Abstract
Two genetic fingerprinting techniques, pulsed-field gel electrophoresis (PFGE) and ribotyping, were used to characterize 207 Escherichia coli O157 isolates from food animals, foods of animal origin, and cases of human disease (206 of the isolates were from the United Kingdom). In addition, 164 of these isolates were also phage typed. The isolates were divided into two general groups: (i) unrelated isolates not known to be epidemiologically linked (n = 154) and originating from food animals, foods and the environment, or humans and (ii) epidemiologically related isolates (n = 53) comprised of four related groups (RGs) originating either from one farm plus the abattoir where cattle from that farm were slaughtered or from one of three different English abattoirs. PFGE was conducted with the restriction endonuclease XbaI, while for ribotyping, two restriction endonucleases (PstI and SphI) were combined to digest genomic DNAs simultaneously. The 207 E. coli O157 isolates produced 97 PFGE profiles and 51 ribotypes. The two genetic fingerprinting methods had similar powers to discriminate the 154 epidemiologically unrelated E. coli O157 isolates in the study (Simpson's index of diversity [D] = 0.98 and 0.94 for PFGE typing and ribotyping, respectively). There was no correlation between the source of an isolate (healthy meat or milk animals, retail meats, or cases of human infection) and either particular PFGE or ribotype profiles or clusters. Combination of the results of both genetic fingerprinting methods produced 146 types, significantly more than when either of the two methods was used individually. Consequently, the superior discriminatory performance of the PFGE-ribotyping combination was proven in two ways: (i) by demonstrating that the majority of the E. coli O157 isolates with unrelated histories were indeed distinguishable types and (ii) by identifying some clonal groups among two of the four RGs of E. coli O157 isolates (comprising PFGE types different by just one or two bands), the relatedness of which would have remained unconfirmed otherwise.
Suspect vehicles of infection in Escherichia coli O157 outbreaks have included foods of animal origin, e.g., milk and dairy products (15, 31, 33) and meat and meat products (6, 7, 8, 9, 24, 34). Meat and milk animals themselves may be a source of E. coli O157, as they can carry the organism in their gastrointestinal tracts (11, 22), resulting in fecal shedding of the pathogen and direct or indirect contamination of meat. When considering meat safety, it is clear that the E. coli O157 isolates that are present on retail meat and that cause meat-borne human disease may also originate from numerous other sources along the meat production chain. E. coli O157 isolates existing along the meat chain should be both identified and targeted by adequate control measures. Good traceability (longitudinal and horizontal) not only of animals and meats but also of E. coli O157 is an essential tool for such identification and control. Subserovar characterization is essential to trace E. coli O157 isolates along the farm-abattoir-meat processing-distribution-retail-consumer chain.
Initial typing methods routinely used during outbreak investigations include phage typing (3, 23, 35) and Shiga toxin (Stx) typing. However, the usefulness of these data can be limited, as the numbers of phage or Stx types identified are often limited (1, 29). In England and Wales, the predominant phage type (PT) isolated from cases of human infection has changed with time, from PT2 to PT21/28 (13). The predominance of particular PTs and the resulting lack of discrimination means that phage typing should not be used as the sole typing technique for investigation of E. coli O157 outbreaks (26).
In addition to these two methods, genetic fingerprinting is used to establish relatedness of E. coli O157 isolates. Pulsed-field gel electrophoresis (PFGE) is the “gold standard” of genetic fingerprinting methods for E. coli O157 and has frequently been used in epidemiological investigations of outbreaks in order to establish relatedness between human clinical isolates and isolates from suspect foods or animals (4, 9, 33). Ribotyping has been used to genetically fingerprint some major human pathogens, including Salmonella enterica serovar Enteritidis (27), Campylobacter jejuni (21), and Listeria monocytogenes (32), among species of other genera. However, there are few reports on ribotyping of E. coli O157, and results have indicated that ribotyping is not a very discriminatory technique (16, 20, 30). Wachsmuth et al. (41) found that ribotyping with NcoI produced maximum discrimination of E. coli O157:H7 and that it may have been more stable than plasmid profiling.
Plasmid profiling has been used to examine E. coli O157 isolates from different sources in the United Kingdom, including meat and meat products, animals, and humans (10, 12). However, the main scope of published genetic fingerprinting studies of E. coli O157 isolates from food animals and foods in the United Kingdom was relatively narrow; i.e., they primarily involved isolates epidemiologically linked to human disease outbreaks. The majority of those isolates were from humans, while the remainder were from suspect food or animal sources (1, 34). Knowledge about the genetic diversity of a wider range of United Kingdom E. coli O157 isolates, as measured by genetic fingerprinting of whole bacterial genomes, is lacking. Also, there are no data on the performance of ribotyping for the discrimination of United Kingdom E. coli O157 isolates. Therefore, the aims of the present study were (i) to examine the diversity of the genetic fingerprints of E. coli O157 isolates from a wider range of food chain-related sources or cases of human disease within the United Kingdom and (ii) to examine the performance of ribotyping (with two restriction enzymes), alone or in combination with PFGE, in differentiating the E. coli O157 isolates.
