Abstract
The attenuated Mycobacterium tuberculosis strain H37Ra is one of the most commonly used controls for M. tuberculosis identification in the clinical laboratory and is a source of false-positive results for M. tuberculosis as a consequence of cross-contamination. Therefore, the ability to discriminate between H37Ra and real clinical isolates has important public health implications. To date, differentiation of H37Ra from M. tuberculosis clinical isolates is possible only by IS6110 genotyping and spoligotyping. In the 1950s, some authors reported that the virulent strain H37Rv and M. tuberculosis clinical isolates were able to fix basic dyes in their anionic forms, while H37Ra was not. We have studied the different techniques described for M. tuberculosis cytochemical staining and have chosen the best of these, introducing certain modifications in order to increase their discriminative power and reproducibility. We describe cytochemical staining of M. tuberculosis cells with neutral red and Nile blue, which differentiates H37Ra from virulent strains. This method could be used as an easy laboratory tool for distinguishing between H37Ra and real M. tuberculosis clinical isolates.
At present, Mycobacterium tuberculosis identification is normally carried out in clinical laboratories by using conventional tests such as niacin, nitrate reduction, pyrazinamidase, thiopen-2-carbonic acid hydrazide (TCH), or catalase (13, 21, 22). In addition, the M. tuberculosis complex can be distinguished from other mycobacteria by commercialized nucleic acid probes (20) and high-performance liquid chromatography analysis of mycolic acid (4). However, it is not possible to distinguish the attenuated strain H37Ra from M. tuberculosis clinical isolates by use of these techniques. Nevertheless, H37Ra is used as a control in identification tests, and false diagnosis of tuberculosis due to cross-contamination of laboratory specimens with H37Ra is both likely and frequent (3, 16). To date, reliable differentiation of H37Ra from virulent strains has been possible only through spoligotyping and IS6110 genotyping (1), laborious and time-consuming procedures.
Cytochemical reactions in tubercle bacilli were studied in great depth in the 1940s and 1950s in order to find phenotypic markers correlated to virulence. The action of many basic dyes, such as Nile blue, methylene blue, neutral red, and methyl violet, on different Mycobacterium strains has been studied (5, 6, 7, 8). The most successful results were obtained with neutral red and Nile blue (6, 8). Dubos and Middlebrook reported that H37Rv, together with 22 M. tuberculosis and Mycobacterium bovis isolates, fixed neutral red in an alkaline aqueous environment and became red in color, whereas H37Ra cells did not (8). Two subsequent studies in which larger numbers of M. tuberculosis clinical isolates were used (about 200 strains in each) corroborated these findings (11, 15). At the same time, Desbordes and coworkers concluded that, unlike attenuated and avirulent strains, the virulent strain H37Rv strongly fixed basic dyes, such as Nile blue and neutral red, in their anionic forms (red and blue, respectively) in very alkaline environments.
In this work, we present an easy test for distinguishing H37Ra from H37Rv and from clinical isolates, developed by modifying previous cytochemical techniques and based on the variant behaviors of H37Ra, H37Rv, and clinical isolates when stained with neutral red or Nile blue.
M. tuberculosis strains, culture media, and growth conditions.
Twenty-eight M. tuberculosis strains, including recent clinical isolates (Laboratory of Clinical Microbiology, Hospital “de la Santa Creu i Sant Pau,” and Microbiology Service, “Germans Trias i Pujol” University Hospital, Barcelona, Spain), and the M. tuberculosis reference strains H37Rv (ATCC 27294) and H37Ra (ATCC 25177), both dissociated from H37 in 1934, were used in this study. M. tuberculosis clinical isolates were identified as M. tuberculosis complex by the AccuProbe assay (Gen-Probe Inc., San Diego, Calif.) (20) and as M. tuberculosis species by standard biochemical procedures (13, 21, 22). For cytochemical staining, colonies were grown on plates of Middlebrook 7H10 Bacto-agar enriched with oleic acid-albumin-dextrose-catalase (Difco Laboratories, Detroit, Mich.) at 37°C for 4 weeks.
Cytochemical staining of M. tuberculosis cells.
