Abstract
The test approved by the U.S. Food and Drug Administration for assessment of the barrier quality of medical exam gloves includes visual inspection and a water leak test. Neither method tests directly the ability of gloves to prevent penetration by microorganisms. Methods that use microorganisms (viruses and bacteria) to test gloves have been developed but require classical culturing of the organism to detect it. We have developed a PCR assay for bacteriophage φX174 that allows the rapid detection of penetration of gloves by this virus. The method is suitable for use with both latex and synthetic gloves. The presence of glove powder on either latex or synthetic gloves had no effect on the ability of the PCR assay to detect bacteriophage DNA. The assay is rapid, sensitive, and inexpensive; requires only small sample volumes; and can be automated.
In response to a growing need to protect health care workers and patients from the possibility of transmission of human immunodeficiency virus through contact with body fluids, the Occupational Safety and Health Administration issued standard precautions that state that gloves must be worn when “touching blood, body fluids, secretions, excretions, and contaminated items” (17). Consequently, the rate of use of exam gloves in clinical practice rose dramatically (7). To ensure consistent quality in the manufacture of gloves, the U.S. Food and Drug Administration (FDA) introduced a two-part testing protocol in which gloves are first visually inspected and then subjected to a water leak test (5). These tests, while convenient and inexpensive, suffer from a number of drawbacks. Both the visual test for defects and the water leak test, in which an inspector looks for leaks in a glove filled with a liter of water, are subject to operator error. It has also been observed that microorganisms can penetrate gloves that do not show visible water leaks (2, 9). Studies that use microorganisms to gauge the propensities of gloves to permit pathogen penetration examine the question more directly than water leak studies. However, the microbial assays described rely on classical microbiological culture techniques to detect microbial penetration and therefore are more labor-intensive, time-consuming, and expensive than the FDA-approved tests. This report describes an assay for the detection of glove penetration by a small virus, bacteriophage φX174. The assay uses PCR to detect the bacteriophage. PCR offers the advantages of being highly sensitive and specific for the agent being detected. With the recent and rapid increase in the availability of laboratory robotics for molecular biology, this PCR assay also has the potential to be automated and therefore made useful in the glove manufacturing setting.
MATERIALS AND METHODS
Bacterial strains and growth media.
Stock cultures of bacteriophage φX174 (ATCC 13706-B1) and its bacterial host, Escherichia coli C (ATCC 13706), were obtained from the American Type Culture Collection (Manassas, Va.). Broth cultures of E. coli were grown in NB medium (8 g of Nutrient Broth [EM Science, Gibbstown, N.J.]), 5 g of KCl, and 100 μl of Tween 80 [EM Science] in 1 liter of distilled H2O). Cultures of E. coli were maintained on solid NB medium, made as described above by the addition of 15 g of agar per liter and 1 ml of CaCl2 and the omission of Tween 80. For plaque analysis of bacteriophage φX174, the top agar composition was identical to that of the plate agar, except that the concentration of agar was 8 g/liter. Stock suspensions of bacteriophage φX174 were prepared by growth in liquid culture with E. coli C (8, 15). E. coli was incubated at 37°C to a density of approximately 108 CFU/ml (optical density at 600 nm, 0.3 to 0.4). The culture was then inoculated with stock bacteriophage φX174 (approximately 108 PFU) and incubated at 37°C with shaking. The optical density of the culture was monitored after the addition of bacteriophage, and incubation was continued until the density of the culture stopped dropping (approximately 4 to 5 h). After incubation, unlysed cells and cell debris were removed by centrifugation at 2,000 × g for 10 min, and the supernatant was filtered through a sterile 0.2-μm-pore-size filter. Suspensions of bacteriophage φX174 were stored at 4°C.
Assay for bacteriophage penetration of punctured gloves.