MATERIALS AND METHODS
E. coli O157 isolates.
The E. coli O157 isolates examined included 206 isolates from the United Kingdom and 1 isolate from the Republic of Ireland. Among them were 154 isolates not known to be related epidemiologically or geographically (called “unrelated isolates”). They originated from the gastrointestinal tracts of healthy meat or milk animals (n = 56, consisting of 36 isolates from cattle, 13 isolates from sheep, 6 isolates from pigs, and 1 isolate from a goat), meats (n = 34, 27 of which were from Chapman et al. [10, 12]), or from cases of human infection (n = 56). Eight isolates originated from other environments: two from retail milk, two from river waters, three from bovine slurries, and one from a bird.
Other E. coli O157 isolates (n = 53) consisted of related groups (RGs) from four sources. RG1 isolates (n = 33) originated from the feces, hides, or carcasses of cattle from one farm or from an abattoir race while those cattle were being slaughtered. RG2, RG3, and RG4 contained 5, 10, and 5 isolates, respectively, from cattle hides (36) and abattoir lairage surfaces (39) collected during 1-day visits to three different abattoirs. Four of the 5 isolates from RG2 and 4 of the 10 isolates from RG3 did not produce Stx, but all other isolates in the study produced Stx during the Vero cell assay (25).
Epidemiological, O-antigen, Stx-type, and PT data were supplied along with the E. coli O157 isolates, where they were known (see the Acknowledgments for the suppliers). Additional phage typing for a selection of the isolates was conducted at the Public Health Laboratory Service, Colindale, United Kingdom, by the method of Frost et al. (18). Isolates were of the following 14 different PTs: 1, 2, 4, 8, 14, 21, 21/28, 32, 34, 43, 49, 54, reacted but did not conform, and untypeable. All isolates were kept on Dorset egg slopes before use.
PFGE.
PFGE was conducted as described previously (2). Briefly, plugs were prepared from heat-inactivated bacterial suspensions mixed with equal volumes of 2% agarose (Clean Cut; Bio-Rad, Hemel Hempstead, United Kingdom), and the cells were lysed in ESP buffer (19) (0.5 M EDTA [pH 9.0], 1% sodium lauryl sarcosine, 1 mg of proteinase K per ml). Washed lysed plugs were digested with XbaI (Promega, Southampton, United Kingdom), and electrophoresis was conducted in 1% Pulsed Field Gel certified agarose (Bio-Rad) with 0.5× TBE (Tris-borate-EDTA) extended-range buffer (Bio-Rad) in a CHEF DRIII apparatus (Bio-Rad), with pulse times ramped from 2.2 to 54.2 s for 22 h at 6 V/cm, a 120o angle, and cooling to 14°C (19). The gels were stained in ethidium bromide, and images were captured under UV light transillumination with Gel Doc (Bio-Rad).
Genomic DNA extraction.
Genomic DNA was extracted from cells after overnight growth at 37°C in 3-ml volumes of Luria-Bertani broth containing 1 g of glucose per liter by the method of Cousins et al. (14). Briefly, cell pellets were resuspended in 350 μl of sterile high-pressure liquid chromatography-grade water, and the suspension was incubated at 37°C for 3 h in 2.5 μg of lysozyme per μl. Sodium dodecyl sulfate (1%) and 0.1 μg of proteinase K per μl were added, and the suspensions were heated at 50°C until they were clear. NaCl (0.735 M) and cetyltrimethylammonium bromide (0.012 μg/μl) were added to remove polysaccharides, and the suspensions were heated at 65°C for 10 min. DNA was extracted with phenol-chloroform, resuspended in 100 μl of TE (Tris-EDTA) buffer, and stored at −70°C.
Ribotyping.
Ribotyping was performed as described previously (27). Briefly, labeled probe was prepared from plasmid pKK3535 containing the rrnB operon purified from E. coli with the QIAfilter plasmid Midi purification kit (Qiagen, Crawley, United Kingdom) and labeled by using the DIG-High prime kit (Roche Molecular Biochemicals, Lewes, United Kingdom) (5).
E. coli O157 DNA (4 μg from each strain) was digested with 20 U of PstI and 10 U of SphI in combination (Promega). Digested DNAs (2 μg) were electrophoresed in 0.8% agarose (Sigma) gels in Tris-acetate-EDTA buffer (Sigma) at 14°C and 45 V for 20 h. DNA fragments were transferred to nylon membranes (Roche) with 0.4 M NaOH under vacuum. The membranes were washed, cross-linked, and prehybridized at 42°C for about 4 h and then hybridized overnight at 42°C in 20 ml of DIG Easy Hyb buffer (Roche) containing 20 ng of prepared probe per ml. The presence of labeled probe was detected with an alkaline phosphatase-conjugated antibody detection kit (Roche) and the chemiluminescent substrate disodium 3-(4-methoxyspiro[1,2-dioxetane-3,2-(5′-chloro)tricyclo{3.3.1.13,7}decan]-4-yl)phenyl phosphate (CSPD; Roche) according to the instructions of the manufacturer. Chemiluminescence, developed at 37°C for about 30 min, was detected on film (Lumi-film; Roche).