H37Ra was distinguished from H37Rv and clinical isolates inside borosilicate glass screw-cap tubes by means of two cytochemical tests with two different basic dyes, neutral red and Nile blue. Neutral red staining was carried out as follows. A few colonies of mycobacteria were placed inside a screw-cap tube containing 5 ml of 50% aqueous methanol, mixed, and incubated for 1 h at 37°C. The fluid was then removed, and cells were washed again, as above, with aqueous methanol. Finally, 5 ml of 0.002% neutral red (Merck AG, Darmstadt, Germany) in barbital buffer (1% sodium barbital in 5% NaCl [pH 9.8]) was added and mixed with the washed cells, and the tubes were kept at room temperature. It is not necessary to centrifuge in order to eliminate the washing fluids, but it is important in each step to mix the cells well with the washing and staining solutions. Deionized water was used throughout the entire process.
Results were read after 1 h, and a definitive reading was performed after 24 h. The reading may be done either in daylight or in artificial light.
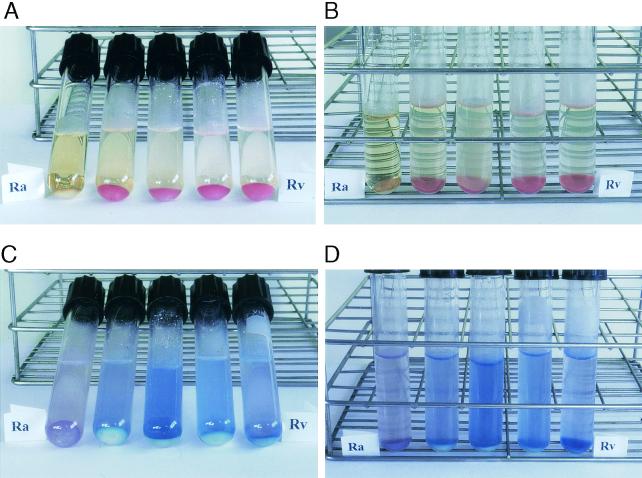
Neutral red became yellow when added to barbital buffer, pH 9.8. However, the H37Rv cells and all 28 clinical isolates immediately took on a red coloration with the staining solution, while the H37Ra cells were not colored (Fig. 1A and B). No appreciable differences in the intensity of red coloration were observed among the clinical isolates. After the staining test was completed, H37Rv and clinical-isolate cells retained their coloration for several days. Special care must be taken to eliminate the maximum amount of washing solution after the second wash, because it is slightly acidic, and if it is not efficiently removed, the H37Ra cells can take on a pink coloration. After 24 h this pink coloration disappears completely. We therefore recommend that the definitive reading of results be done after 24 h. Another way of avoiding false positives due to the acidity of the washing solution is to carry out an additional wash with barbital buffer and without neutral red before adding the staining solution,. We studied the stability of the neutral red reaction over a 2-year period; during this time, the strains used in this work were passed in culture medium several times and retested every 4 weeks. H37Rv has proved to be very stable for this characteristic, and we observed no loss in the intensity of staining throughout the 2 years that the study lasted. Likewise, H37Ra showed itself consistently unable to take on the dye over the same period. However, in six clinical strains, after 7 or 8 passages, we observed a decrease in the intensity of neutral red fixation compared with that in H37Rv and the other clinical isolates. The cells of the strains that lost intensity became more pink than red when stained with neutral red and kept this coloration until the end of the study. In the remaining clinical isolates, no loss of intensity in the staining was observed. Therefore, this technique cannot be used on clinical isolates kept for long periods in the laboratory through several passages in culture medium. We recommend using H37Ra as the negative control and H37Rv or a recently tested clinical isolate as the positive control when testing clinical isolates by this technique.
FIG. 1.
Cytochemical staining of M. tuberculosis H37Ra (Ra), H37Rv (Rv), and three clinical isolates (the three tubes between the tubes marked “Ra” and “Rv”). M. tuberculosis cells were harvested and stained either with 0.002% neutral red (A and B) or with 0.0006% Nile blue (C and D) in barbital buffer (see the text for details).
