among the P. multocida isolates obtained by AFLP was 0.93, slightly higher than that obtained from rep-PCR fingerprints (0.89), and some P. multocida isolates that were indistinguishable by rep-PCR were identified as distinct clones by AFLP. More importantly, the results of the AFLP analysis correlated well with those of rep-PCR.
In contrast, the concordances between somatic types and rep-PCR types or somatic types and AFLP types were relatively low, indicating that somatic serotypes, in general, do not correlate well with molecular subtyping systems. These results indicate that although there was fair agreement between the somatic serotype and rep-PCR or AFLP type of the isolates examined in this investigation, somatic serotypes are, in general, poor indicators of overall genetic relatedness among isolates of P. multocida recovered from avian sources. These findings reflect the fact that the somatic antigens are likely to be adaptive traits and hence subject to strong evolutionary pressure and therefore poor indicators of genetic (rather than phenotypic) similarity. As has been shown for other species of gram-negative and gram-positive bacteria (14, 19), P. multocida somatic antigens may be horizontally transferred among genetically divergent clones of the bacterium.
The results of these studies show that rep-PCR and AFLP are useful techniques for indexing genetic variation in this bacterium. There are two major advantages of AFLP. First, the internal size standard included in each lane obviates ambiguities due to interlane variation in electrophoretic mobility, allowing facile intergel comparisons. Second, a large number of isolates can be examined in a short period of time. However, this method has the limitation of being more expensive than rep-PCR in terms of reagent and equipment costs. Overall, our studies show that depending on the scope of the study, investigators have two powerful molecular genetic methods, rep-PCR and AFLP, at their disposal for investigating fowl cholera outbreaks.
The occurrence of host and disease specificity among bacterial clones has been well described for a variety of pathogenic bacteria (19). For instance, host specificity is found among isolates of Bordetella bronchiseptica, for which clones or clone families are strongly associated with either pigs or dogs (19). A similar host specificity is seen among clones of Staphylococcus aureus, in which certain clones are preferentially associated with either humans or cows (10).
The results of the present investigation show that some P. multocida clones may colonize several avian species. For instance, clones belonging to rep-PCR type 7 and AFLP type 10 were recovered from chickens, ducks, and turkeys (Table 1). These data suggest that certain clones of P. multocida are able to colonize a wide variety of avian species. However, a striking example of host specificity of P. multocida isolates is seen among in cluster B (Fig. 1), in which all four clones (representing 20 isolates) were recovered only from turkeys. Hence, the results of this investigation are fully consistent with the general concept of host specificity among clones, and we hypothesize that the distinctive host range of a bacterial clone is due to innate differences in the ability to successfully colonize a specific host. This hypothesis needs to be tested by constructing a population genetic framework for a larger sample of P. multocida isolates recovered from several avian species and from different geographic areas.
In conclusion, the results of our investigation show that rep-PCR and AFLP fingerprinting techniques are useful for the rapid DNA fingerprinting of P. multocida isolates. These techniques enable straightforward genetic typing of P. multocida isolates and provide a facile means of conducting molecular epidemiologic analyses and outbreak investigations. In addition, the data also provide evidence for host specificity of certain P. multocida clones.
Acknowledgments
Research in the laboratory of V. Kapur is funded by grants from the Minnesota Agricultural Experiment Station, the Minnesota Turkey Growers Association, the U.S. Department of Agriculture's National Research Initiative, and the National Institutes of Health. A. Amonsin gratefully acknowledges the award of a doctoral fellowship from Chulalongkorn University, Bangkok, Thailand.
We thank Caroline Hoellrich, Gireesh Rajashekara, and Kathy Tune for comments and suggestions for improving the manuscript.
