Abstract
To examine the transmission of drug-resistant (DR) tuberculosis between Texas and Mexico, Mycobacterium tuberculosis isolates resistant to one or more of the first-line antimycobacterial drugs were obtained from 606 patients who resided in Texas and 313 patients who resided in Mexico, primarily within the state of Tamaulipas. The isolates were genotyped by IS6110-based restriction fragment length polymorphism (RFLP) analysis and spoligotyping. Of the 919 isolates genotyped, 413 (45%) grouped into 105 clusters containing 2 or more isolates with identical genotypes. In addition to having identical genotypes, identical drug resistance patterns were identified in 250 isolates in 78 clusters (DR clusters). Twenty DR clusters, containing isolates from 32% of the total number of patients infected with DR strains, were geographically distributed across Mexico and Texas. Within this population of 919 patients infected with DR isolates, the probability of being in a DR cluster was the same for residents of Mexico and Texas. In Texas, the significant independent predictors of clustering within DR clusters as opposed to genotype clusters were found to be race, age, country of birth, human immunodeficiency virus (HIV) infection status, and resistance to more than one drug. Specifically, isolates from African Americans, individuals under age 65, individuals born in the United States, and HIV-positive individuals were each more likely to be associated with a DR cluster. By contrast, no significant independent predictors of clustering in a DR cluster were identified in Mexico. Although some DR M. tuberculosis strains are geographically restricted, this study suggests that a number of strains are transmitted between Mexico and the United States.
Immigration is changing the epidemiology of tuberculosis (TB) in the United States. The number of TB cases in the population born in the United States has declined as the number of cases in the foreign-born population has increased. In 1986, 21.6% of the cases reported in the United States were among foreign-born individuals (14). In 1992, the proportion had risen to 61% (4). Mexico is the country of origin for the largest proportion (23%) of foreign-born U.S. patients (3, 14). In 1996, 83% of the cases of TB among foreign-born Hispanics in the United States were reported in the states bordering Mexico (23). Within the U.S. border state of Texas, the incidence of TB is higher in counties along the border than elsewhere in the state (8, 19).
The higher incidence of TB in Mexico compared with that in the United States, particularly within the northern Mexican border area, and the continuous, cyclical migration of Mexican nationals to and from the United States (23) suggest that strains of Mycobacterium tuberculosis may be transmitted across the border. However, differences in language, culture, and TB control programs make it difficult to conduct contact investigations that identify internationally related cases of TB (1, 17, 18). In a comparative study of the genotypes of M. tuberculosis isolates from 43 Mexican-born persons in a San Francisco population of 1,074 TB patients, Jasmer et al. (10) concluded that transmission from people born in Mexico leading to culture-positive TB in non-Mexican-born San Franciscans was uncommon. Those results do not, however, address ongoing transmission. DNA fingerprinting analysis was therefore done for 919 drug-resistant (DR) isolates collected between 1992 and 2000 from patients residing in Mexico and the U.S. state of Texas to characterize the extent of transmission of DR TB strains between Texas and Mexico.
MATERIALS AND METHODS
Patient consent.
The collection of epidemiological data used for this study was approved by the Institutional Review Boards of the Texas Department of Health (protocol 980025) and the University of Texas Health Science Center at San Antonio (protocol 001-6000-105).
Study populations. (i) Drug-resistant TB patients from Texas.
Demographic information, county of residence, and serologic evidence of human immunodeficiency virus (HIV) infection were obtained for patients in Texas reported to have TB from 1992 through 2000 by reviewing records at the Tuberculosis Elimination Division, Texas Department of Health. Patients infected with isolates found to be resistant to isoniazid (INH), rifampin (RIF), ethambutol (EMB), or streptomycin (STR) by culture were identified; and the isolates were obtained from the Bureau of Laboratories, Texas Department of Health. All isolates for which restriction fragment length polymorphism (RFLP) analysis was completed by 1 January 2001 were included in the study.
