Abstract
The causes of sarcoidosis are unknown. In this study, we report the presence of Mycobacterium tuberculosis complex and Propionibacterium granulosum DNA in a significant proportion of Greek patients with sarcoidosis. Human herpesvirus 8 DNA was not detected in sarcoid tissues from Greek patients. Our findings are discussed.
Sarcoidosis has been described as a systemic disorder characterized by the presence of noncaseating epithelioid cell granulomas in multiple tissues (21). Although, the etiology of sarcoidosis remains unclear, a number of putative etiopathogenic factors have been proposed. Various infectious agents have been suggested as being involved, of which mycobacteria seem to be the most important. Even though a considerable number of investigators have failed to detect mycobacteria in clinical samples from patients with sarcoidosis (28, 33), several studies have suggested the exclusive involvement of Mycobacterium avium complex members in sarcoidosis (8, 13, 20, 24, 25), while other studies have proposed Mycobacterium tuberculosis complex to be responsible (9, 22, 27, 29) (Table 1). Therefore, a convincing association between sarcoidosis and mycobacterial infections has yet to be established.
TABLE 1.
Summary of studies dealing with detection of Mycobacterium spp. in biopsy specimens from patients with sarcoidosisaa
| Result and reference | Source of samples | Sequence amplified | Sarcoidosis result
|
|
|---|---|---|---|---|
| No. tested | No. positive | |||
| Positive | ||||
| 1 | Various biopsies | 65-kDa antigen | 14 | 3 |
| 27 | Lavage | IS6110 | 20 | 10 |
| 13 | Various biopsies | NR | NR | All |
| 22 | NR | Hybridization | 5 | 5 |
| 9 | Various biopsies | IS6110, 65-kDa antigen | 16 | 7 |
| 25 | Serologic | 28 | 23 | |
| 23 | Various biopsies | 65-kDa antigen | 15 | 2 |
| 8 | Skin biopsies | IS902, IS900 | 6 | 4 |
| 26 | Various biopsies | 16S rRNA | 24 | 1 |
| 24 | Various biopsies | 65-kDa antigen | 35 | 11 |
| 29 | Lymph nodes | IS6110 | 30 | 2 |
| 20 | Skin biopsies | 65-kDa antigen | 20 | 16 |
| 18 | Lymph nodes | IS6110 | 15 | 3 |
| 16 | Various biopsies | MBP64 | 25 | 9 |
| 17 | FFPE tissues | Various primers | 25 | 6 |
| 15 | FFPE tissues | IS986, IS6110 | 50 | 32 |
| 7 | Lymph nodes | IS6110 | 103 | 5 |
| Negative | ||||
| 10 | Various biopsies | 16S rRNA | 14 | 0 |
| 28 | FFPE tissues | 65-kDa antigen | 14 | 0 |
| 1 | Various biopsies | IS6110 | 8 | 0 |
| 11 | FFPE tissues | 65-kDa antigen | 10 | 0 |
| 32 | Various biopsies | IS6110 | 18 | 0 |
| 33 | Various biopsies | IS6110 | 23 | 0 |
NR, no reference.
Ishige et al. (18) have proposed members of Propionibacteria species may also be involved in the pathogenesis of sarcoidosis. Recently, Eishi et al. (7) suggested that Propionibacterium spp. are more likely to be the cause of sarcoidosis than Mycobacteria spp. Finally, Di Alberti et al. (5), using a PCR technique, detected the presence of human herpesvirus 8 (HHV-8) DNA sequences exclusively in sarcoid tissue and not in specimens from healthy individuals. From the studies presented above, it is clear that no single agent can be held solely responsible for all cases of sarcoidosis. The intense controversy concerning the role of mycobacteria and other infectious agents in sarcoidosis points out the necessity for further epidemiological studies in order to shed more light on the possible role of these agents in the development of sarcoidosis.
Until now, there have been no reports investigating the presence of propionibacterial and HHV-8 DNA in sarcoidosis in Greek patients, whereas data concerning mycobacterial DNA detection are limited (16, 17).
The goal of the present study was to determine the frequency of detection of mycobacterial, propionibacterial, and HHV-8 DNA from sarcoid patients and thus to evaluate the possible involvement of these agents in sarcoidosis.