Data analysis.
Images from PFGE and ribotyping were saved and scanned, respectively, into software (GelCompar II, version 2.0; Applied Maths, Kortrijk, Belgium), enabling estimation of fragment molecular weights and normalization before analysis. Bands were assigned initially by using the autosearch facility of the software and were later assigned by eye. Isolates were allocated a different fingerprint type (an X type for the PFGE profile and a PS type for the ribotype profile) when a genetic difference was detected. Cluster analysis was performed by using the Jaccard coefficient and the unweighted pair group method with arithmetic averages. The discriminatory powers of both genetic fingerprinting methods were calculated only for the 154 unrelated E. coli O157 isolates by using Simpson's index of diversity (D), as it is inappropriate to measure diversity with epidemiologically related isolates (their diversity may be significantly limited by their epidemiological relatedness):
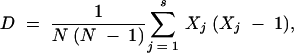 |
where s is the number of types recognized by a particular technique, Xj is the number of isolates identical to the jth isolate, and N is the total number of unrelated isolates examined. PFGE and ribotype profiles were analyzed together to produce final combined profiles.
RESULTS AND DISCUSSION
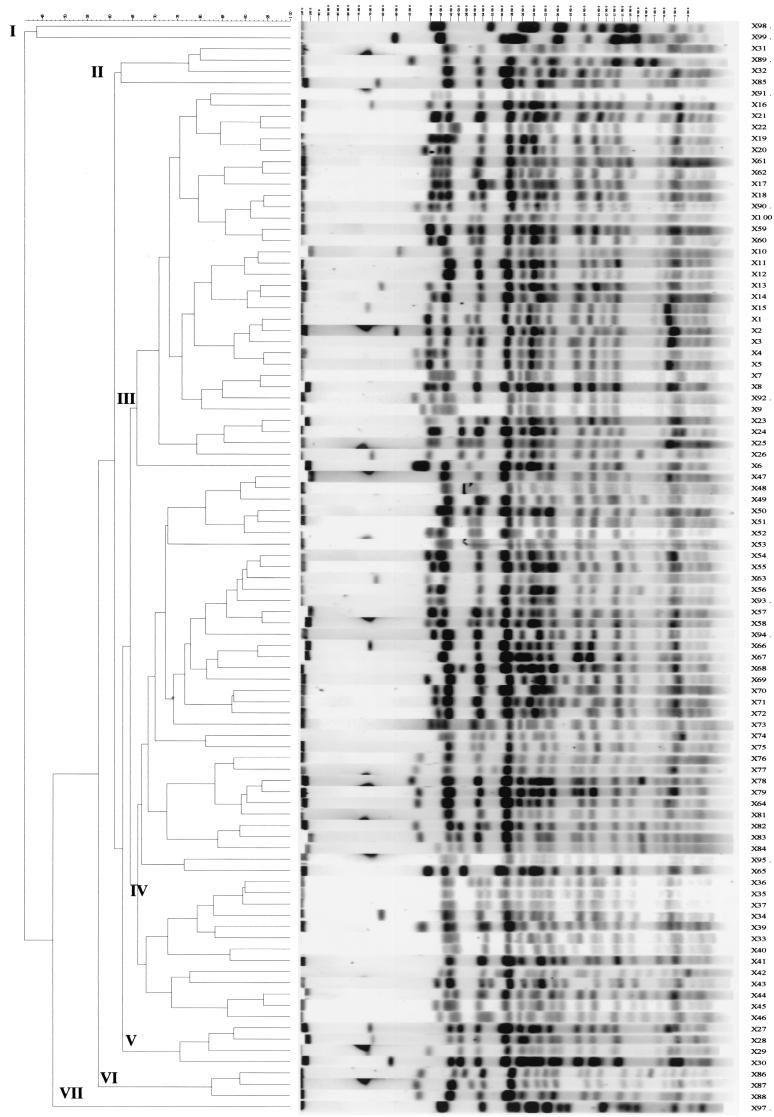
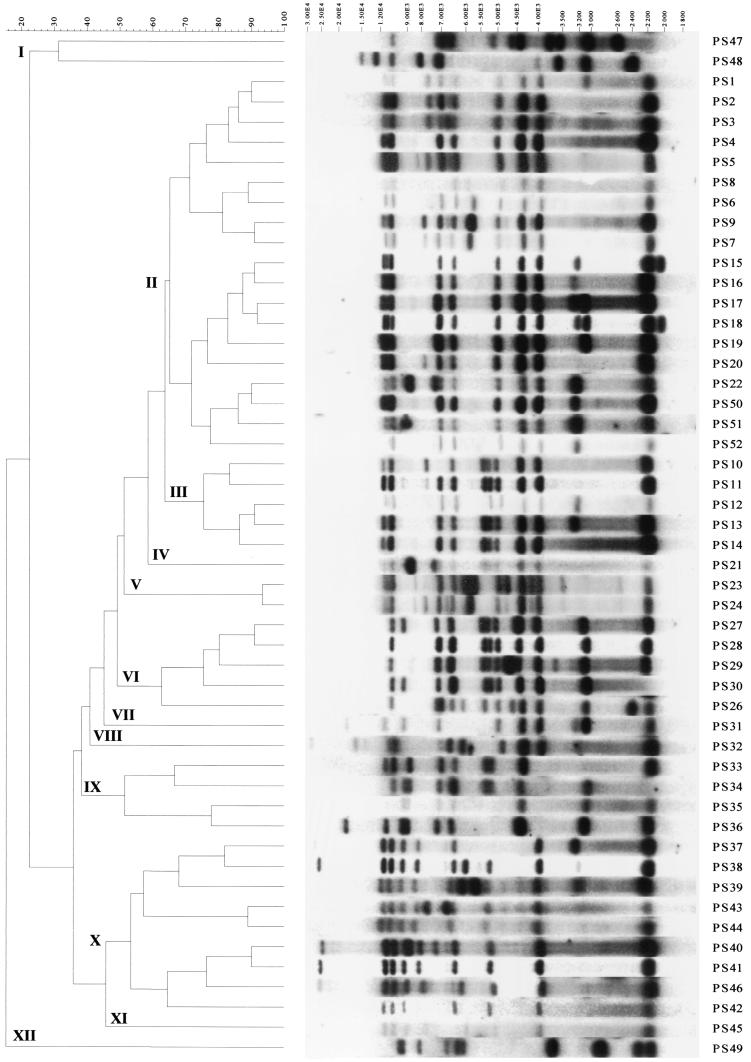
When all 207 E. coli O157 isolates in the study were examined, 97 PFGE profiles (Fig. 1) and 51 ribotypes (Fig. 2) grouped into 7 and 12 main clusters, respectively, which were separated at 67% similarity or less, were produced. The PTs were known for 164 of the 207 isolates. There was no correlation of the PT (where it was known) with either the PFGE cluster or the ribotype cluster. For example, PFGE type X55 contained isolates of PTs 1, 2, 4, 8, 21, 21/28, 32, and 34, while three PT54 isolates had distinct PFGE profiles, X1, X32, and X49 (Fig. 1). Also, two isolates indistinguishable by both genetic fingerprinting methods (both were X32 and PS30) were different PTs (PT43 and PT54). One of these isolates (isolate PT43) originated from a healthy pig during 1999, while the other (isolate PT54) originated from retail beef mince during 1997. There was no correlation of isolate source (healthy meat or milk animals, retail meats, or cases of human infection) with genetic fingerprint profiles or clusters.
FIG. 1.
Dendrogram generated with GelCompar II software showing the relationship of 97 representative fingerprints (XbaI-PFGE or X types) for 207 E. coli O157 isolates. The analysis of the bands generated was performed by using the Jaccard coefficient and the unweighted pair group method with arithmetic averages.
FIG. 2.