REFERENCES
- 1.Amonsin, A., J. F. Wellehan, L.-L. Li, P. Vandamme, C. Lindeman, M. Edman, R. A. Robinson, and V. Kapur. 1997. Molecular epidemiology of Ornithobacterium rhinotracheale. J. Clin. Microbiol. 35:2894-2898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Blackall, P. J., J. L. Pahoff, D. Marks, N. Fegan, and C. J. Morrow. 1995. Characterisation of Pasteurella multocida isolated from fowl cholera outbreaks on turkey farms. Aust. Vet. J. 72:135-138. [DOI] [PubMed] [Google Scholar]
- 3.Brogden, K. A., and R. A. Packer. 1979. Comparison of Pasteurella multocida serotyping systems. Am. J. Vet. Res. 40:1332-1335. [PubMed] [Google Scholar]
- 4.Diallo, I. S., J. C. Bensink, A. J. Frost, and P. B. Spradbrow. 1995. Molecular studies on avian strains of Pasteurella multocida in Australia. Vet. Microbiol. 46:335-342. [DOI] [PubMed] [Google Scholar]
- 5.Duim, B., C. W. Ang, A. van Belkum, A. Rigter, N. W. van Leeuwen, H. P. Endtz, and J. A. Wagenaar. 2000. Amplified fragment length polymorphism analysis of Campylobacter jejuni strains isolated from chickens and from patients with gastroenteritis or Guillain-Barre or Miller Fisher syndrome. Appl. Environ. Microbiol. 66:3917-3923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gibson, J. R., E. Slater, J. Xerry, D. S. Tompkins, and R. J. Owen. 1998. Use of an amplified-fragment length polymorphism technique to fingerprint and differentiate isolates of Helicobacter pylori. J. Clin. Microbiol. 36:2580-2585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gunawardana, G. A., K. M. Townsend, and A. J. Frost. 2000. Molecular characterisation of avian Pasteurella multocida isolates from Australia and Vietnam by REP-PCR and PFGE. Vet. Microbiol. 72:97-109. [DOI] [PubMed] [Google Scholar]
- 8.Hunter, P. R., and M. A. Gaston. 1988. Numerical index of the discriminatory ability of typing systems: an application of Simpson's index of diversity. J. Clin. Microbiol. 26:2465-2466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kainz, A., W. Lubitz, and H. J. Busse. 2000. Genomic fingerprints, ARDRA profiles and quinone systems for classification of Pasteurella sensu stricto. Syst. Appl. Microbiol. 23:494-503. [DOI] [PubMed] [Google Scholar]
- 10.Kapur, V., W. M. Sischo, R. S. Greer, T. S. Whittam, and J. M. Musser. 1995. Molecular population genetic analysis of Staphylococcus aureus recovered from cows. J. Clin. Microbiol. 33:376-380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Keim, P., A. Kalif, J. Schupp, K. Hill, S. E. Travis, K. Richmond, D. M. Adair, M. Hugh-Jones, C. R. Kuske, and P. Jackson. 1997. Molecular evolution and diversity in Bacillus anthracis as detected by amplified fragment length polymorphism markers. J. Bacteriol. 179:818-824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Koeleman, J. G., M. W. van der Bijl, J. Stoof, C. M. Vandenbroucke-Grauls, and P. H. Savelkoul. 2001. Antibiotic resistance is a major risk factor for epidemic behavior of Acinetobacter baumannii. Infect. Control Hosp. Epidemiol. 22:284-288. [DOI] [PubMed] [Google Scholar]
- 13.Lindstedt, B. A., E. Heir, T. Vardund, K. K. Melby, and G. Kapperud. 2000. Comparative fingerprinting analysis of Campylobacter jejuni subsp. jejuni strains by amplified-fragment length polymorphism genotyping. J. Clin. Microbiol. 38:3379-3387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Musser, J. M. 1996. Molecular population genetic analysis of emerged bacterial pathogens: selected insights. Emerg. Infect. Dis. 2:1-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nair, S., E. Schreiber, K. L. Thong, T. Pang, and M. Altwegg. 2000. Genotypic characterization of Salmonella typhi by amplified fragment length polymorphism fingerprinting provides increased discrimination compared to pulsed-field gel electrophoresis and ribotyping. J. Microbiol. Methods 41:35-43. [DOI] [PubMed] [Google Scholar]
- 16.Price, S. B., M. D. Freeman, and M. W. MacEwen. 1993. Molecular analysis of a cryptic plasmid isolated from avian strains of Pasteurella multocida. Vet. Microbiol. 37:31-43. [DOI] [PubMed] [Google Scholar]
- 17.Rhoades, K. R., and R. B. Rimler. 1991. Pasteurellosis, p. 145-171. In B. W. Calnek, H. J. Barnes, C. W. Beard, W. R. Reid, and H. W. Yoder (ed.), Diseases of poultry, 9th ed. Iowa State University Press, Ames, Iowa.
- 18.Rimler, R. B., and K. R. Rhoades. 1987. Serogroup F, a new capsule serogroup of Pasteurella multocida. J. Clin. Microbiol. 25:615-618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Selander, R. K., and J. M. Musser. 1990. Population genetics of bacterial pathogenesis, p. 11-36. In B. H. Iglewski and V. L. Clark (ed.), Molecular basis of bacterial pathogenesis, Academic Press Inc., San Diego, Calif.