The types of gloves used in this study are listed in Table 1. A polypropylene beaker was filled with 200 ml of sterile nanopure water. For proof-of-principle testing, each finger of the gloves was punctured five times with an 18-gauge needle and then held over the water-filled beaker and filled with a solution containing bacteriophage (105 to 106 PFU/ml in sterile H2O), ensuring that all five fingers were filled. The glove was placed into the water-filled beaker and left at room temperature for 30 min, after which samples were drawn from inside and outside the glove.
TABLE 1.
Penetration of punctured synthetic and latex gloves by bacteriophage detected by plate enumeration and PCR
| Glove material | Inner titera (PFU/μl) | Outer titera (PFU/μl) | Ratio of outer titer to inner titer | PCR detection (no. of samples positive/total no. tested)b
|
|
|---|---|---|---|---|---|
| Inside | Outside | ||||
| Latex, powderedc | 11,900 ± 795 | 72 ± 23 | 0.0059 | 6/6 | 5/6 |
| Latex, nonpowdered | 10,237 ± 726 | 77 ± 22 | 0.0075 | 8/8 | 6/8 |
| PVC,d powdered | 3,098 ± 375 | 806 ± 109 | 0.260 | 8/8 | 8/8 |
| Synthetic (unknown material), nonpowdered | 4,162 ± 1,036 | 1,271 ± 259 | 0.305 | 8/8 | 8/8 |
Mean ± standard error of measurements from inside and outside eight gloves (except where noted). Titers were determined by sampling 100 μl from inside and outside each glove and calculating the number of PFU per microliter.
Samples of 1 μl were drawn and placed in a 50-μl PCR mixture. Ten microliters of each reaction mixture was analyzed for the presence of a 1,500-bp band by agarose electrophoresis.
Measurements from six powdered latex gloves.
PVC, polyvinyl chloride.
Samples were serially diluted in 10-fold increments and tested for the presence of bacteriophage by coculture in top agar with the bacterial host. After diluted samples (100 μl) were mixed with E. coli and the top agar and poured onto NB medium plates, the plates were incubated for 3 to 4 h at 37°C and the plaques were counted. Diluted samples were also tested for the presence of bacteriophage DNA by PCR. Controls without template were set up in parallel with all PCRs and showed no evidence of contamination.
Preparation of bacteriophage φX174 genomic DNA.
Bacteriophage φX174 DNA was prepared from a culture of E. coli infected with φX174 as described above by using a QIAprep Spin Miniprep kit (QIAGEN, Valencia, Calif.) and quantified by UV spectrometry at 260 and 280 nm.
Bacteriophage DNA detection by PCR amplification.
PCR was performed with diluted samples of the samples used for the method described above. PCR mixtures (total volume, 50 μl) consisted of 45 μl of PCR Supermix (Invitrogen Life Technologies, Carlsbad, Calif.), 1 μl of each primer solution (16 to 18 pmol/μl), 1 μl of diluted sample, and 2 μl of nuclease-free water. The primers (primers 5′-GCTTGCGTTTATGGTACG-3′ and (5′-ATACGAAGGCGCATAACG-3′) were designed on the basis of the bacteriophage φX174 genomic sequence (GenBank accession no. NC_001422) by using the World Wide Web-based GeneFisher software package (6) (the software package is no longer available at the time of this writing). The reaction mixtures were incubated in a RoboCycler Gradient 96 temperature cycler (Stratagene, La Jolla, Calif.): 5 min of preincubation at 94°C; 35 cycles of 94°C for 45 s, 54°C for 60 s, and 72°C for 60 s; and holding of the mixture for 10 min at 72°C postincubation. The PCR products were visualized in 1% agarose gels stained with ethidium bromide.
RESULTS
Detection of purified bacteriophage DNA from a stock preparation.