Dendrogram generated with GelCompar II software showing the relationship of 51 representative fingerprints (PstI and SphI ribotypes or PS types) for 207 E. coli O157 isolates. The analysis of the bands generated was performed by using the Jaccard coefficient and the unweighted pair group method with arithmetic averages.
The results of the two methods of genetic typing were frequently not in agreement. In several instances, multiple ribotypes were found among particular PFGE types, and similarly, multiple PFGE profiles were found among some ribotypes.
Combination of both genetic (PFGE and ribotyping) typing methods produced 146 final types, significantly more than when either PFGE or ribotyping alone was used. Therefore, many of the epidemiologically unrelated isolates which were not distinguishable when only one of the genetic fingerprinting methods was used could be distinguished when both methods were used. This may be significant, as frequently, only one fingerprinting method (typically PFGE) is used to differentiate E. coli O157 isolates. In that case, isolates may be classified as indistinguishable, even though use of another typing method, as we have demonstrated using ribotyping, could show that this is not the case. On the other hand, we observed that it was sometimes difficult to assign bands to PFGE profiles, particularly for the lower fragment sizes (about 50 kbp). This, coupled with the fact that we did not assign band differences on the basis of intensity (the amount of DNA in each plug was not standardized), could result in allocation of one PFGE type to multiple isolates that may not, in fact, be identical.
The 154 unrelated E. coli O157 isolates were very diverse when diversity was measured by either genetic fingerprinting method, producing 84 PFGE profiles and 46 ribotypes (D = 0.98 and 0.94 for PFGE and ribotyping, respectively). This high degree of diversity was observed regardless of the source of the isolates. Among the 154 unrelated E. coli O157 isolates, the most prevalent PFGE profile was X55 (Fig. 1), with 12 isolates originating from meat, cases of human disease, a goat, or the gastrointestinal tracts of cattle having this profile. Six E. coli O157 isolates from cases of human disease, bovine slurry, or the gastrointestinal tracts of cattle on-farm were PFGE type X11; all six isolates had the same ribotype profile, and five of them were PT8 (the PT of the sixth isolate was not known). The six isolates were geographically spread, originating from Scotland and the north, the midlands, and the south of England and were originally cultured over a 16-month period, from March 1999 to June 2000. Among the unrelated isolates, three ribotype profiles, PS14, PS41, and PS50 (Fig. 2), containing 19, 22, and 15 isolates, respectively, were the most prevalent.