For Nile blue (Merck AG), cells were washed twice with 50% aqueous methanol, as described above for neutral red staining, and then washed again with 5 ml of water. Finally, 5 ml of barbital buffer (1% sodium barbital in 5% KCl [pH 11.5]) was added to the cells, and aqueous Nile blue was then added to each tube to reach a final concentration of 0.0006%. Tubes were kept at room temperature. Deionized water was used throughout the entire process. As with neutral red staining, it is not necessary to centrifuge in order to eliminate the washing fluids, but it is important in each step to mix the cells well with the washing and staining solutions. Reading of results was performed after 1 h and can be carried out either in daylight or in artificial light.
When Nile blue was added to the very alkaline barbital buffer (pH 11.5), the staining solution took on a pink coloration. Subsequently, the staining solution became blue within a few minutes of reacting with H37Rv or clinical-isolate cells, while the solution reacting with H37Ra cells retained the pink coloration (Fig. 1C and D). No differences in the intensity of the reaction were observed among the clinical isolates. Unlike the neutral red staining, Nile blue did not stain the cells completely, and differences between positive and negative strains were better observed in the staining solution.
The six strains that after several passages showed a lower capacity to fix neutral red were the same ones that also turned the Nile blue staining solution to a weaker blue color. The other clinical strains, H37Ra, and H37Rv were stable to Nile blue staining over the 2-year period, as they were to neutral red staining.
The two stainings are equally clear and reproducible. To the naked eye the difference between the colors is very clear in both stainings, but when a photograph is taken, the difference between pink and blue is more difficult to capture correctly (Fig. 1). However, if we had to choose between the two stains, perhaps we would choose neutral red, because the cells become completely stained quite quickly and the difference in color between a positive and a negative strain lasts for several days. On the other hand, when Nile blue is used, the cells become only slightly stained, and the color difference between a positive and a negative isolate is most apparent in the supernatant during the first hour. After the first hour, the cells take on a more intense coloring but the staining loses its discriminative power.
Our results agree with those obtained 50 years ago by several authors (5-8, 11, 15): H37Rv and M. tuberculosis clinical isolates are able to absorb basic dyes (neutral red and Nile blue) in their anionic forms (red and blue, respectively), whereas H37Ra is not.
We have made two contributions to these cytochemical staining techniques: first, by developing reproducible protocols and second, by rescuing them from oblivion. The first step in our study was to reproduce the previously reported protocols for neutral red and Nile blue staining (5-8). When these procedures were carried out, we encountered certain technical problems and made a series of observations. First, we noted that H37Ra differentiation by neutral red staining was not favorable at a pH of <9. At such pH values, the H37Ra strain fixed the dye and the cells took on a pale pink color. When staining was carried out at a pH of >9, H37Rv and clinical-isolate cells took on a purple-red coloration, whereas H37Ra cells became yellow, like the staining solution. We tried different concentrations of neutral red and we found that 0.002% was the most discriminative. On the other hand, when reproducing the neutral red staining reaction described by Desbordes et al. (7), we observed that the staining solution, made in saturated Na2CO3 (27.5 g of Na2CO3 in 96 ml of water) (10), was unstable. A colored precipitate was obtained a few minutes after the addition of neutral red. However, the neutral red staining solution in barbital buffer remained stable during the staining procedure. With the Nile blue test, the same problem occurred: when Desbordes' reaction was applied, a precipitate was formed in the staining solution when the dye was added to saturated Na2CO3. Dye precipitation was prevented by using a different alkaline environment (1% sodium barbital in 5% KCl adjusted at pH 11.5) and a very low dye concentration (0.0006%). pH was a critical parameter for optimal Nile blue staining results; the best differences in stain coloration were obtained at pH 11.5. With the modifications that we have introduced, both in the neutral red and in the Nile blue staining, there were no problems of precipitation, and clear differentiation of colors was observed both between H37Ra and H37Rv and between H37Ra and the clinical isolates.