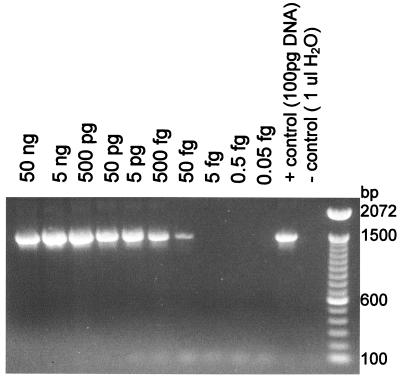
To ensure the ability of the selected primer pair to amplify bacteriophage φX174 DNA, the limit of detection for purified φX174 DNA was first determined. After a stock of purified φX174 DNA was serially diluted 10-fold, the lowest detectable amount of bacteriophage φX174 DNA was approximately 50 fg (Fig. 1). Since the mass of one double-stranded copy of the φX174 genome is approximately 3.2 × 106 Da (5.5 × 10−18 g), the detectable amount of DNA corresponds to approximately 9,000 double-stranded copies of the bacteriophage φX174 genome.
FIG. 1.
Limit of detection of purified bacteriophage φX174 DNA by PCR. Bacteriophage φX174 DNA was purified from infected E. coli cells with a QIAprep spin column and diluted in sterile water. One microliter of each dilution was subjected to PCR amplification. The results shown are representative of those for two independent DNA preparations.
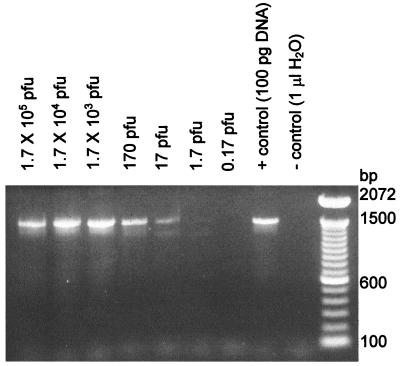
Before attempting to detect bacteriophage particles in the presence of glove materials, the ability of the assay to detect bacteriophage DNA directly from a host cell lysate containing bacteriophage φX174 was examined. Tenfold dilutions of a lysate were added to the PCR mixtures (Fig. 2). The titers of aliquots of each dilution were also determined by growing diluted bacteriophage on the E. coli host in top agar. On the basis of the titers in the plates, the limit of detection for bacteriophage DNA by PCR was found to correlate with the presence of approximately 1 to 100 PFU per μl of sample (Fig. 2).
FIG. 2.
Detection of bacteriophage φX174 DNA in a host cell lysate. A stock suspension of bacteriophage was prepared by growth in liquid culture with E. coli C, which was then serially diluted. One-microliter aliquots of each dilution were subjected to PCR amplification and to determination of phage titers by plate counting. The number of PFU in each PCR mixture is indicated above each lane. The results shown are representative of those from eight experiments.
Detection of viral penetration of punctured gloves.
A total of 30 medical examination gloves from various manufacturers were punctured, filled with a suspension of bacteriophage, and tested as described in Materials and Methods. The gloves were intentionally punctured to ensure their penetration by viruses in a sample of a reasonable size. A separate test by classical microbiological methods with undamaged gloves indicated that large numbers of gloves would have to be tested before gloves that leaked virus could be located (K. P. O'Connell et al., unpublished data).
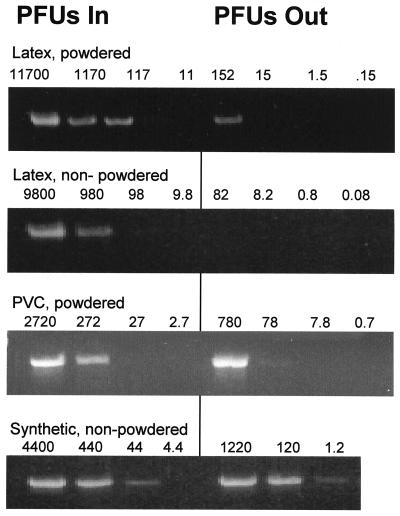
The number of bacteriophage in water inside and outside each punctured glove was enumerated by plate counting (100-μl samples), and 1-μl samples were drawn for PCR analysis. One-fifth (10 μl) of each PCR mixture was applied to an agarose gel to determine whether amplified bacteriophage DNA was present. Representative results of the PCR analyses are shown in Fig. 3. In all cases, the limit of detection of phage by PCR assays in the presence of gloves was higher than that in the absence of gloves. We seldom detected a PCR product from an assay with gloves when the titer in the sample was less than 100 PFU/μl (Fig. 3), while PCR products were routinely detected by PCR with lysates when the titer in the sample was less than 100 PFU/μl (Fig. 2). This may be due to the interference of glove residues with the PCRs.