Use of the combination of both genetic fingerprinting methods for the 154 unrelated E. coli O157 isolates produced 130 types, confirming the high degree of diversity. However, use of the combination of both genetic fingerprinting methods for the unrelated isolates also produced five groups that each contained three or more isolates, i.e., the isolates appeared to be clonal. For three of these groups, phage typing data (the PT was not known for one isolate) were in agreement, as the isolates were PT 2, 4, or 8; but two of the groups contained isolates of more than one PT. The isolates in one of the clonal groups initially originated from a cow, a case of human disease, and meat but were spread temporally from 1994 to 2000 and geographically from Scotland to the English midlands. Data showing similar temporal and geographic spread were observed for isolates in other clonal groups (data not shown). These data show that some clonal groups (indistinguishable by both PFGE and ribotyping) were found among the E. coli O157 isolates studied, even though the isolates are not known to be linked. The significance of such clones within E. coli O157 populations originating from either animals, foods, or humans is unknown, even though outbreaks of human disease in Scotland not known to be linked geographically or temporally have been caused by clones of E. coli O157 that cannot be distinguished (1). It is vital to establish whether clonal E. coli O157 isolates occur within likely sources, including foods and animals, and at what frequency, before the impacts of these clones on human health can be fully understood, and before their behavior and pathogenicity and their relative importance in the food chain can be compared with those of other, nonclonal E. coli O157 isolates.
The 53 E. coli O157 isolates that comprised the four RGs were of 20 PFGE profiles but 10 ribotypes. For those isolates, ribotyping was useful in establishing their relatedness when small numbers of band differences were observed by PFGE. For example, the 33 isolates in RG1, originating from cattle from one farm, produced 11 PFGE profiles but four ribotypes, which enabled us to identify four clonal groups among these isolates. PFGE profiles X1, X3, X5, X14, and X15 differed by one or two bands (Fig. 1), but all isolates with these PFGE profiles were ribotype PS41. Similarly, PFGE profiles X34, X35, X36, and X37 were different by one or two bands (Fig. 1), but all isolates with these profiles had the same ribotype (PS31). Similarly, the use of PFGE and ribotyping in combination showed that the 10 E. coli O157 isolates from one abattoir (RG3) were from three different clonal groups. The use of PFGE and ribotyping in combination was not necessary to differentiate isolates from either RG2 or RG4, as the results of both genetic fingerprinting methods were in agreement for these two sets of isolates.
PFGE typing is frequently used to determine the sources of outbreaks by comparing human isolates with those from foods and, occasionally, animals. PFGE was more discriminatory than both phage typing (1, 29) and differentiation based on the presence of virulence genes (38). However, it is unclear when differences in PFGE profiles should be regarded as significant genetic differences (20). Tenover et al. (40) proposed that for small (n = <31) collections of microorganisms related to outbreaks of human disease, minor band differences should not be regarded as major genetic shifts and that only isolates differing by seven or more bands should be regarded as unrelated, while those differing by four to six bands should be regarded as possibly related. Louie et al. (29) considered that for their collection of isolates, those differing by three or more bands were different. In the present study, among the 154 unrelated E. coli O157 isolates, we found isolates that could not be distinguished on the basis of PFGE typing alone or on the basis of PFGE and ribotyping used in combination. These may be clonal groups even if they are not epidemiologically related at present (we do not know if they were linked in the past), or they may, in fact, be unrelated genetically. This affirms both the importance of the availability of good-quality epidemiological data to assess the results of any genetic fingerprinting and the importance of using genetic fingerprinting methods with suitable discriminatory powers in epidemiological studies.
The present study shows the power of the ribotyping technique with PstI and SphI in combination, as ribotyping and PFGE had similar abilities to discriminate the 154 unrelated E. coli O157 isolates (D = 0.94 and 0.98, respectively). This contrasts with the results of other studies. Martin et al. (30) found that ribotyping did not distinguish types among their pool of 54 E. coli O157 isolates, as all had identical ribotypes after digestion with NcoI. Dalla-Costa et al. (16), using separate BglI and HindIII digestions of one E. coli O157:H−, nine E. coli O157:H7, and three E. coli O157:H43 isolates, found that the 11 motile isolates had identical ribotypes, while the sole E. coli O157:H− isolate had another, closely related profile. Grif et al. (20) used EcoRI and PvuII in separate digestions to automatically ribotype (riboprint) 47 E. coli O157 isolates originating from human patients, food (unrelated to the human cases), or healthy calves and produced five different ribotypes. In contrast, in the present study, 154 unrelated E. coli O157 isolates produced 46 different ribotypes. We offer two explanations for this. First, the greater degree of diversity observed in the present study may be due to the greater degree of genetic diversity of our E. coli O157 isolate collection compared with the degrees of diversity of the isolates used in other studies. Second, as we used different restriction enzymes and also combined them during genome digestion, this particular ribotyping technique may produce a greater power of differentiation than other methods. Further studies are necessary to establish the reason(s) for the greater discriminatory power of ribotyping in the present study compared with those in with previous studies.
