Abstract
We compared the performance of two PCR assays, an IS6110-based in-house protocol and the COBAS AMPLICOR MTB PCR (COBAS MTB) system, for the detection of Mycobacterium tuberculosis complex in 43 human lymph node samples from 40 patients. For the in-house PCR and the COBAS MTB assays, respectively, sensitivities were 87.5% versus 45.5% (P < 0.05), specificities were 100.0% versus 91.3% (P > 0.05), and inhibition rates were 4.8% versus 19.5% (P < 0.05). For the COBAS MTB system, additional N-acetyl-l-cysteine-NaOH pretreatment of the samples changed neither the inhibition rate nor the sensitivity significantly.
In Germany, tuberculous lymphadenitis is the most common form of extrapulmonary tuberculosis, comprising about 7 to 8% of all cases of tuberculosis (19). Rapid and accurate diagnosis is important for the effective treatment of lymph node tuberculosis. Conventional methods for detecting Mycobacterium tuberculosis complex include acid-fast staining and culture. The sensitivity of microscopy in lymph node samples of human immunodeficiency virus noninfected patients is low because of the low numbers of mycobacteria present. Furthermore, M. tuberculosis complex cannot be differentiated from mycobacteria other than tuberculosis (MOTT) by acid-fast staining. Culturing methods, including those in solid and liquid media, detect M. tuberculosis complex with reasonable sensitivity. They are a prerequisite for accurate identification to species level and susceptibility testing. However, culture requires several weeks for completion due to the slow growth of mycobacteria. Nucleic acid amplification assays have been developed for detecting M. tuberculosis complex directly from clinical specimens. However, commercially available automated methods like the COBAS AMPLICOR MTB PCR (COBAS MTB) assay (Roche Diagnostics) have been approved for pulmonary specimens only. Several authors have reported acceptable sensitivity and specificity rates for extrapulmonary specimens by the COBAS MTB assay (8, 18), but so far, lymph node samples have not been specifically examined.
The purpose of this study was to evaluate the performance of the COBAS MTB assay for the detection of M. tuberculosis complex in lymph node samples and to compare it with an IS6110 in-house PCR assay. The results of acid-fast smear and culture and the clinical data of the patients were used for reference.
Patients and samples.
From January 1998 to December 1999, a total of 43 lymph node samples were surgically removed from 40 patients with possible lymph node tuberculosis. The specimens were sent in sterile tubes containing saline to the Hygiene Institute of the University of Heidelberg for the detection of mycobacteria. The patients were 18 females and 22 males with a median age of 25 years (range, 1 to 91 years).
Processing of lymph node specimens.
The specimens were carefully cut into 1- to 2-mm3 pieces in sterile Petri dishes with sterile scalpels before being divided into three portions of equal size. The three aliquots were homogenized in 1 ml of sterile saline each. (i) A sterile glass homogenizer was used for processing aliquot 1 for microscopy and culture. (ii) Plastic homogenizers (Destroy sticks; Biozym, Hessisch Oldendorf, Germany) in 1.5-ml tubes were used for processing aliquots 2 and 3 for the PCR assays. (iii) After homogenization, aliquot 3 was processed by pretreatment with N-acetyl-l-cysteine-NaOH (NALC [12]) and kept at −20°C for a backup PCR assay in case of inhibition of the first assay and for evaluation of NALC effectiveness.
Microscopy.
Heat-fixed smears were stained with auramine fluorochrome as a screening method for acid-fast bacteria (AFB). Positive slides of AFB were confirmed to be positive by Kinyoun staining (12).
Culture.
Half of the aliquot 1 processed (500 μl) was used for liquid culture in an MGIT 960 incubator (Becton Dickinson, Heidelberg, Germany). Vials were incubated at 37°C for up to 8 weeks, and growth was measured automatically by detection of fluorescence. In addition, two solid media, (i) Löwenstein-Jensen medium supplemented with glycerol and PACT (polymyxin B, amphotericin B, carbenicillin, trimethoprim) and (ii) Stonebrink medium supplemented with PACT, were inoculated with 200 μl each of aliquot 1. The slant media were incubated at 37°C for up to 8 weeks and inspected for growth twice a week for the first 2 weeks and weekly thereafter. All isolates were identified by acid-fast staining and IS6110 PCR assay (9), M. tuberculosis complex strains were identified by standard biochemical tests (15), and MOTT were identified by restriction enzyme analysis of the 65-kDa heat shock protein gene (22).