We would encourage clinical laboratories currently using H37Rv or other M. tuberculosis strains as controls for identification of M. tuberculosis isolates to use H37Ra, since the techniques described here will allow them to carry out rapid screening and detect possible cross-contamination. Naturally, any clinical isolate giving negative results with neutral red or Nile blue staining would have to undergo spoligotyping or IS6110 genotyping in order to confirm the cross-contamination.
Another aspect to bear in mind is that the variant H37Ra was obtained from strain H37 in 1934 and then distributed to different laboratories throughout the world. During these 68 years, many variants may have been generated. Each laboratory will, therefore, have to acquire a strain of H37Ra which responds correctly to the cytochemical tests described.
One study correlated neutral red staining with the sulfolipid content in virulent M. tuberculosis strains (14). This correlation was doubtful, since some virulent strains were devoid of sulfolipids but were able to fix neutral red (14). Phospholipids and other acidic lipids have been proposed as additional possible structural components responsible for neutral red staining in strains lacking sulfolipids (9, 14). No clear explanation of this cytochemical staining has been obtained, and the component or components responsible for neutral red staining of virulent M. tuberculosis strains are not known with any certainty. Nile blue probably reacts to the same components as neutral red in the cell wall of M. tuberculosis, but no study has been published on this subject.
The technique described here would be useful in searching for the component that fixes neutral red and Nile blue. On the other hand, many studies are concerned with identifying the genomic differences between the virulent H37Rv strain and its attenuated variant H37Ra (2, 17-19). Furthermore, it is also common to study the differences between the abilities of virulent and attenuated (H37Rv and H37Ra) M. tuberculosis strains to invade and survive within cells of the immune system (12, 23). Many laboratories, therefore, work with these two variants, dissociated from the original M. tuberculosis H37 strain. In such laboratories, the possible exchange between these two strains requires evaluation. Thus, the application of the techniques described in this work could also be a very useful tool for control in mycobacterial research laboratories. Finally, this method of differentiating H37Ra from H37Rv and from clinical isolates is easy, inexpensive, reliable, and currently capable of being carried out in any laboratory.
Acknowledgments
This work was supported by EEC grant QLK2-CT-1999-01093 and by ML-98 grant from the Sociedad Española de Enfermedades Infecciosas y Microbiología Clínica.
REFERENCES
- 1.Bifani, P., S. Moghazeh, B. Shopsin, J. Driscoll, A. Ravikovitch, and B. N. Kreiswirth. 2000. Molecular characterization of Mycobacterium tuberculosis H37Rv/Ra variants: distinguishing the mycobacterial laboratory strain. J. Clin. Microbiol. 38:3200-3204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brosch, R., S. V. Gordon, A. Billault, T. Garnier, K. Eiglmeier, C. Soravito, B. G. Barrel, and S. T. Cole. 1998. Use of a Mycobacterium tuberculosis H37Rv bacterial artificial chromosome library for genome mapping, sequencing, and comparative genomics. Infect. Immun. 66:2221-2229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Burman, W. J., B. L. Stone, R. R. Reves, M. L. Wilson, Z. Yang, H. El-Hajj, J. H. Bates, and M. D. Cave. 1997. The incidence of false-positive cultures for Mycobacterium tuberculosis. Am. J. Respir. Crit. Care Med. 155:321-326. [DOI] [PubMed] [Google Scholar]
- 4.Butler, W. R., and L. S. Guthertz. 2001. Mycolic acid analysis by high-performance liquid chromatography for identification of Mycobacterium species. Clin. Microbiol. Rev. 14:704-726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Desbordes, J. 1952. Mise en évidence in vitro de la “nonvirulence” de germes a. a. r. par une réaction au bleu de nil. Ann. Inst. Pasteur 83:809-810. [Google Scholar]
- 6.Desbordes, J., and E. Fournier. 1954. Action des substances colorantes sur les mycobactéries. I. Colorants basiques. Étude de la cinétique de la réaction. Ann. Inst. Pasteur 86:657-660. [PubMed] [Google Scholar]
- 7.Desbordes, J., E. Fournier, and D. Alix. 1955. Action des substances colorantes sur les mycobactéries. II. Colorants basiques. Influence du pH. Ann. Inst. Pasteur 88:120-124. [PubMed] [Google Scholar]
- 8.Dubos, R. J., and G. Middlebrook. 1948. Cytochemical reaction of virulent tubercle bacilli. Am. Rev. Tuberc. 58:698-699. [DOI] [PubMed] [Google Scholar]
- 9.Goren, M. B., O. Brokl, and W. B. Schaefer. 1974. Lipids of putative relevance to virulence in Mycobacterium tuberculosis: correlation of virulence with elaboration of sulfatides and strongly acidic lipids. Infect. Immun. 9:142-149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gurr, E. 1962. Section 3, p. 471-574. In Leonard Hill (ed.), Staining animal tissues. Practical and theoretical. J. W. Arrowsmith, Bristol, United Kingdom.