A total of 50 formalin-fixed paraffin-embedded (FFPE) tissues (lung and lymph nodes) from patients suffering from sarcoidosis were collected from “Sotiria” Chest Hospital in Athens, Greece, and were analyzed. In addition, FFPE material from 16 patients with tuberculosis and 20 patients with non-small cell lung carcinomas (NSCLC) were analyzed in parallel as controls. The diagnosis of sarcoidosis was established by the compatible clinical picture: the absence of evidence of current infection by organisms known to produce granulomatous disease, such as M. tuberculsosis, as assessed by culture; and histologic demonstration of noncaseating granulomas in biopsy specimens of involved tissues. The diagnosis of tuberculosis was based on the compatible clinical findings, chest radiographs, positive results of culture of sputum of bronchoalveolar lavage samples, and the histologic findings of caseous necrosis. DNA extraction from FFPE samples was performed as previously described (17). To confirm the integrity of DNA, a 430-bp sequence in the human GAPDH (glyceraldehyde-3-phosphate dehydrogenase) gene was amplified (12). The primers for the PCRs and the corresponding annealing temperatures are listed in Table 2. All of the primers used were synthesized and purchased by MWG-Biotech AG, Ebersberg, Germany. To determine the specificity of the primers, PCR was also performed with DNA from different bacterial species (Escherichia coli, Klebsiella pneumoniae, Proteus mirabilis, and Salmonella enterica), from sterile paraffin samples, and from mycobacterium-, propionibacterium-, and HHV-8-negative FFPE samples. For determination of primer sensitivity, M. tuberculosis complex, M. avium complex, Propionibacterium acnes, and P. granulosum strains, as well as DNA from skin samples from a patient with classic Kaposi's sarcoma, were used. The minimum amount of DNA necessary for a positive result was determined by absorbance measurements of serial DNA dilutions as previously described (17). We used sterile water as a negative control in each PCR in order to ensure that the reagents used were not contaminated, and an adequate number of certified negative control samples were processed in parallel with clinical specimens in order to ensure that no contamination was occurring prior to amplification. In all cases, extracted genomic DNA (1 μg from each sample) was amplified in a final volume of 30 μl containing 20 mM Tris-Cl (pH 8.3), 50 mM KCl, 0.2 mM (each) deoxynucleoside triphosphate, 2 mM MgCl2, 10 pmol of each primer, and 0.2 U of Taq DNA polymerase (Promega). The thermocycling profile consisted of 40 cycles, with 1 cycle for 1 min at 95°C, 1 min at the annealing temperature (Table 2), and 2 min at 72°C, followed by a final extension step of 6 min at 72°C. The PCR products were analyzed on 2% agarose. The results were confirmed by DNA sequencing of representative PCR products (Taq DyeDeoxy Terminator Cycle Sequencing kit and ABI 373A DNA Sequencer; Perkin-Elmer, Forest City, Calif.).
TABLE 2.