PCR.
Precautions to prevent PCR assay contamination, including the use of separate rooms for PCR setup and product detection, disposable plastic ware supplies, aliquoted reagents, and aerosol-resistant pipette tips, were observed (13).
In-house PCR.
Five hundred microliters of aliquot 2 was centrifuged at 14,000 × g for 10 min. The supernatant was discarded, and 50 μl of 1 N NaOH and 50 μl of 2% sodium dodecyl sulfate were added to the pellet. The mixture was thoroughly mixed, and alkaline lysis was performed by boiling for 5 min in a water bath. After neutralization with 50 μl of 1 N HCl and 50 μl of 1 M Tris (pH 8.0), the samples were processed by a purification and concentration procedure for DNA with the GeneClean II kit (Bio 101, La Jolla, Calif.) according to the recommendations of the manufacturer. The DNA preparation was finally dissolved in 25 μl of sterile water. One tube containing 500 μl of sterile saline as a negative sample control was processed the same way as the samples each time a set of sample preparations was processed.
For M. tuberculosis complex-specific amplification, primers IS1 and IS2 were used to amplify a 123-bp segment of IS6110 (9). PCR amplifications were carried out in 100-μl volumes with 10 μl of prepared sample added. The mixture contained 0.5 μM concentrations of each primer (TIB Molbiol, Berlin, Germany); 200 μM each of dATP, dCTP, dTTP, and dGTP; 2 mM MgCl2; 50 mM KCl; 10 mM Tris HCl (pH 9); and 1 U of Thermoprime Plus (ABGene, Hamburg, Germany) DNA polymerase. PCR runs were performed in a thermocycler (PerkinElmer model 9600) employing a protocol with an initial denaturation step for 2 min at 95°C; 35 cycles each for 0.5 min at 95°C, 1 min at 68°C, and 1 min at 72°C; and a final extension segment for 3 min at 72°C. The products were refrigerated until used. Controls for each PCR run included (i) a positive control, 100 fg of M. tuberculosis H37Rv DNA; (ii) two negative controls, one fully processed DNA-free sample preparation and one DNA-free PCR mixture (90 μl of reaction mixture plus 10 μl of sterile water instead of samples); and (iii) one inhibition control for each patient sample, which consisted of a second PCR tube containing a 10-μl sample DNA preparation spiked with 1 pg of M. tuberculosis H37Rv DNA. When the inhibition control failed to be positive, the PCR assay was repeated with NALC-treated aliquot 3.
PCR products were subjected to electrophoresis in 8% polyacrylamide gels with 1× Tris-borate-EDTA as running buffer, visualized by ethidium bromide staining and UV light detection, and documented with an MP4+ Polaroid camera.
The amplified M. tuberculosis complex DNA was hybridized and detected by the GEN-ETI-K (BykDiaSorin, Düsseldorf, Germany) DNA enzyme immunoassay (DEIA [14]) according to the instructions of the manufacturer.
Briefly, 100 μl of the biotinylated oligonucleotide probe IS3 (50 ng/ml) was added to each well of a streptavidin-coated microtiter plate and incubated at 4°C for 18 h. The wells were washed five times with washing buffer, and 100 μl of hybridization buffer was added to each well. Amplicons were denatured at 95°C for 15 min and placed on ice immediately. Twenty microliters of each sample was added to the wells, and hybridization was performed at 55°C for 60 min. After washing, 100 μl of anti-double-stranded DNA antibody was pipetted into each well, followed by incubation at room temperature for 30 min. After washing, 100 μl of the enzyme tracer (protein A conjugated with horseradish peroxidase) was added and kept at room temperature for 30 min. All wells were washed, and 100 μl of chromogen substrate was added. The colorimetric reaction was allowed to develop for 30 min in the dark at room temperature before being stopped with 200 μl of 1 N sulfuric acid. The absorbance at 450 and 630 nm was determined with a microtiter plate reader. The optical density was calculated after subtraction of A630 from A450. The DEIA cutoff value was defined as 0.15 optical density units above the mean value of the negative controls.
COBAS MTB assay.
The COBAS MTB assay was performed with 100 μl of homogenized aliquot 2 according to the instructions of the manufacturer for respiratory tract samples without modifications. This has been previously described in detail (11). If there was repeated inhibition of the processed aliquot 2 in the COBAS MTB assay, aliquot 3 of the NALC-treated lymph node was tested.