Data from the literature show that E. coli O157 isolates that caused human disease in Chile (38) and Germany (28) or that originated from animals in the United States (17, 37) are genetically very diverse. Previous studies conducted in the United Kingdom (10, 12) indicated that E. coli O157 isolates from meat and meat products, animals, and humans in England were diverse when assessed by a combination of plasmid profiling, Stx typing, and phage typing. Nevertheless, one should keep in mind that plasmid profiling shows differences in the sizes and numbers of plasmids, which may be highly mobile between bacterial isolates. In the present study, the diversity of the E. coli O157 isolates evaluated was measured by the use of different tools, i.e., by analyzing differences in the DNAs of whole bacterial genomes (PFGE and ribotyping), and also included a wider range of food chain-related isolates. The results of both PFGE and ribotyping from the present study individually confirm that a high degree of genetic diversity exists in E. coli O157 populations in the United Kingdom, not only among human isolates but also among isolates from a range of other, previously underexamined sources. No particular isolate source in the present study was linked with particular genetic fingerprint types or clusters. However, the present study clearly shows the superior performance of the PFGE-ribotyping combination in the discrimination of otherwise indistinguishable E. coli O157 clones, which has significant implications for both evaluation of the “farm-to-fork” epidemiology of the pathogen and the development of related integrated control systems.
Acknowledgments
We thank the Food Standards Agency of the United Kingdom, the Department for the Environment, Food and Rural Affairs of the United Kingdom, and the University of Bristol for funding this study.
We also gratefully thank M. Altwegg, who supplied plasmid pKK3535, and the staff in the abattoirs. A number of the E. coli O157 isolates were kindly supplied by Peter Chapman, Mike Hutchison, Iain Ogden, Giles Paiba, and Barti Synge; the Department of Enteric Pathogens, Public Health Laboratory Service; the Food Microbiology Research Unit, Pubic Health Laboratory Service, Exeter, United Kingdom; and Veterinary Laboratories Agency, Weybridge, United Kingdom.
REFERENCES
- 1.Allison, L. J., P. E. Carter, and F. M. Thomson-Carter. 2000. Characterization of a recurrent clonal type of Escherichia coli O157:H7 causing major outbreaks of infection in Scotland. J. Clin. Microbiol. 38:1632-1635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Avery, S. M., A. Small, C.-A. Reid, and S. Buncic. Pulsed-field gel electrophoresis characterization of Shiga toxin-producing Escherichia coli O157 from hides of cattle at slaughter. J. Food Prot., in press. [DOI] [PubMed]
- 3.Barrett, T. J., H. Lior, J. H. Green, R. Khakhria, J. G. Wells, B. P. Bell, K. D. Greene, J. Lewis, and P. M. Griffin. 1994. Laboratory investigation of a multistate food-borne outbreak of Escherichia coli O157:H7 by using pulsed-field gel electrophoresis and phage typing. J. Clin. Microbiol. 32:3013-3017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bell, B. P., M. Goldoft, P. M. Griffin, M. A. Davis, D. C. Gordon, P. I. Tarr, C. A. Bartleson, J. H. Lewis, T. J. Barrett, J. G. Wells, R. Baron, and J. Kobayashi. 1994. A multistate outbreak of Escherichia coli O157:H7-associated bloody diarrhea and hemolytic uremic syndrome from hamburgers. JAMA 272:1349-1353. [PubMed] [Google Scholar]
- 5.Brosious, J., A. Ullrich, M. A. Raker, A. Gray, T. J. Dull, R. R. Gutell, and H. F. Noller. 1981. Construction and fine mapping of recombinant plasmids containing the rrnB operon of Escherichia coli. Plasmid 6:112-118. [DOI] [PubMed] [Google Scholar]
- 6.Centers for Disease Control and Prevention. 1993. Update: multistate outbreak of Escherichia coli O157:H7 infections from hamburgers—Western United States, 1992-1993. Morb. Mortal. Wkly. Rep. 42:258-263. [PubMed] [Google Scholar]
- 7.Centers for Disease Control and Prevention. 1994. Escherichia coli O157:H7 outbreak linked to home-cooked hamburger—California, July 1993. Morb. Mortal. Wkly. Rep. 43:213-216. [PubMed] [Google Scholar]
- 8.Centers for Disease Control and Prevention. 1995. Escherichia coli O157:H7 outbreak linked to commercially distributed dry-cured salami—Washington and California, 1994. Morb. Mortal. Wkly. Rep. 44:157-160. [PubMed] [Google Scholar]