FIG. 3.
PCR amplification of bacteriophage DNA. Water from inside and outside of punctured gloves was sampled and serially diluted 10-fold. The titers in each of the dilutions were then determined by plate counting, and the dilutions were used as templates in the PCRs. The numbers above each lane indicate the numbers of PFU per microliter in each dilution; the left-most lane contains undiluted sample. One microliter of each dilution was used as the template for PCR. Data are representative of those for six to eight gloves for each type of glove material. Complete data for the experiment are summarized in Table 1. As a percentage of the total number of phage inside each glove, more bacteriophage were found to have leaked from punctured synthetic gloves than from latex gloves. PVC, polyvinyl chloride.
The number of phage leaking out of the holes in synthetic gloves was approximately 800 to 1,220 PFU/μl, corresponding to an amount of PCR-detectable material well above our limit of detection. Bacteriophage DNA was detected, as anticipated, outside each of the 16 gloves tested (Table 1).
We detected far fewer PFUs outside the punctured latex gloves by plate counting. The average was approximately 75 PFU/μl (range, 20 to 196 PFU/μl), which is close to or somewhat below our limit of detection of phage from gloves by PCR. This result was surprising, given that the amount of bacteriophage inside the gloves was up to three times greater than the amount inside the synthetic gloves (Table 1). While 75 PFU/μl is above the limit of detection of our assay in the absence of gloves, we detected bacteriophage DNA outside only 11 of the 14 latex gloves tested. In an attempt to explain this discrepancy, we examined the recovery of phage from beakers containing water, bacteriophage, and gloves that had been sliced open. In three experiments, as much as 40% of the PFU added to the water was not recoverable, independent of whether the glove was powdered and independent of the glove material (data not shown), suggesting that both PFU and PCR-detectable material are adhering to the glove surfaces and are being sequestered from the assay.
DISCUSSION
Several studies have been published on the use of viruses (animal viruses as well as bacteriophage) for the testing of latex and other elastomeric materials as microbial barriers (9-12, 14). While the use of viruses or bacteria in barrier testing provides a means of testing barrier quality that addresses the core issue (the effectiveness of the barrier against microorganisms) more directly than the FDA water test does, microbiological methods are far less convenient. Our goal was to develop a PCR assay for bacteriophage detection with a view to streamlining the use of viruses in a quality control laboratory setting.
Our data demonstrated that the use of PCR to detect leakage of virus from gloves is at least as sensitive as conventional plate assays for the detection of bacteriophage. Conventional microbiological assays for the detection of bacteria or bacteriophage require a microbiology laboratory, preparation and/or storage of bacterial growth media, an incubation period of several hours, and the inspection of plates by a trained worker. While culture-based assays are sensitive and quantifiable, they are also time-consuming and have limited use in industrial settings that require the rapid testing of large numbers of samples. PCR offers the advantages of being faster (generally 1 to 3 h), potentially automatable through the use of laboratory robots, and more sensitive per unit of sample. While the dilution endpoints for detection of phage by PCR and the titers determined in the plates were approximately equivalent (Fig. 2), the sample size required for PCR is 1% that required for plate counting. In addition, only one-fifth of each PCR sample was analyzed by agarose gel electrophoresis, and the detection of PCR products by electrophoresis is inefficient relative to that by newer DNA detection technologies. For example, although it was not used in this study, real-time PCR technology with fluorescence detection of the product allows the detection of tens to hundreds of target DNA copies (1, 16), compared to the thousands detected in this work. The method is fast, relatively simple, and inexpensive on a per-assay basis. The use of PCR with fluorescence detection has already been demonstrated for influenza virus types A and B (18). Similar methods have been established for the detection of Chlamydia trachomatis and Mycobacterium tuberculosis and for the quantification of human immunodeficiency virus (13) in clinical settings. It is therefore logical that PCR detection of microorganisms could be useful in the quality assurance setting.