Sensitivity levels of the PCR assays.
Serial 10-fold dilutions of the M. tuberculosis H37Rv strain were prepared in sterile saline, and 0.1-ml aliquots were plated onto Löwenstein-Jensen agar. Plates were incubated for 4 weeks at 37°C before colonies were counted. Serial dilutions ranging from 106 to 10−1 CFU per ml were used for the PCR assays. The samples were pretreated and tested in the same way as the patient specimens.
Analysis of results.
Culture was considered the gold standard for proven lymph node tuberculosis. Differences between sensitivities, specificities, and inhibition rates were compared by using Fisher's exact test. A P value of <0.05 indicated statistical significance.
Patient groups and samples.
Fourteen patients with a total of 17 samples suffered from lymph node tuberculosis as proven by at least one positive culture (Table 1, patients 1 through 14). One patient suffered from lymphadenitis caused by M. genavense (Table 1, patient 15). Twenty-five patients had no evidence of mycobacterial lymphadenopathy (data not shown).
TABLE 1.
Performance of laboratory tests in lymph node samples from patients with culturally proven lymphadenitis due to mycobacteria
| Sample no. | Patient no. | Age (yr) | Sexa | Results of tests in lymph node samplesb
|
||||
|---|---|---|---|---|---|---|---|---|
| Microscopy (AFB) | Culture grown | Assay
|
||||||
| In-house PCR | COBAS AMPLICOR MTB
|
|||||||
| Without NALC | With NALC | |||||||
| 1 | 1c | 40 | M | ++++ | M. tuberculosis | + | Inhibition | + |
| 2 | 2 | 43 | F | + | M. tuberculosis | + | − | − |
| 3 | 3 | 9 | F | − | M. tuberculosis | + | Inhibition | − |
| 4 | 4 | 20 | M | − | M. tuberculosis | + | + | ND |
| 5 | 5 | 27 | F | − | M. tuberculosis | + | Inhibition | Inhibition |
| 6 | 6 | 28 | M | − | M. tuberculosis | + | + | − |
| 7 | 7 | 38 | F | − | M. tuberculosis | + | + | ND |
| 8d | 7 | 38 | F | − | M. tuberculosis | + | + | ND |
| 9 | 8 | 54 | F | − | M. tuberculosis | + | − | − |
| 10 | 9 | 61 | M | − | M. tuberculosis | + | ND | − |
| 11 | 10 | 69 | F | − | M. tuberculosis | + | − | − |
| 12 | 11 | 91 | M | − | M. tuberculosis | + | − | − |
| 13 | 12 | 22 | M | − | Negative | − | − | − |
| 14d | 12 | 22 | M | − | M. africanum | + | − | − |
| 15 | 13 | 87 | F | − | M. bovis | − | Inhibition | − |
| 16 | 14 | 23 | F | − | M. tuberculosis | ND | ND | ND |
| 17d | 14 | 23 | F | − | Negative | + | + | ND |
| 18 | 15 | 61 | M | +++ | M. genavense | − | − | ND |
F, female; M, male.
ND, not done.
Human immunodeficiency virus-infected patient.
Obtained from the same patient 10, 15, and 21 days after the first sample, respectively.
Microscopy and culture.
AFB were detected in 3 of the 18 samples from proven cases, including the case of atypical mycobacteriosis (sensitivity, 16.7%). The specificity was 100.0% for mycobacteria and 96.2% for tuberculosis.
Sixteen samples from 15 patients grew mycobacteria: 15 M. tuberculosis complex samples (13 M. tuberculosis, 1 M. africanum, 1 M. bovis) and 1 MOTT sample (M. genavense).
Characterization of the PCR assays.
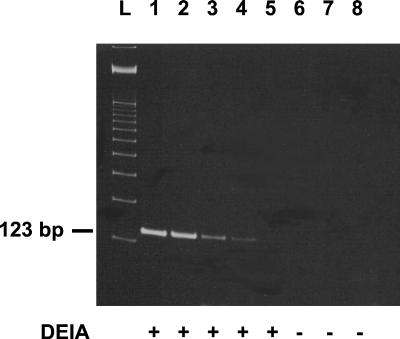
In each run, the positive and negative controls yielded the correct results. The in-house PCR assay detected 10 M. tuberculosis CFU per ml of saline (Fig. 1); the COBAS MTB assay detected 100 CFU per ml.