(ii) Drug-sensitive TB patients from Texas.
As part of their participation in the National Tuberculosis Genotyping and Surveillance Network, sponsored by the Centers for Disease Control and Prevention (CDC; Atlanta, Ga.), the University of Texas Health Science Center at San Antonio (UTHSCSA) and the Texas Department of Health (TDH) conducted a population-based study in two northern Texas counties bordering Mexico (Cameron and Hidalgo) and two urban northern Texas counties (Dallas and Tarrant). The UTHSCSA laboratory served as a reference genotyping laboratory for M. tuberculosis isolates collected from each TB patient residing in the four counties from 1996 through 2000. The RFLP fingerprint patterns of isolates were submitted to the CDC and assigned a National Fingerprint Pattern, and the patterns were compared with those for isolates obtained from other sites in United States (Arkansas, California, Massachusetts, Maryland, Michigan, and New Jersey) to examine the interstate transmission of M. tuberculosis strains. By 1 January 2001, the evaluations of isolates from 1,691 patients in the CDC surveillance areas had been completed and the isolates were included in this study. The isolates from an additional 393 TB patients residing in other areas of Texas were genotyped during the study period to assist local health departments in investigations of TB outbreaks or recurrences. These 393 isolates were also included in this study.
(iii) Drug-resistant TB patients from Tamaulipas.
TDH manages four binational projects in Texas-Mexico border cities. These cities include Matamoros, Reynosa, and Nuevo Laredo in the Mexican state of Tamaulipas and Cuidad Juarez in the Mexican state of Chihuahua. One aspect of these projects is to provide mycobacteriology testing of clinical specimens from TB patients managed by governmental and private medical providers in the four cities. This testing is performed at TDH. Laboratory records were reviewed to identify any binational patients infected with M. tuberculosis isolates resistant to INH, RIF, STR, or EMB.
Drug susceptibility testing.
Drug susceptibility testing of the M. tuberculosis isolates was performed with the rapid BACTEC 460TB radiometric system (Becton Dickinson, Paramus, N.J.). The anti-TB agents and the concentrations tested by the BACTEC 460TB system were as follows: INH, 0.4 μg/ml; RIF, 2.0 μg/ml; and EMB, 2.5 μg/ml. Resistance to any of the anti-TB agents detected by the BACTEC 460TB system was confirmed by the Middlebrook 7H10 agar proportion method (15). The anti-TB agents and concentrations tested by the agar proportion method were as follows: INH, 1.0 μg/ml; RIF, 1.0 μg/ml; EMB, 5.0 μg/ml; and STR, 2.0 μg/ml.
Genotyping.
IS6110-based RFLP analysis was performed by Southern blotting of PvuII-digested genomic DNA with a 523-bp right-handed IS6110 probe (20). Two sets of DNA molecular size markers were used for IS6110-based fingerprinting. The first marker set consisted of PvuII-restricted chromosomal DNA of M. tuberculosis H37Rv and two additional DNA fragments that hybridize to IS6110. The molecular sizes of the additional fragments were 13.5 and 690 kb. The second set consisted of standardized IS6110 molecular weight markers provided by Jack Crawford (CDC). Autoradiographs were developed with an enhanced chemiluminescence detection system (Amersham, Piscataway, N.J.). Images of the resulting autoradiographs were analyzed with a BioImage Whole Band Analyzer (version 3.4.2; Genomic Solutions, Ann Arbor, Mich.).