Nucleotide sequences and annealing temperatures of oligonucleotides for amplification of mycobacterial and propionibacterial DNA
| Bacterial species or virus | Target | Primer sequence (5′→3′)
|
Product size (bp) | Tm (°C) | Refer- ence | |
|---|---|---|---|---|---|---|
| Forward | Reverse | |||||
| M. tuberculosis complex | IS6110 | CGTGAGGGCATCGAGGTGGC | GCGTAGGCGTCGGTGACAAA | 245 | 68 | 30 |
| MBP64 | GCTCTGTTGTTCGGGTGTGGCCA | GATATTCAATTCGTAGGGGGCTT | 243 | 65 | 17 | |
| mtp40 | CAACGCGCCGTCGGTGG | CCCCCCACGGCACCGC | 396 | 65 | 3 | |
| M. avium complex | 16S rRNA | GTGGGCAATCTGCCCTGCACTTCGG | GCCCGCACGCTCACAGTTAAGCCGT | 504 | 60 | 17 |
| IS1245 | GCCGCCGAAACGATCTAC | AGGTGGCGTCGAGGAAGAC | 427 | 65 | 31 | |
| IS901 | GCAACGGTTGTTGCTTGAAA | TGATACGGCAATCGCGT | 1,108 | 65 | 19 | |
| M. paratuberculosis | IS900 | ACGCCGCGGGTAGTTA | GGGGCGTTTGAGGTTTC | 707 | 60 | 14 |
| P. acnes | 16S rRNA | GGCACACCCATCTCTGAGCAC | GGGTTGTAAACCGCTTTCGCTG | 587 | 54 | 25 |
| P. granulosum | 16S rRNA | GGAAACCTGATGCTTAACGT | GGCACCAACCATCTCTGGAA | 426 | 58 | 18 |
| HHV-8 | KS330233 | AGCCGAAAGGATTCCACCAT | TCCGTGTTGTCTACGTCCAG | 233 | 58 | 2 |
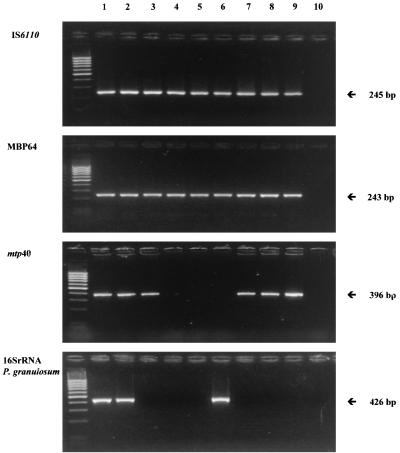
Forty-six samples with sarcoid lesions as well as all of the controls had intact DNA for PCR analysis. The results are summarized in Table 3, and representative patterns of PCR application are shown in Fig. 1. We observed a positive signal in 33 of 46 of the histopathologically proven sarcoid patients (71.7%) by using the M. tuberculosis IS6110 complex-specific primers (30). Given that there are some M. tuberculosis strains that do not possess the IS6110 sequence in their genome (34), we proceeded with a further confirmatory PCR for the gene coding for the immunogenic M. tuberculosis complex-specific protein MBP64 (17) (Table 3). In addition, since diagnosis based on IS6110 and MPB64 cannot discriminate between M. tuberculosis and other species from the M. tuberculosis complex (M. bovis, M. africanum, and M. microti), we examined our samples with species-specific mtp40 primers, which do not recognize M. bovis, M. africanum, and M. microti (4). Twenty-seven of the 33 M. tuberculosis complex-positive samples were found to be mtp40 positive, indicating that the majority of our samples had evidence of M. tuberculosis. Six of the samples were negative for mtp40, suggesting that possibly other members of M. tuberculosis complex could also be present in our samples. All of the samples of the positive control group (patients with tuberculosis) had evidence of M. tuberculosis (IS6110, MBP64, and mtp40 positive). No members of M. avium complex were detected, and no mycobacterial DNA was detected in the negative control group.
TABLE 3.
Results from PCR assays for mycobacterial and propionibacterial DNA detectiona
| Clinical diagnosis (total no. of samples) | No. (%) of samples with:
|
||
|---|---|---|---|
| M. tuberculosis complex | P. granulosum | P. granulosum and M. tuberculosis complex | |
| Sarcoidosis (46) | 33 (71.7) | 20 (43.47) | 20 (43.47) |
| Tuberculosis (16) | 16 (100) | 0 (0) | 0 (0) |
| NSCLCs (20) | 0 (0) | 0 (0) | 0 (0) |
No DNA of M. avium complex, P. acnes, and HHV-8 was detected.
FIG. 1.
Representative patterns of PCR detection of an M. tuberculosis complex strain or strains and P. granulosum. Lanes: 1 to 9, representative specimens; 10, negative control without DNA. Molecular size markers (100-bp DNA ladder from New England BioLabs) are in the unmarked lane on the left.
Consequently, examining the presence of propionibacteria, we found P. granulosum in 20 of 46 patients (43.47%; Table 3). The patients with NSCLC and those with clinically and histologically documented tuberculosis were found to be negative for the presence of P. granulosum. Interestingly, the 20 P. granulosum-positive samples were M. tuberculosis complex positive as well (Table 3). Finally, no HHV-8 DNA sequences were detected in our series.