The ability to detect phage DNA was not obviously hindered by the presence of glove powder, as data for powdered and nonpowdered gloves among the gloves evaluated were comparable (Table 1), an important finding given the current prevalence of powdered gloves in use. There is at least one report that glove powder can inhibit PCR (4). In that study powder from a laboratory worker's gloves evidently entered PCR tubes during handling and subsequently inhibited DNA amplification. The lack of effect of glove powder in our study is probably the result of dilution, as the powder from each glove was dispersed in 300 ml of water in each experiment, and only 1 μl of sample was drawn for PCR analysis.
Glove material had a significant impact on the efficacy of the assay. A far lower proportion of bacteriophage inside punctured latex gloves leaked into the water outside the gloves. The reason for this has not been rigorously established; in fact, the latex gloves were more resilient than the synthetic gloves. Several workers have observed that some punctured latex gloves fail to leak water in a visual test, although microbial penetration is still detectable by culturing (3, 9; J. M. Broyles and D. Korniewicz, unpublished observations). This phenomenon has been termed “resealing.” It is possible that, because of their greater resiliency, latex gloves contract when the puncturing object is withdrawn, leaving a hole smaller than the diameter of the puncturing object. In contrast, synthetic material may be tearing at the site of puncture or failing to retract around the hole when the puncturing object is removed. In any case, it is evident that higher titers of bacteriophage may be required to test latex gloves reliably when samples volumes are as small as those used in this study, or the method will have to be modified to place latex gloves under a stress that will result in the stretching and widening of the holes.
A factor possibly complicating the interpretation of our results is the likely presence of free phage DNA in the host cell lysates used in this study. While we sampled 100-fold less material for PCR than for plate counting, we found that both methods gave approximately equal limits of detection according to their dilution endpoints. Because the likelihood that the 1 μl of sample drawn for PCR contained all the PFU detected in 100 μl of the same dilution of a phage preparation is exceedingly low, we conclude that phage lysates contained either free DNA or noninfective phage particles, or both. Gloves could be tested by using calibrated phage preparations purified by density gradient centrifugation and particles enumerated by electron microscopy; this would avoid uncertainty in identifying the PCR target. To increase the likelihood of detecting penetrating phage, the size of the PCR mixture can also be scaled up to allow use of a larger sample size, higher titers of bacteriophage may be added to the assay, or, as described above, more sensitive methods of performing the PCR and detecting amplified DNA can be used. Future studies should address standardization of viral preparations, their interactions with glove material, the use of real-time fluorogenic PCR methods, and the use of engineering modifications either to adapt the current glove testing apparatus to PCR testing or to develop entirely new automated glove testing systems.
REFERENCES
- 1.Aberham, C., C. Pendl, P. Gross, G. Zerlauth, and M. Gessner. 2001. A quantitative, internally controlled real-time PCR assay for the detection of parvovirus B19 DNA. J. Virol. Methods 92:183-191. [DOI] [PubMed] [Google Scholar]
- 2.Arnold, S. G., J. E. Whitman, Jr., C. H. Fox, and M. H. Cottler-Fox. 1988. Latex gloves not enough to exclude viruses. Nature 335:19.. [DOI] [PubMed] [Google Scholar]
- 3.Brough, S. J., T. M. Hunt, and W. W. Barrie. 1988. Surgical glove perforations. Br. J. Surg. 75:317.. [DOI] [PubMed] [Google Scholar]
- 4.de Lomas, J. G., F. J. Sunzeri, and M. P. Busch. 1992. False-negative results by polymerase chain reaction due to contamination by glove powder. Transfusion 32:83.. [DOI] [PubMed] [Google Scholar]
- 5.Federal Register. 2001. Patient examination and surgeons' gloves. Title 21 C.F.R. Part 800.20. Fed. Regist. 8:7-10.