While IS6110-based RFLP analysis has been shown to be the most discriminatory genotyping method overall, for strains with less than six IS6110 bands, typing by a secondary typing method further distinguishes strain relationships (12). The genetic relatedness of the isolates was examined by spoligotype analysis (11). Locally synthesized and biotinylated PCR primers (Advanced Nucleic Acids Technology Core Facility, UTHSCSA) were used with commercially available spoligotyping membranes (Isogen, Maarssen, The Netherlands). Spoligotyping PCRs were done with 50-μl volumes containing 100 ng of template DNA, 50 pmol of each primer, and 0.4 U of Taq polymerase in (final concentrations) 0.2 mM deoxynucleoside triphosphates, 1× Taq polymerase buffer A, and 2.0 mM MgCl2 (Promega, Madison, Wis.). Alternatively, for those isolates from which DNA had not yet been isolated, small colonies were touched with a sterile loop, the isolates on the loop were suspended in 200 μl of sterile double-distilled H2O, the mixture was diluted 1:1 with 0.5% bovine serum albumin, and the isolates were placed in lid-locked microcentrifuge tubes and heat killed at 105°C for 5 min. Ten microliters of this suspension was then used in the PCR. The thermocycler (MJ Research, Watertown, Mass.) was programmed as follows: step 1, 96°C for 3 min; step 2, 96°C for 1 min; step 3, 55°C for 1 min; step 4, 72°C for 30 s; step 5, 30 cycles of steps 2 through 4; step 5, 72°C for 5 min; step 6, hold at 4°C. Hybridization was performed as described by Isogen. In addition to isolates having less than six IS6110 bands, spoligotype analysis was also done for all isolates in groups with two or more isolates with the same IS6110-based RFLP pattern. The resulting spoligotype patterns were given numerical designations that can be translated by using a nomenclature system with an established rationale (7). DNA from M. tuberculosis H37Ra and DNA from the M. bovis bacillus Calmette-Guérin Tice strain were used as reference controls. Negative controls consisted of one lane for a PCR performed in the absence of template and another lane loaded with buffer only to control for adequate stripping of the reusable membranes.
Cluster definitions.
Genotype analysis was done with isolates collected between 1992 and 2000 from 2,399 individuals. Nine hundred nineteen of the isolates were resistant to one of more of the following: INH, RIF, STR, and/or EMB. Since we were interested in the possibility of the direct transmission of a DR strain rather than the identification of multiple instances of acquired drug resistance after transmission of a drug-susceptible strain, we restricted the cluster definition to include two or more patients infected with isolates with identical IS6110-based RFLP patterns, identical spoligotype patterns, and matching drug resistance profiles. These clusters were designated DR clusters.
Statistical analysis.
Differences in proportions were tested by Fisher's exact test. Maximum-likelihood estimates of odds ratios (ORs) and 95% confidence intervals (CIs) were computed by logistic regression to analyze factors associated with clustering. Multiple logistic regression was used to determine factors independently associated with clustering by a stepwise approach. All tests were two-sided with a 0.05 significance level.
RESULTS
Demographics of the study population.
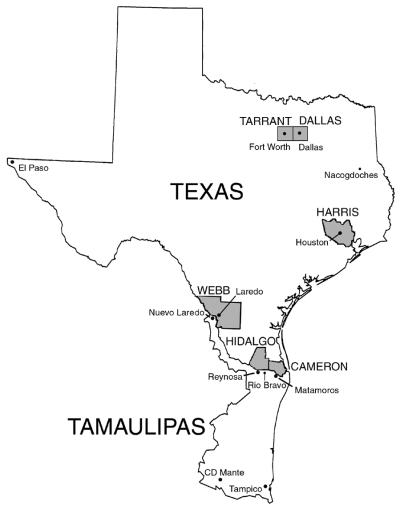
DNA fingerprinting analysis was done for a total of 919 DR M. tuberculosis isolates from 606 patients residing in Texas and 313 patients residing in Mexico (Table 1). Fifty-four percent of the Texan patients were foreign born, and of these, 57% were born in Mexico and 16% were from Vietnam. Hispanics predominated in the patient population, comprising 51% of the patients from Texas and 100% of the patients from Mexico. The remaining Texan patient population consisted of 19% African Americans, 17% Asians, and 12% whites. The majority of Texan patients resided in Harris (22%), Dallas (16%), Tarrant (10%), Hidalgo (9%), and Cameron (6%) Counties (Fig. 1). Of the patients from Mexico, the city of residence was known for 305 (97%) patients. Two hundred seventy-five (88%) of the Mexican patients resided in the state of Tamaulipas. Of the Tamaulipas residents, 25% resided in Reynosa, 28% resided in Matamoros, 16% resided in Nuevo Laredo, 10% resided in Tampico, 4% resided in Ciudad Mante, 4% resided in Rio Bravo, 3% resided in Ciudad Victoria, and 2% resided in San Fernando.