FIG. 1.
Sensitivity levels of the in-house PCR assay with detection of amplicons by using an ethidium bromide-stained polyacrylamide gel and DEIA. Lanes: L, 100-bp ladder; 1 through 7, respectively, 105 to 10−1 M. tuberculosis CFU per ml of saline; 8, negative control. +, DEIA positive; −, DEIA negative.
Rate of inhibition of the PCR assays and effectiveness of NALC for the removal of PCR assay inhibitors.
The in-house PCR assay was inhibited for 2 of 42 samples (4.8%). The inhibitory substances were not removable by NALC. The inhibition rate of the COBAS MTB assay was significantly higher (19.5%, 8 of 41 samples, P < 0.05). In a subgroup of 23 samples, in which sufficient volume was available for retesting in the COBAS MTB assay, NALC removed inhibitory substances in five of the eight primarily inhibited samples and one originally noninhibitory sample attained inhibitory qualities, resulting in a total inhibition rate of 21.7%. This did not differ significantly from the results of the assay without NALC added (21.7% versus 19.5%, P > 0.05). On the basis of the noninhibitory of the nontreated and NALC-treated samples, the COBAS MTB assay inhibition rate was reduced to 4 of 42 samples, although not significantly (9.5% versus 19.5%, P > 0.05).
PCR assays of patient lymph node samples.
The in-house PCR assay was positive in 14 of 16 samples of proven cases of tuberculosis and did not show a single false-positive result. The COBAS MTB assay was positive in 6 of 15 samples of proven cases of infection due to members of the M. tuberculosis complex and in two patients without evidence of mycobacterial disease. The sensitivities of the in-house PCR and COBAS MTB assays differed significantly (87.5% versus 45.5 and 40.0% for nontreated and the best of the nontreated and NALC-treated samples, respectively, P < 0.05). In addition, in the NALC subgroup, the sensitivity of the COBAS MTB assay was further reduced from 45.5 to 9.1% (1 of 11 samples), although not significantly (P > 0.05). The specificities of the in-house PCR assay and the COBAS MTB assay were similar (100.0% versus 91.3%, P > 0.05).
Detection of specific DNA fragments in tissue samples has been valuable for the diagnosis of nonculturable or fastidious organisms. For lymph node tuberculosis, several authors have reported on in-house PCR assays with sensitivities between 68 and 100%, and specificities between 86 and 100%, respectively (16, 21, 23). Furthermore, the commercially available manual AMPLICOR PCR assay has been tested for extrapulmonary specimens with sensitivities ranging from 33.3% for smear-negative specimens to 85.7% for all specimens and specificities ranging from 93.1 to 100% (2, 3, 5, 7, 20). Automated PCR assays have the great advantage of processing a large number of samples in shorter amount of hands-on time (11). The COBAS MTB assay showed good performance with respiratory tract samples (4, 8, 17, 18). Two authors also tested extrapulmonary specimens (8, 18) and reported sensitivities and specificities of 66.3 to 83.5% and 98.8 to 99.7%, respectively.
The presented in-house PCR assay is based on the amplification of a fragment of the IS6110, which is specific for the M. tuberculosis complex (9, 10). In our hands, it detected as few as 10 mycobacterial CFU per ml and showed both high sensitivity and specificity for the detection of M. tuberculosis complex DNA in human lymph node samples. Only one culturally positive sample with M. bovis tested negative in the in-house PCR assay. This could be due to the low copy number of IS6110 in M. bovis compared to that in M. tuberculosis (6).
We compared the performance of the in-house PCR assay with that of the automated COBAS MTB assay for the detection of lymph node tuberculosis. The COBAS MTB assay showed a lower sensitivity and, simultaneously, a higher inhibition rate. The COBAS MTB assay detected 100 M. tuberculosis CFU per ml. This value is similar to that given in a previous paper (24) in which the authors reported a detection limit of 42 CFU per ml. However, the sensitivity of the COBAS MTB assay was 10-fold lower than that for the in-house PCR assay. The inferior sensitivity could be due to the small sample volume of 100 μl used for the COBAS MTB assay, compared to the 500 μl used for the in-house PCR assay.
The higher inhibition rate associated with COBAS MTB measurements could be due to the different sample pretreatment. In the COBAS MTB assay, there is no DNA purification step. Therefore, potential inhibitory substances in the sample are not removed before adding the processed sample into the master mix. These inhibitory substances could originate from the blood present in lymph node samples. It has been shown that hemoglobin and lactoferrin inhibit PCR assays (1).