Data from other groups using PCR techniques for the detection of mycobacterial DNA in sarcoidosis are inconsistent (Table 1). The significant proportion (71.7%) of M. tuberculosis complex found in our patients supports the hypothesis that this pathogen may contribute to the development of sarcoidosis.
In our series, a notable percentage (43.47%) of patients were positive for propionibacterial genomes. In contrast to recent epidemiological (7, 18) and immunological (6) studies, which suggest a central role of propionibacteria in the pathogenesis of sarcoidosis, our data cannot support this assumption, because propionibacterium-positive samples were a subgroup of the M. tuberculosis complex-positive ones. (Twenty out of 33 mycobacterium-positive sarcoid patients were also positive for propionibacteria.)
Our results imply, at least in part, a possible mycobacterial etiology for sarcoidosis in Greece. However, other bacteria, especially propionibacteria or their antigens, may also play an as-yet-undefined etiologic role. It is possible that sarcoidosis may result from a delayed hypersensitivity immune response initiated by different bacterial antigens (mycobacteria and/or propionibacteria), leading to granuloma formation. Finally, HHV-8 is not implicated in sarcoidosis in Greek patients, confirming the conclusions of a recent multination study on sarcoidosis.
Acknowledgments
We thank L. Arvanitakis (Virology Department, Shering-Plough, Greece) for providing the primers for HHV-8 detection, S. Kokotas for technical assistance, and N. Rezaei for editing the manuscript.
This work was supported by the EU Project “Sacrohn” N. QLK2-CT-2000-00928.
REFERENCES
- 1.Bocart, D., D. Lecossier, A. De Lassence, D. Valeyre, J. P. Battesti, and A. J. Hance. 1992. A search for mycobacterial DNA in granulomatous tissues from patients with sarcoidosis using the polymerase chain reaction. Am. Rev. Respir. Dis. 145:1142-1148. [DOI] [PubMed] [Google Scholar]
- 2.Chang, Y., E. Cesarman, M. S. Pessin, F. Lee, J. Culpepper, D. M. Knowles, and P. S. Moore. 1994. Identification of herpesvirus-like DNA sequences in AIDS-associated Kaposi's sarcoma. Science 266:1865-1869. [DOI] [PubMed] [Google Scholar]
- 3.Del Portillo, P., L. A. Murillo, and M. E. Patarroyo. 1991. Amplification of a species-specific DNA fragment of Mycobacterium tuberculosis and its possible use in diagnosis. J. Clin. Microbiol. 29:2163-2168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Del Portillo, P., M. C. Thomas, E. Martinez, C. Marañón, B. Valladares, M. E. Patarroyo, and M. C. Lopez. 1996. Multiprimer PCR system for differential identification of mycobacteria in clinical samples. J. Clin. Microbiol. 34:324-328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Di Alberti, L., A. Piattelli, L. Artese, G. Favia, S. Patel, N. Saunders, S. R. Porter, C. M. Scully, S. L. Ngui, and C. G. Teo. 1997. Human herpesvirus 8 variants in sarcoid tissues. Lancet 350:1655-1661. [DOI] [PubMed] [Google Scholar]
- 6.Ebe, Y., S. Ikushima, T. Yamagushi, K. Kohno, A. Azuma, K. Sato, I. Ishige, Y. Usui, T. Takemura, and Y. Eishi. 2000. Proliferative response or peripheral blood mononuclear cells and levels of antibody to recombinant protein from Propionibacterium acnes DNA expression library in Japanese patients with sarcoidosis. Sarcoidosis Vasc. Diffuse Lung Dis. 3:256-265. [PubMed] [Google Scholar]