TABLE 1.
Demographic data for the study populationa
| Characteristic | All residents
|
Texas residents
|
Mexico residents
|
||||||
|---|---|---|---|---|---|---|---|---|---|
| All | Genotype cluster | DR cluster | All | Genotype cluster | DR cluster | All | Genotype cluster | DR cluster | |
| No. of patients | 919 | 413 | 250 | 606 | 256 | 171 | 313 | 157 | 79 |
| Sex | |||||||||
| Male (no.) | 639 | 297 | 189 | 422 | 181 | 130 | 217 | 116 | 59 |
| Female (no.) | 280 | 116 | 61 | 184 | 75 | 41 | 96 | 41 | 20 |
| Male:female | 2.3 | 2.6 | 3.1 | 2.3 | 2.4 | 3.2 | 2.3 | 2.8 | 3.0 |
| No. of individuals with the following: | |||||||||
| Ethnicity | |||||||||
| White | 73 | 33 | 24 | 73 | 33 | 24 | 0 | 0 | 0 |
| African American | 114 | 70 | 61 | 114 | 70 | 61 | 0 | 0 | 0 |
| Hispanic | 627 | 294 | 155 | 314 | 137 | 76 | 313 | 157 | 79 |
| Asian | 102 | 15 | 9 | 102 | 15 | 9 | 0 | 0 | 0 |
| Other or unknown | 3 | 0 | 0 | 3 | 1 | 1 | 0 | 0 | 0 |
| Age (yr) | |||||||||
| <15 | 17 | 6 | 5 | 11 | 4 | 4 | 6 | 2 | 1 |
| 15-24 | 108 | 49 | 26 | 69 | 30 | 17 | 39 | 19 | 9 |
| 25-44 | 430 | 207 | 130 | 292 | 135 | 95 | 138 | 72 | 35 |
| 45-60 | 216 | 91 | 66 | 131 | 52 | 41 | 85 | 39 | 25 |
| >60 | 145 | 58 | 22 | 103 | 35 | 14 | 42 | 23 | 8 |
| Unknown | 3 | 2 | 1 | 0 | 0 | 0 | 3 | 2 | 1 |
| Country of birth | |||||||||
| United States | 258 | 143 | 111 | 258 | 143 | 111 | 0 | 0 | 0 |
| Mexico | 500 | 240 | 122 | 187 | 83 | 43 | 313 | 157 | 79 |
| Vietnam | 52 | 14 | 8 | 52 | 14 | 8 | 0 | 0 | 0 |
| Other | 88 | 7 | 5 | 88 | 4 | 5 | 0 | 3 | 0 |
| Unknown | 21 | 9 | 4 | 21 | 9 | 4 | 0 | 0 | 0 |
| HIV infection status | |||||||||
| Positive | 89 | 57 | 46 | 88 | 57 | 46 | 1 | 0 | 0 |
| Negative | 469 | 173 | 104 | 427 | 154 | 95 | 42 | 19 | 9 |
| Unknown | 361 | 183 | 100 | 91 | 45 | 30 | 270 | 138 | 70 |
| No. of individuals infected with isolates resistant to: | |||||||||
| INH | 661 | 273 | 151 | 405 | 148 | 89 | 256 | 125 | 62 |
| INH only | 197 | 73 | 52 | 158 | 57 | 44 | 39 | 16 | 8 |
| RIF | 406 | 206 | 112 | 213 | 104 | 62 | 193 | 102 | 50 |
| RIF only | 93 | 58 | 42 | 78 | 49 | 39 | 15 | 9 | 3 |
| STM only | 129 | 62 | 47 | 101 | 48 | 37 | 28 | 14 | 10 |
| EMB only | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| One drug only | 419 | 193 | 141 | 337 | 154 | 120 | 82 | 39 | 21 |
| INH and RIF | 278 | 129 | 60 | 113 | 44 | 17 | 165 | 85 | 43 |
| INH, RIF, STM, and EMB | 93 | 47 | 25 | 34 | 17 | 7 | 59 | 30 | 18 |
Genotype clusters consist of two or more individuals whose isolates share identical IS6110-based RFLP and spoligotype patterns. DR clusters consist of two or more individuals whose isolates share identical IS6110-based RFLP and spoligotype patterns, as well as identical profiles of resistance to INH, RIF, STR, and/or EMB.