Other authors described similarly high inhibition rates with gastric aspirates (8) and other clinical samples (S. J. Kohlhepp, C. Gleaves, R. Casciato, C. Kviz, M. Campbell, J. Welle, and P. Metz, Abstr. 100th Gen. Meet. Am. Soc. Microbiol., abstr. C-167, 2000) and significantly reduced the inhibition rates by the Qiagen DNA purification procedure. We tested the potential benefit of NALC in lymph node samples.
When nontreated and NALC-treated samples were compared, there was no difference between the inhibition rates and an insignificant reduction in the sensitivity was observed. Therefore, general NALC cannot be recommended for lymph node samples being processed for the COBAS MTB assay. Furthermore, when the nontreated group was compared to the best group of nontreated and NALC-treated samples, the inhibition rate and the sensitivity were not significantly changed.
In conclusion, PCR assays proved to be suitable for detecting lymph node tuberculosis. The IS6110 assay was found to be more sensitive than the COBAS MTB assay. For the COBAS MTB assay, NALC pretreatment of the samples did not significantly reduce the high inhibition rate.
Acknowledgments
We thank H.-G. Sonntag, Director of the Hygiene Institute, Ruprecht-Karls-University, Heidelberg, Germany, for providing the laboratory equipment. We thank Sabine Zäuner and Heike Haag for excellent technical assistance with the PCR assays and A. Podbielski for critical reading of the manuscript.
S. Tyagi was supported by the IAESTE (International Association for Exchange of Students for Technical Experience) program of the German Academic Exchange Service (DAAD) and by the GNW (Gemeinnützige Gesellschaft zur Förderung von wissenschaftlichen Nachwuchskräften m.b.H., Heidelberg, Germany).
REFERENCES
- 1.Al-Soud, W. A., and P. Radstrom. 2001. Purification and characterization of PCR-inhibitory components in blood cells. J. Clin. Microbiol. 39:485-493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baek, C.-H., S.-I. Kim, Y.-H. Ko, and K.-C. Chu. 2000. Polymerase chain reaction detection of Mycobacterium tuberculosis from fine-needle aspirate for the diagnosis of cervical tuberculous lymphadenitis. Laryngoscope 110:30-34. [DOI] [PubMed] [Google Scholar]
- 3.Bemer-Melchior, P., P. Germaud, and H. B. Drugeon. 1998. Diagnosis of extrapulmonary tuberculosis by a commercial polymerase chain reaction kit. Pathol. Biol. 46:597-603. [PubMed] [Google Scholar]
- 4.Bodmer, T., A. Gurtner, M. Scholkmann, and L. Matter. 1997. Evaluation of the COBAS AMPLICOR MTB system. J. Clin. Microbiol. 35:1604-1605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brown, T. J., E. G. M. Power, and G. L. French. 1999. Evaluation of three commercial detection systems for Mycobacterium tuberculosis where clinical diagnosis is difficult. J. Clin. Pathol. 52:193-197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cave, M. D., K. D. Eisenach, P. F. McDermott, J. H. Bates, and J. T. Crawford. 1991. IS6110: Conservation of sequence in the Mycobacterium tuberculosis complex and its utilization in DNA fingerprinting. Mol. Cell. Probes 5:73-80. [DOI] [PubMed] [Google Scholar]
- 7.D'Amato, R. F., L. H. Hochstein, P. M. Colaninno, M. Scardamaglia, K. Kim, A. J. Mastellone, R. C. Patel, S. Alkhuja, V. J. Tevere, and A. Miller. 1996. Application of the Roche AMPLICOR Mycobacterium tuberculosis (PCR) test to specimens other than respiratory secretions. Diagn. Microbiol. Infect. Dis. 24:15-17. [DOI] [PubMed] [Google Scholar]
- 8.Eing, B. R., A. Becker, A. Sohns, and R. Ringelmann. 1998. Comparison of Roche Cobas Amplicor Mycobacterium tuberculosis assay with in-house PCR and culture for detection of M. tuberculosis. J. Clin. Microbiol. 36:2023-2029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Eisenach, K. D., M. D. Cave, J. H. Bates, and J. T. Crawford. 1990. Polymerase chain reaction amplification of a repetitive DNA sequence specific for Mycobacterium tuberculosis. J. Infect. Dis. 161:977-981. [DOI] [PubMed] [Google Scholar]
- 10.Hellyer, T. J., L. E. DesJardin, M. K. Assaf, J. H. Bates, M. D. Cave, and K. D. Eisenach. 1996. Specificity of IS6110-based amplification assays for Mycobacterium tuberculosis complex. J. Clin. Microbiol. 34:2843-2846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jungkind, D., S. DiRenzo, K. G. Beavis, and N. S. Silverman. 1996. Evaluation of automated COBAS AMPLICOR PCR system for detection of several infectious agents and its impact on laboratory management. J. Clin. Microbiol. 34:2778-2783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kent, P. T., and G. Kubica. 1985. Public health mycobacteriology. A guide for the level III laboratory. Centers for Disease Control, Atlanta, Ga.