- 11.Hughes, D. E., E. S. Moss, P. D. Maigon Hood, and M. Henson. 1954. Virulence of Mycobacterium tuberculosis. Evaluation of a test, using neutral red indicator. Am. J. Clin. Pathol. 24:621-625. [DOI] [PubMed] [Google Scholar]
- 12.Keane, J., H. G. Remold, and H. Kornfeld. 2000. Virulent Mycobacterium tuberculosis strains evade apoptosis of infected alveolar macrophages. J. Immunol. 164:2016-2020. [DOI] [PubMed] [Google Scholar]
- 13.Kent, P. T., and G. P. Kubica. 1995. Public health mycobacteriology: a guide for the level III laboratory. U.S. Department of Health and Human Services publication (CDC) 86-8230. Centers for Disease Control, Atlanta, Ga.
- 14.Middlebrook, G., C. M. Coleman, and W. B. Schaefer. 1959. Sulfolipid from virulent tubercle bacilli. Proc. Nat. Acad. Sci. USA 45:1801-1804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Morse, W. C., M. C. Dail, and I. Olitzky. 1953. A study of the neutral red reaction for determining the virulence of mycobacteria. Am. J. Public Health 43:36-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nivin, B., P. I. Fujiwara, J. Hannifin, and B. N. Kreiswirth. 1998. Cross-contamination with Mycobacterium tuberculosis: an epidemiological and laboratory investigation. Infect. Control Hosp. Epidemiol. 19:500-503. [DOI] [PubMed] [Google Scholar]
- 17.Pascopella, L., F. M. Collins, J. M. Martin, M. H. Lee, G. F. Hatfull, C. K. Stover, B. R. Bloom, and W. R. Jacobs. 1994. Use of in vivo complementation in Mycobacterium tuberculosis to identify a genomic fragment associated with virulence. Infect. Immun. 62:1313-1319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rindi, L., N. Lari, and C. Garzelli. 1999. Search for genes potentially involved in Mycobacterium tuberculosis virulence by mRNA differential display. Biochem. Biophys. Res. Commun. 258:94-101. [DOI] [PubMed] [Google Scholar]
- 19.Rivera-Marrero, C. A., M. A. Burroughs, R. A. Masse, F. O. Vannberg, D. L. Leimbach, J. Roman, and J. J. Murtagh, Jr. 1998. Identification of genes differentially expressed in Mycobacterium tuberculosis by differential display PCR. Microb. Pathog. 25:307-316. [DOI] [PubMed] [Google Scholar]
- 20.Stockman, L. 1992. DNA probes for the identification of mycobacteria, p.3.15.1-3.15.4. In H. D. Isenberg (ed.), Clinical microbiology procedures handbook. American Society for Microbiology, Washington, D.C.
- 21.Tsukamura, M. 1967. Identification of Mycobacteria. Tubercle 48:311-338. [DOI] [PubMed] [Google Scholar]
- 22.Vestal, A. 1975. Procedures for the isolation and identification of mycobacteria. U.S. Public Health Service publication 65-90. Centers for Disease Control, Atlanta, Ga.
- 23.Zhang, M., J. Gong, Y. Lin, and P. Barnes. 1998. Growth of virulent and avirulent Mycobacterium tuberculosis strains in human macrophages. Infect. Immun. 66:794-799. [DOI] [PMC free article] [PubMed] [Google Scholar]