FIG. 1.
Map of the U.S. state of Texas and the Mexican state of Tamaulipas. The names of counties are in capital letters.
The age associations of patients from Mexico and Texas were virtually identical (Table 1). The mean age for patients residing in Texas and for patients residing in Mexico was 43 years, with a range of <1 to 97 years among the Texan patients and a range of <1 to 87 years among the Mexican patients. In both populations, the highest percentages of cases (48% in Texas and 44% in Mexico) were found among individuals in the age range of 25 to 44 years. Males outnumbered females by a greater than 2:1 ratio among both residents of Texas and residents of Mexico (Table 1).
Analysis of resistance to the first-line drugs INH, RIF, STR, and EMB showed that 74% of the isolates from residents of Mexico were resistant to two or more drugs, whereas 44% of the isolates from residents of Texas were resistant to two or more drugs (P < 0.0001). There was a threefold greater prevalence of multi-DR (MDR) M. tuberculosis isolates, defined as resistance to at least INH and RIF, among isolates from Mexico compared with that among isolates from Texas. Similar differences were also reflected in the proportion of isolates resistant to all four drugs among isolates from Mexico compared with that among isolates from Texas. Fifty-nine (19%) of the DR isolates from Mexico were resistant to all four drugs, whereas 34 (6%) of the DR isolates from Texas were resistant to all four drugs (P < 0.0001) (Table 1). It bears comment, however, that the incidence of resistance to more than one of the first-line drugs may have been overrepresented for the Mexican isolates, in that specimens for culture were not routinely collected from patients in the binational treatment programs; rather, patients who were not responding to drug therapy were primarily targeted for specimen collection. The latter criterion would likely, therefore, favor the inclusion of isolates resistant to more than one drug.
DR clusters.
Genotyping showed that 506 of the 919 patients were infected with unique DR isolates, in that they were genotypically different from any of the other isolates. One hundred ten patients were in clusters of 2 individuals whose isolates had identical IS6110-based RFLP and spoligotype patterns, and 303 patients were in clusters with more than 2 patients whose drug-resistant isolates had identical IS6110-based RFLP and spoligotype patterns. Two hundred fifty (27% of the overall DR population) of the 413 patients whose isolates were included in genotype clusters remained in the more narrowly defined DR cluster population; viz., they were infected with isolates that were identical by IS6110-based RFLP analysis and spoligotyping and according to their antimycobacterial drug resistance patterns (Table 2). Recognizing that the majority of reports in the literature include patient isolates with IS6110-based RFLP patterns that differ by the presence or the absence of a single band when secondary typing methods yield identical results, the effect of using this cluster definition on the size and the number of clusters was similarly determined. In addition, assuming that it is probable that a strain may continue to acquire resistance to additional drugs while being transmitted within a population, the effect of including isolates with a common core drug resistance pattern rather than identical drug resistance patterns on the proportion of clustered patients was also determined (Table 3).
TABLE 2.