- 13.Kwok, S., and R. Higuchi. 1989. Avoiding false positives with PCR. Nature 339:237-238. [DOI] [PubMed] [Google Scholar]
- 14.Mantero, G., A. Zonaro, A. Albertini, P. Bertolo, and D. Primi. 1991. DNA enzyme immunoassay: general method for detecting products of polymerase chain reaction. Clin. Chem. 37:422-429. [PubMed] [Google Scholar]
- 15.Metchock, B., F. S. Nolte, and R. J. Wallace, Jr. 1999. Mycobacterium, p. 399-437. In P. R. Murray, E. J. Baron, M. A. Pfaller, F. C. Tenover, and R. H. Yolken (ed.), Manual of clinical microbiology, 7th ed. ASM Press, Washington, D.C.
- 16.Portillo-Gomez, L., S. L. Morris, and A. Panduro. 2000. Rapid and efficient detection of extra-pulmonary Mycobacterium tuberculosis by PCR analysis. Int. J. Tuberc. Lung Dis. 4:361-370. [PubMed] [Google Scholar]
- 17.Rajalahti, I., P. Vuorinen, M. M. Nieminen, and A. Miettinen. 1998. Detection of Mycobacterium tuberculosis complex in sputum specimens by the automated Roche Cobas Amplicor Mycobacterium tuberculosis test. J. Clin. Microbiol. 36:975-978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Reischl, U., N. Lehn, H. Wolf, and L. Naumann. 1998. Clinical evaluation of the automated COBAS AMPLICOR MTB assay for testing respiratory and nonrespiratory specimens. J. Clin. Microbiol. 36:2853-2860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Robert Koch Institut. 2001. Zur Struktur der Tuberkulosemorbidität in Deutschland. Epidemiol. Bull. 11:78-80. [Google Scholar]
- 20.Shah, S., A. Miller, A. Mastellone, K. Kim, P. Colaninno, L. Hochstein, and R. D'Amato. 1998. Rapid diagnosis of tuberculosis in various biopsy and body fluid specimens by the AMPLICOR Mycobacterium tuberculosis polymerase chain reaction test. Chest 113:1190-1194. [DOI] [PubMed] [Google Scholar]
- 21.Singh, K. K., M. Muralidhar, A. Kumar, T. K. Chattopadhyaya, K. Kapila, M. K. Singh, S. K. Sharma, N. K. Jain, and J. S. Tyagi. 2000. Comparison of in house polymerase chain reaction with conventional techniques for the detection of Mycobacterium tuberculosis DNA in granulomatous lymphadenopathy. J. Clin. Pathol. 53:355-361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Telenti, A., F. Marchesi, M. Balz, F. Bally, E. C. Böttger, and T. Bodmer. 1993. Rapid identification of mycobacteria to the species level by polymerase chain reaction and restriction enzyme analysis. J. Clin. Microbiol. 31:175-178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Vitale, F., G. Capra, L. Maxia, S. Reale, G. Vesco, and S. Caracappa. 1998. Detection of Mycobacterium tuberculosis complex in cattle by PCR using milk, lymph node aspirates, and nasal swabs. J. Clin. Microbiol. 36:1050-1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yajko, D. M., C. Wagner, V. J. Tevere, T. Kocagöz, W. K. Hadley, and H. F. Chambers. 1995. Quantitative culture of Mycobacterium tuberculosis from clinical sputum specimens and dilution endpoint of its detection by the Amplicor PCR assay. J. Clin. Microbiol. 33:1944-1947. [DOI] [PMC free article] [PubMed] [Google Scholar]