- 9.Centers for Disease Control and Prevention. 1997. Escherichia coli O157:H7 infections associated with eating a nationally distributed commercial brand of frozen ground beef patties and burgers—Colorado, 1997. Morb. Mortal. Wkly. Rep. 46:777-778. [PubMed] [Google Scholar]
- 10.Chapman, P. A., A. T. Cerdan Malo, M. Ellin, R. Ashton, and M. A. Harkin. 2001. Escherichia coli O157 in cattle and sheep at slaughter, on beef and lamb carcasses and in raw beef and lamb products in South Yorkshire, UK. Int. J. Food Microbiol. 64:139-150. [DOI] [PubMed] [Google Scholar]
- 11.Chapman, P. A., C. A. Siddons, A. T. Cerdan Malo, and M. A. Harkin. 1997. A 1-year study of Escherichia coli O157 in cattle, sheep, pigs and poultry. Epidemiol. Infect. 119:245-250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chapman, P. A., C. A. Siddons, A. T. Cerdan Malo, and M. A. Harkin. 2000. A one year study of Escherichia coli O157:H7 in raw beef and lamb products. Epidemiol. Infect. 124:207-213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Communicable Disease Surveillance Report. 1999. Vero cytotoxin producing Escherichia coli O157: phage types reported in 1998. Communic. Dis. Rep. Wkly. 9:291.. [PubMed] [Google Scholar]
- 14.Cousins, D. V., S. N. Williams, B. C. Ross, and T. M. Ellis. 1993. Use of a repetitive element isolated from Mycobacterium tuberculosis in hybridisation studies with Mycobacterium bovis: a new tool for epidemiological studies of bovine tuberculosis. Vet. Microbiol. 37:1-17. [DOI] [PubMed] [Google Scholar]
- 15.Curnow, J. 10June1999, posting date. Escherichia coli O157 outbreak in Scotland linked to unpasteurised goat's milk. Eurosurveill. Wkly. Issue 24. [Online.] http://www.eurosurv.org/1999/pfp/990610_pfp.html#2.
- 16.Dalla-Costa, L. M., K. Irino, J. Rodrigues, I. N. G. Rivera, and L. R. Trabulsi. 1998. Characterisation of diarrhoeagenic Escherichia coli clones by ribotyping and ERIC-PCR. J. Med. Microbiol. 47:227-234. [DOI] [PubMed] [Google Scholar]
- 17.Faith, N. G., J. A. Shere, R. Brosch, K. W. Arnold, S. E. Ansay, M. S. Lee, J. B. Luchansky, and C. W. Kaspar. 1996. Prevalence and clonal nature of E. coli O157:H7 on dairy farms in Wisconsin. Appl. Environ. Microbiol. 62:1519-1525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Frost, J. A., H. R. Smith, G. A. Willshaw, S. M. Scotland, R. J. Gross, and B. Rowe. 1989. Phage-typing of Vero-cytotoxin producing Escherichia coli O157 isolated in the United Kingdom. Epidemiol. Infect. 103:73-81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gautom, R. K. 1997. Rapid pulsed-field gel electrophoresis protocol for typing of Escherichia coli O157:H7 and other gram-negative organisms in 1 day. J. Clin. Microbiol. 35:2977-2980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Grif, K., H. Karch, C. Schneider, F. D. Daschner, L. Beutin, T. Cheasty, H. Smith, B. Rowe, M. P. Dierich, and F. Allerberger. 1998. Comparative study of five different techniques for epidemiological typing of Escherichia coli O157. Diagn. Microbiol. Infect. Dis. 32:165-176. [DOI] [PubMed] [Google Scholar]
- 21.Hanninen, M. L., P. Perko-Makela, H. Rautelin, B. Duim, and J. A. Wagenaar. 2001. Genomic relatedness within five common Finnish Campylobacter jejuni pulsed-field gel electrophoresis genotypes studied by amplified fragment length polymorphism analysis, ribotyping, and serotyping. Appl. Environ. Microbiol. 67:1581-1586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Heuvelink, A. E., F. L. A. M. van den Biggelaar, E. de Boer, R. G. Herbes, W. J. G. Melchers, J. H. J. Huis in't Veld, and L. A. H. Monnens. 1998. Isolation and characterization of verocytotoxin-producing Escherichia coli O157 strains from Dutch cattle and sheep. J. Clin. Microbiol. 36:878-882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Izumiya, H., T. Masuda, R. Ahmed, R. Khakhria, A. Wada, J. Terajima, K. Itoh, W. M. Johnson, H. Konuma, K. Shinagawa, K. Tamura, and H. Watanabe. 1998. Combined use of bacteriophage typing and pulsed-field gel electrophoresis in the epidemiological analysis of Japanese isolates of enterohemorrhagic Escherichia coli O157:H7. Microbiol. Immunol. 42:515-519. [DOI] [PubMed] [Google Scholar]
- 24.Keene, W. E., E. Sazie, J. Kok, D. H. Rice, D. D. Hancock, V. K. Balan, T. Zhao, and M. P. Doyle. 1997. An outbreak of Escherichia coli O157:H7 infections traced to jerky made from deer meat. JAMA 277:1229-1231. [DOI] [PubMed] [Google Scholar]