Cluster identification, drug resistance, and geographic distributions of isolates
| IS6110-based RFLP analysis
|
No. of patientsa
|
Drug resistance | Residenceb
|
Present in adjoining counties | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Mexico
|
Texas
|
|||||||||||
| No. of bands | Patternc | Spoligotyped | DS | DR | DR match | Interior | Border | Border | TDCJ | Interior | ||
| 1 | 1.001 | 777777776413771 | 9 | 6 | 4 | INH, STM | X | Yes | ||||
| 2 | 2.001 | 617776777760601 | 5 | 6 | 2 | INH, RIFe | X | Yes | ||||
| 2 | 2.001 | 617776777760601 | 51 | 61 | 2 | INH | X | Yes | ||||
| 2 | 2.001 | 777776777760771 | 52 | 27 | 4 | INH, RIF, STM, EMB | X | X | X | |||
| 2 | 2.001 | 777776777760771 | 52 | 27 | 2 | INH, RIF, STM | X | X | ||||
| 2 | 2.001 | 777776777760771 | 52 | 27 | 3 | INH, STM | X | X | ||||
| 2 | 2.001 | 777776777760771 | 52 | 27 | 5 | INH | X | X | X | |||
| 2 | 2.001 | 777776777760771 | 52 | 27 | 4 | RIF, STM | X | Yes | ||||
| 2 | 2.001 | 777776777760771 | 52 | 27 | 3 | RIF | X | |||||
| 2 | 2.001 | 777776777760771 | 52 | 27 | 4 | STM | X | X | ||||
| 2 | 2.001 | 777776777760601 | 48 | 15 | 4 | RIF | X | |||||
| 2 | 2.001 | 777776777760601 | 48 | 15 | 9 | STM | X | |||||
| 2 | 2.006 | 400037777413771 | 51 | 6 | 2 | RIF | X | Yes | ||||
| 2 | 2.006 | 400037777413771 | 51 | 16 | 3 | STM | X | |||||
| 3 | 3.004 | 777776777760601 | 5 | 3 | 2 | INH, RIF | X | Yes | ||||
| 3 | 3.009 | 777776777760601 | 9 | 2 | 2 | RIF | X | |||||
| 3 | 3.010 | 777776777760601 | 0 | 7 | 2 | RIF, STM | X | Yes | ||||
| 3 | 3.010 | 777776777760601 | 01 | 7 | 5 | STM | X | |||||
| 3 | 3.026 | 777000377760771 | 2 | 2 | 2 | INH | X | X | ||||
| 4 | 4.001 | 777776777760771 | 22 | 12 | 3 | INH, RIF, STM, EMB | X | X | ||||
| 4 | 4.001 | 777776777760771 | 22 | 12 | 2 | INH, RIF, STM | X | X | ||||
| 4 | 4.001 | 777776777760771 | 22 | 12 | 2 | INH, STM | X | X | ||||
| 4 | 4.001 | 777776777760771 | 22 | 12 | 3 | INH | X | X | X | |||
| 4 | 4.002 | 777776777760771 | 60 | 29 | 4 | INH, RIF, STM, EMB | X | |||||
| 4 | 4.002 | 777776777760771 | 60 | 29 | 5 | INH, RIF | X | |||||
| 4 | 4.002 | 777776777760771 | 60 | 29 | 8 | INH, STM | X | X | X | |||
| 4 | 4.002 | 777776777760771 | 60 | 29 | 3 | INH | X | X | ||||
| 4 | 4.002 | 777776777760771 | 60 | 29 | 4 | STM | X | X | ||||
| 4 | 4.002 | 700076777760771 | 7 | 5 | 2 | INH, RIF | X | X | ||||
| 4 | 4.004 | 777776777760771 | 0 | 7 | 4 | INH, STM, EMB | X | X | Yes | |||
| 4 | 4.011 | 777776777760771 | 1 | 4 | 2 | RIF | X | Yes | ||||
| 6 | 6.003 | 777777557760771 | 0 | 4 | 2 | RIF, STM | X | Yes | ||||
| 7 | 7.040 | 777777760000611 | 0 | 4 | 2 | INH, STM | X | X | ||||
| 8 | 8.005 | 777777760000611 | 0 | 2 | 2 | STM | X | Yes | ||||