- 20.Sneath, P. H. A., and R. R. Sokal. 1973. Numerical taxonomy, p. 230-234. W. H. Freeman and Co., San Francisco, Calif.
- 21.Snipes, K. P., D. C. Hirsh, R. W. Kasten, T. E. Carpenter, D. W. Hird, and R. H. McCapes. 1990. Differentiation of field isolates of Pasteurella multocida serotype 3,4 from live vaccine strain by genotypic characterization. Avian Dis. 34:419-424. [PubMed] [Google Scholar]
- 22.Sokahl, R. R., and F. J. Rohlf. 1995. Analysis of frequencies, p. 685-793. In R. R. Sokal and F. J. Rohlf (ed.), Biometry. Freeman, New York, N.Y.
- 23.Swaminathan, B., and G. M. Matar. 1994. Molecular typing methods, p. 26-50. In D. H. Persing, T. F. Smith, F. C. Tenover, and T. J. White (ed.), Diagnostic molecular microbiology: principles and applications. American Society for Microbiology, Washington, D.C.
- 24.Townsend, K. M., H. J. Dawkins, and J. M. Papadimitriou. 1997. REP-PCR analysis of Pasteurella multocida isolates that cause haemorrhagic septicaemia. Res. Vet. Sci. 63:151-155. [DOI] [PubMed] [Google Scholar]
- 25.Townsend, K. M., D. O'Boyle, T. T. Phan, T. X. Hanh, T. G. Wijewardana, I. Wilkie, N. T. Trung, and A. J. Frost. 1998. Acute septicaemic pasteurellosis in Vietnamese pigs. Vet. Microbiol. 63:205-215. [DOI] [PubMed] [Google Scholar]
- 26.Townsend, K. M., T. X. Hanh, D. O'Boyle, I. Wilkie, T. T. Phan, T. G. Wijewardana, N. T. Trung, and A. J. Frost. 2000. PCR detection and analysis of Pasteurella multocida from the tonsils of slaughtered pigs in Vietnam. Vet. Microbiol. 72:69-78. [DOI] [PubMed] [Google Scholar]
- 27.van der Zwet, W. C., G. A. Parlevliet, P. H. Savelkoul, J. Stoof, A. M. Kaiser, J. G. Koeleman, and C. M. Vandenbroucke-Grauls. 1999. Nosocomial outbreak of gentamicin-resistant Klebsiella pneumoniae in a neonatal intensive care unit controlled by a change in antibiotic policy. J. Hosp. Infect. 42:295-302. [DOI] [PubMed] [Google Scholar]
- 28.Versalovic, J., V. Kapur, T. Koeuth, G. H. Mazurek, T. S. Whittam, J. M. Musser, and J. R. Lupski. 1995. DNA fingerprinting of pathogenic bacteria by fluorophore-enhanced repetitive sequence-based polymerase chain reaction. Arch. Pathol. Lab. Med. 119:23-29. [PubMed] [Google Scholar]
- 29.Vos, P., R. Hogers, M. Bleeker, M. Reijans, T. van de Lee, M. Hornes, A. Frijters, J. Pot, J. Peleman, M. Kuiper, and M. Zabeau. 1995. AFLP: a new technique for DNA fingerprinting. Nucleic Acids Res. 23:4407-4414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Welsh, J., and M. McClelland. 1990. Fingerprinting genomes with PCR with arbitrary primers. Nucleic Acids Res. 18:7213-7218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wilson, M. A., M. J. Morgan, and G. E. Barger. 1993. Comparison of DNA fingerprinting and serotyping for identification of avian Pasteurella multocida isolates. J. Clin. Microbiol. 31:255-259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wilson, M. A., R. M. Duncan, G. E. Nordholm, and B. M. Berlowski. 1995. Serotypes and DNA fingerprint profiles of Pasteurella multocida isolated from raptors. Avian Dis. 39:94-99. [PubMed] [Google Scholar]
- 33.Wilson, M. A., R. M. Duncan, G. E. Nordholm, and B. M. Berlowski. 1995. Pasteurella multocida isolated from wild birds of North America: a serotype and DNA fingerprint study of isolates from 1978 to 1993. Avian Dis. 39:587-593. [PubMed] [Google Scholar]
- 34.Woods, C. R., J. Versalovic, T. Koeuth, and J. R. Lupski. 1993. Whole-cell repetitive element sequence-based polymerase chain reaction allows rapid assessment of clonal relationships of bacterial isolates. J. Clin. Microbiol. 31:1927-1931. [DOI] [PMC free article] [PubMed] [Google Scholar]