- 25.Konawalchuk, J., J. I. Speir, and S. Stavric. 1977. Vero response to a cytokine of Escherichia coli. Infect. Immun. 18:775-779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Krause, U., F. M. Thomson-Carter, and T. H. Pennington. 1996. Molecular epidemiology of Escherichia coli O157:H7 by pulsed-field gel electrophoresis and comparison with that by bacteriophage typing. J. Clin. Microbiol. 34:959-961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liebana, E., L. Garcia-Migura, M. F. Breslin, R. H. Davies, and M. J. Woodward. 2001. Diversity of strains of Salmonella enterica serotype Enteritidis from English poultry farms assessed by multiple genetic fingerprinting. J. Clin. Microbiol. 39:1-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lisegang, A., U. Sachse, R. Prager, H. Claus, H. Steinrück, S. Aleksic, W. Rabsch, W. Voigt, A. Fruth, H. Karch, J. Bockemühl, and H. Tschäpe. 2000. Clonal diversity of Shiga toxin-producing E. coli O157:H7/H− in Germany—a ten year study. Int. J. Med. Microbiol. 290:269-278. [DOI] [PubMed] [Google Scholar]
- 29.Louie, M., S. Read, L. Louie, K. Ziebell, K. Rahn, A. Borczyk, and H. Lior. 1999. Molecular typing methods to investigate transmission of Escherichia coli O157:H7 from cattle to humans. Epidemiol. Infect. 123:17-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Martin, I. E., S. D. Tyler, K. D. Tyler, R. Khakhria, and W. M. Johnson. 1996. Evaluation of ribotyping as epidemiologic tool for typing Escherichia coli serogroup O157 isolates. J. Clin. Microbiol. 34:720-723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Morgan, D., C. P. Newman, D. N. Hutchinson, A. M. Walker, B. Rowe, and F. Majid. 1993. Verotoxin-producing Escherichia coli O157 infections associated with the consumption of yoghurt. Epidemiol. Infect. 111:181-187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Norton, D. M., J. M. Scarlett, K. Horton, D. Sue, J. Thimothe, K. J. Boor, and M. Wiedmann. 2001. Characterization and pathogenic potential of Listeria monocytogenes isolates from the smoked fish industry. Appl. Environ. Microbiol. 67:646-653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.O'Brien, S., and H. Smith. 18March1999, posting date. Outbreak of vero cytotoxin producing Escherichia coli O157 infection in northern England: update. Eurosurveill. Wkly. Issue 12. [Online.] http://www.eurosurv.org/1999/pfp/990318_pfp.html#3.
- 34.Pennington, H. 1998. Factors involved in recent outbreaks of Escherichia coli O157:H7 in Scotland and recommendations for its control. J. Food Safety 18:383-391. [Google Scholar]
- 35.Preston, M. A., W. Johnson, R. Khakhria, and A. Borczyk. 2000. Epidemiologic subtyping of Escherichia coli serogroup O157 strains isolated in Ontario by phage typing and pulsed-field gel electrophoresis. J. Clin. Microbiol. 38:2366-2368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Reid, C.-A., S. M. Avery, A. Small, and S. Buncic. Presence of food-borne pathogens on cattle hides. Food Control, in press.
- 37.Rice, D. H., K. M. McMenamin, L. C. Pritchett, D. D. Hancock, and T. E. Besser. 1999. Genetic subtyping of Escherichia coli O157 isolates from 41 Pacific Northwest USA cattle farms. Epidemiol. Infect. 122:479-484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rios, M., V. Prado, M. Trucksis, C. Arellano, C. Borie, M. Alexandre, A. Fica, and M. M. Levine. 1999. Clonal diversity of Chilean isolates of enterohemorrhagic Escherichia coli from patients with hemolytic-uremic syndrome, asymptomatic subjects, animal reservoirs, and food products. J. Clin. Microbiol. 37:778-781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Small, A., C.-A. Reid, S. Avery, N. Karabasil, C. Crowley, and S. Buncic. 2002. Potential for the spread of Escherichia coli O157, Salmonella, and Campylobacter in the lairage environment at abbatoirs. J. Food Prot. 65:931-936. [DOI] [PubMed] [Google Scholar]
- 40.Tenover, F. C., R. D. Arbeit, R. V. Goering, P. A. Mickelsen, B. E. Murray, D. H. Persing, and B. Swaminathan. 1995. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J. Clin. Microbiol. 33:2233-2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wachsmuth, I. K., J. A. Kiehlbauch, C. A. Bopp, D. N. Cameron, N. A. Stockbrine, J. G. Wells, and P. A. Blake. 1991. The use of plasmid profiles and nucleic acid probes in epidemiologic investigation of foodborne, diarrheal diseases. Int. J. Food Microbiol. 12:77-90. [DOI] [PubMed] [Google Scholar]