| 8 | 8.036 | 777777777760771 | 0 | 3 | 2 | INH, RIF | X | X | ||||
| 8 | 8.060 | 737777423560771 | 0 | 4 | 2 | INH, RIF, STM, EMB | X | Yes | ||||
| 9 | 9.001 | 000000004020771 | 2 | 8 | 3 | INH, RIF, STM | X | |||||
| 9 | 9.001 | 000000004020771 | 2 | 8 | 4 | INH, STM | X | X | X | |||
| 9 | 9.029 | 777777777760771 | 0 | 4 | 2 | INH, RIF, STM | X | Yes | ||||
| 9 | 9.031 | 777777644720771 | 0 | 5 | 5 | INH | X | Yes | ||||
| 9 | 9.081 | 777776770000000 | 0 | 2 | 2 | INH, RIF, STM, EMB | X | Yes | ||||
| 9 | 11.008 | 777777777760771 | 0 | 4 | 2 | STM | X | X | ||||
| 10 | 9.001 | 000000007720771 | 1 | 12 | 10 | INH, STM | X | X | X | |||
| 10 | 9.002 | 777777774020771 | 0 | 3 | 3 | RIF | X | X | ||||
| 10 | 10.003 | 777777607760771 | 2 | 4 | 3 | RIF | X | |||||
| 10 | 10.006 | 777777777760771 | 1 | 3 | 2 | INH, RIF, STM, EMB | X | X | ||||
| 10 | 10.012 | 000000000003771 | 56 | 7 | 5 | INH | X | X | ||||
| 10 | 10.141 | 777777774020731 | 0 | 2 | 2 | STM | X | Yes | ||||
| 10 | 11.008 | 777777777760771 | 4 | 2 | 2 | INH | X | X | ||||
| 11 | 11.114 | 777777774020771 | 0 | 7 | 7 | INH | X | Yes | ||||
| 12 | 12.024 | 777777607760771 | 1 | 5 | 3 | INH, RIF | X | Yes | ||||
| 12 | 12.048 | 376377777760771 | 0 | 2 | 2 | INH | X | |||||
| 12 | 12.092 | 657737607760771 | 2 | 2 | 2 | INH, RIF, STM, EMB | X | |||||
| 13 | 12.004 | 777777774020771 | 0 | 10 | 10 | STM | X | X | Yes | |||
| 13 | 13.002 | 777777777600341 | 0 | 6 | 2 | INH, RIF, EMB | X | X | ||||
| 13 | 13.011 | 777777607760771 | 8 | 11 | 2 | INH, RIF, STM, EMB | X | |||||
| 13 | 13.011 | 777777607760771 | 8 | 11 | 2 | INH, RIF | X | Yes | ||||
| 13 | 13.011 | 777777607760771 | 8 | 11 | 3 | INH | X | X | Yes | |||
| 13 | 13.082 | 017777607760771 | 0 | 2 | 2 | INH, RIF, STM | X | Yes | ||||
| 13 | 13.121 | 776377777760771 | 16 | 13 | 13 | RIF | X | X | Yes/PICK>{tt} | |||
| 14 | 13.024 | 776137607760771 | 0 | 4 | 2 | INH, RIF, STM, EMB | X | X | ||||
| 14 | 13.024 | 776137607760771 | 0 | 4 | 2 | INH, RIF, STM | X | X | ||||
| 14 | 13.030 | 777777607760771 | 0 | 5 | 2 | INH | X | X | ||||
| 14 | 14.041 | 777777607560771 | 0 | 3 | 2 | RIF | X | X | ||||
| 14 | 15.024 | 000000000003771 | 0 | 2 | 2 | INH | X | |||||
| 15 | 15.024 | 000000000003771 | 9 | 6 | 2 | INH, RIF | X | |||||
| 15 | 15.024 | 000000000003771 | 9 | 6 | 3 | INH | X | |||||
| 15 | 15.067 | 776137607760771 | 0 | 2 | 2 | STM | X | Yes | ||||
| 16 | 16.010 | 677777477413771 | 2 | 6 | 4 | RIF | X | |||||
| 17 | 17.003 | 776137607760771 | 0 | 2 | 2 | STM | X | Yes | ||||
| 17 | 17.013 | 777777777760031 | 0 | 2 | 2 | INH, RIF, STM, EMB | X | |||||
| 17 | 17.027 | 776137607760771 | 0 | 2 | 2 | INH, STM | X | X | ||||
| 18 | 18.010 | 000000000003771 | 0 | 2 | 2 | INH | X | |||||