- 7.Eishi, Y., M. Suga, I. Ishige, D. Kobayashi, T. Yamada, T. Takemura, T. Takizawa, M. Koike, S. Kudoh, U. Castabel, J. Guzman, G. Rizzato, M. Gambacorta, R. du Bois, A. G. Nicholson, O. P. Sharma, and M. Ando. 2002. Quantitative analysis of mycobacterial and propionibacterial DNA in lymph nodes of Japanese and European patients with sarcoidosis. J. Clin. Microbiol. 40:198-204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.El-Zaatari, F. A. K., S. A. Naser, D. C. Markesich, D. C. Kalter, L. Engstand, and D. Y. Graham. 1996. Identification of Mycobacterium avium complex in sarcoidosis. J. Clin. Microbiol. 34:2240-2245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fidler, H. A., G. S. Rook, N. M. Johnson, and J. McFadden. 1993. Mycobacterium tuberculosis DNA in tissue affected by sarcoidosis. Br. Med. J. 306:546-549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gerdes, J., E. Richter, S. Rusch-Gerdes, U. Greinert, J. Galle, M. Schlaak, H. D. Flad, D. Kirsten, and H. Magnussen. 1992. Mycobacterial nucleic acids in sarcoid lesions. Lancet 339:1536-1537. [DOI] [PubMed] [Google Scholar]
- 11.Ghossein, R. A., D. G. Ross, R. N. Salomon, and A. R. Rabson. 1994. A search for mycobacterial DNA in sarcoidosis using the polymerase chain reaction. Am. J. Clin. Pathol. 101:733-737. [DOI] [PubMed] [Google Scholar]
- 12.Goidin, D., A. Mamessier, M. J. Staquet, D. Schmitt, and O. Bethier-Verges. 2001. Ribosomal 18S RNA prevails over glyceraldehyde-3-phosphate dehydrogenase and beta-actin genes as internal standard for quantitative comparison of mRNA levels in invasive and noninvasive human melanoma cell subpopulations. Anal. Biochem. 295:17-21. [DOI] [PubMed] [Google Scholar]
- 13.Graham, D. Y., D. C. Markesich, D. C. Kalter, M. T. Moss, J. Hermon-Taylor, and A. K. El-Zaatari. 1992. Mycobacterial aetiology of sarcoidosis. Lancet 340:52-53. [DOI] [PubMed] [Google Scholar]
- 14.Green, E. P., M. L. V. Tizard, M. T. Moss, J. Tompson, D. J. Winterbourne, J. J. McFadden, and J. Hermon-Taylor. 1989. Sequence and characteristics of IS900, an insertion element identified in a human Crohn's disease isolate of Mycobacterium paratuberculosis. Nucleic Acids Res. 17:9063-9073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Grosser, M., D. D. Dittert, and T. Luther. 2001. Molecular detection of M. tuberculosis DNA in tuberculosis and sarcoidosis. Diagn. Mol. Pathol. 10:66-67. [DOI] [PubMed] [Google Scholar]
- 16.Ikonomopoulos, J. A., V. G. Gorgoulis, P. V. Zacharatos, E. N. Manolis, P. Kanavaros, A. Rassidakis, and C. Kittas. 1999. Multiplex polymerase chain reaction for the detection of mycobacterial DNA in cases of tuberculosis and sarcoidosis. Mod. Pathol. 9:854-862. [PubMed] [Google Scholar]
- 17.Ikonomopoulos, J. A., V. G. Gorgoulis, N. G. Kastrinakis, P. V. Zacharatos, S. N. Kokotas, K. Evagelou, A. G. Kotsinas, A. G. Tsakris, E. N. Manolis, and C. N. Kittas. 2000. Sensitive differential detection of genetically related mycobacterial pathogens in archival material. Am. J. Clin. Pathol. 114:940-950. [DOI] [PubMed] [Google Scholar]
- 18.Ishige, I., Y. Usui, T. Takemura, and Y. Eishi. 1999. Quantitative PCR of mycobacterial and propionibacterial DNA in lymph nodes of Japanese patients with sarcoidosis. Lancet 354:120-123. [DOI] [PubMed] [Google Scholar]
- 19.Kunze, Z. M., F. Portaels, and J. J. McFadden. 1992. Biologically distinct subtypes of Mycobacterium avium differ in possession of insertion sequence IS901. J. Clin. Microbiol. 30:2366-2372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li, N., A. Bojoghli, A. Kubba, and J. Bhawan. 1999. Identification of mycobacterial DNA in cutaneous lesions of sarcoidosis. J. Cutan. Pathol. 6:271-278. [DOI] [PubMed] [Google Scholar]