- 6.Giegerich, R., F. Meyer, and C. Schleiemacher. 1996. GeneFisher software. Support for the detection of postulated genes. American Association of Artificial Intelligence Press, Menlo Park, Calif. [PubMed]
- 7.Hunt, L. W., A. F. Fransway, C. E. Reed, L. K. Miller, R. T. Jones, M. C. Swanson, and J. W. Yunginger. 1995. An epidemic of occupational allergy to latex involving health care workers. J. Occup. Environ. Med. 37:1204-1209. [DOI] [PubMed] [Google Scholar]
- 8.Hutchison, C. A., III, and R. L. Sinsheimer. 1966. The process of infection with bacteriophage phi-X174. X. Mutations in a phi-X lysis gene. J. Mol. Biol. 18:429-447. [DOI] [PubMed] [Google Scholar]
- 9.Korniewicz, D. M., B. E. Laughon, A. Butz, and E. Larson. 1989. Integrity of vinyl and latex procedure gloves. Nurs. Res. 38:144-146. [PubMed] [Google Scholar]
- 10.Korniewicz, D. M., B. E. Laughon, W. H. Cyr, C. D. Lytle, and E. Larson. 1990. Leakage of virus through used vinyl and latex examination gloves. J. Clin. Microbiol. 28:787-788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Neal, J. G., E. M. Jackson, F. Suber, and R. F. Edlich. 1998. Latex glove penetration by pathogens: a review of the literature. J. Long-Term Effects Med. Implants 8:233-240. [PubMed] [Google Scholar]
- 12.Nelson, J. R., T. A. Roming, and J. K. Bennett. 1999. A whole-glove method for the evaluation of surgical gloves as barriers to viruses. Am. J. Contact Dermatol. 10:183-189. [DOI] [PubMed] [Google Scholar]
- 13.Pfaller, M. A. 2001. Molecular approaches to diagnosing and managing infectious diseases: practicality and costs. Emerg. Infect. Dis. 7:312-318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rodeheaver, G. T., D. B. Drake, J. G. Neal, E. M. Jackson, F. Suber, P. A. Foresman, and R. F. Edlich. 1998. Influence of latex glove hydration on bacteriophage penetration. J. Long-Term Effects Med. Implants 8:241-248. [PubMed] [Google Scholar]
- 15.Sinsheimer, R. L., M. Lawrence, and C. Nagler. 1965. The process of infection with bacteriophage phi-X174. 8. Centrifugal analysis in alkaline media of the RF DNA at various stages of infection. J. Mol. Biol. 14:348-360. [DOI] [PubMed] [Google Scholar]
- 16.Tang, K. F., and D. V. Lightner. 2001. Detection and quantification of infectious hypodermal and hematopoietic necrosis virus in penaeid shrimp by real-time PCR. Dis. Aquat. Organisms 44:79-85. [DOI] [PubMed] [Google Scholar]
- 17.U.S. Department of Labor, Occupational Safety and Health Administration. 2001. Bloodborne pathogens. Title 29 C.F.R. Part 1910.1030. Fed. Regist. 6:260-273.
- 18.van Elden, L. J., M. Nijhuis, P. Schipper, R. Schuurman, and A. M. van Loon. 2001. Simultaneous detection of influenza viruses A and B using real-time quantitative PCR. J. Clin. Microbiol. 39:196-200. [DOI] [PMC free article] [PubMed] [Google Scholar]