- 21.Mitchell, D. N., J. G. Scadding, B. E. Heard, and K. F. W. Hinson. 1977. Sarcoidosis: histopathological definition and clinical diagnosis. J. Clin. Pathol. 30:395-408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mitchell, I. C., J. L. Turk, and D. N. Mitchell. 1992. Detection of mycobacterial rRNA in sarcoidosis with liquid-phase hybridization. Lancet 339:1015-1017. [DOI] [PubMed] [Google Scholar]
- 23.Popper, H. H., E. Winter, and G. Hofler. 1994. DNA of Mycobacterium tuberculosis in formalin-fixed, paraffin-embedded tissue in tuberculosis and sarcoidosis detected by polymerase chain reaction. Am. J. Clin. Pathol. 101:738-741. [DOI] [PubMed] [Google Scholar]
- 24.Popper, H. H., H. Klemen, G. Hoefler, and E. Winter. 1997. Presence of mycobacterial DNA in sarcoidosis. Hum. Pathol. 7:796-800. [DOI] [PubMed] [Google Scholar]
- 25.Reid, J. D., and R. J. Chiodini. 1993. Serologic reactivity against Mycobacterium paratuberculosis antigens in patients with sarcoidosis. Sarcoidosis 10:32-35. [PubMed] [Google Scholar]
- 26.Richter, E., U. Greinert, D. Kirsten, S. Rusch-Gerdes, C. Shluter, M. Duchrow, J. Galle, H. Magnussen, M. Schlaak, H. D. Flad, and J. Gerdes. 1996. Assessment of mycobacterial DNA in cells and tissues of mycobacterial and sarcoid lesions. Am. J. Respir. Crit. Care Med. 153:375-380. [DOI] [PubMed] [Google Scholar]
- 27.Saboor, S. A., N. M. Johnson, and J. McFadden. 1992. Detection of mycobacterial DNA in sarcoidosis and tuberculosis with polymerase chain reaction. Lancet 339:1012-1015. [DOI] [PubMed] [Google Scholar]
- 28.Thakker, B., M. Black, and A. K. Foulis. 1992. Mycobacterial nucleic acids in sarcoid lesions. Lancet 339:1537.. [PubMed] [Google Scholar]
- 29.Vago, L., M. Barberis, A. Gori, P. Scarpellini, E. Sala, M. Nebuloni, S. Bonetto, M. Cannone, G. Marchetti, F. Franzetti, and G. Costanzi. 1998. Nested polymerase chain reaction for Mycobacterium tuberculosis IS6110 sequence on formalin-fixed paraffin-embedded tissues with granulomatous diseases for rapid diagnosis of tuberculosis. Am. J. Clin. Pathol. 4:411-415. [DOI] [PubMed] [Google Scholar]
- 30.van Embden, J. D. A., M. D. Cave, J. T. Crawford, J. W. Dale, K. D. Eisenbach, B. Gicquel, P. Hermans, C. Martin, R. McAdam, T. M. Shinnick, and P. M. Small. 1993. Strain identification of Mycobacterium tuberculosis by DNA fingerprinting: recommendations for a standardized methodology. J. Clin. Microbiol. 31:406-409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Van Soolingen, D., J. Bauer, V. Ritacco, S. Cardosos Leão, I. Pavlik, V. Vincent, N. Rastogi, A. Gori, T. Bodmer, C. Garzelli, and M. J. Garcia. 1998. IS1245 restriction fragment length polymorphism typing of Mycobacterium avium isolates: proposal for standardization. J. Clin. Microbiol. 36:3051-3054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vokurka, M., D. Lecossier, R. M. du Bois, B. Wallaert, M. A. Kambouchner, and A. J. Hance. 1997. Absence of DNA from mycobacteria of the M. tuberculosis complex in sarcoidosis. Am. J. Respir. Crit. Care Med. 156:1000-1003. [DOI] [PubMed] [Google Scholar]
- 33.Wilsher, M. L., R. E. Menzies, and M. C. Croxson. 1998. Mycobacterium tuberculosis DNA in tissues affected by sarcoidosis. Thorax 53:871-874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yuen, L. K. W., B. C. Ross, K. M. Jackson, and B. Dwyer. 1993. Characterization of Mycobacterium tuberculosis strains from Vietnamese patients by Southern blot hybridization. J. Clin. Microbiol. 31:1615-1618. [DOI] [PMC free article] [PubMed] [Google Scholar]