| 20 | 21.001 | 000000000003771 | 14 | 7 | 2 | INH | X | X | ||||
| 20 | 21.001 | 000000000003771 | 14 | 7 | 4 | RIF | X | |||||
| 20 | 21.003 | 000000000003771 | 0 | 2 | 2 | RIF, STM | X | |||||
| 22 | 22.006 | 000000000003771 | 0 | 2 | 2 | INH | X | Yes | ||||
| 22 | 22.008 | 000000000003771 | 0 | 2 | 2 | STM | X | Yes | ||||
DS, patients infected with drug-susceptible isolates with the designated genotype; DR, patients infected with DR isolates with the designated genotype; DR match, patients infected with DR isolates with the designated genotype and with identical drug resistance results for INH, RIF, STR, and/or EMB.
Mexico interior, patients residing in Mexican jurisdictions not adjoining the U.S. border; Mexico border, patients residing in Mexican jurisdictions adjoining the U.S. border; Texas border, patients residing in Texas counties adjoining the Mexican border, Texas interior, patients residing in Texas counties not adjoining the Mexican border.
Pattern designations are based on the number of bands and the sequence in which the pattern was identified.
Spoligotype designations were determined by using the nomenclature of Dale et al. (7).
Boldface indicates MDR clusters.
TDCJ, Texas Department of Criminal Justice.
TABLE 3.
Definitions of clusteringa
| No. of clusters | No. of clusters (no. of patients) among patients infected with DR isolates (n = 919)a
|
|||
|---|---|---|---|---|
| Identical genotype (n = 413)
|
Plus or minus one IS6110 band (n = 444)
|
|||
| DR matchb | DR core | DR match | DR core | |
| 2 | 44 (88) | 54 (108) | 52 (104) | 46 (92) |
| > 2 | 34 (162) | 48 (301) | 43 (191) | 52 (341) |
| Total | 78 (250) | 102 (409) | 95 (295) | 98 (433) |
| % of DR patient isolates in clusters | 27 | 45 | 32 | 47 |
Identical genotype indicates identical numbers and pattern of bands by IS6110-based RFLP analysis and identical spoligotype patterns. Isolates with the same patterns of bands by IS6110-based RFLP analysis plus or minus a single band had the same spoligotype pattern. DR match, the profiles for resistance to INH, RIF, STR, and EMB were identical; DR core, all isolates in the cluster were resistant to at least one of the drugs to which the other isolates within the genotypically matched set of isolates were resistant.
The cluster definition used for statistical analyses.
Of note, the isolates from 129 (46%) of the 278 patients infected with MDR isolates were in genotype clusters. When grouped into clusters on the basis of their DR profiles, the isolates from 60 (22%) patients remained in the narrowly defined DR clusters. Of the other 614 DR isolates, 284 (46%) were in genotype clusters and 190 (31%) remained in the narrowly defined DR clusters. MDR isolates were significantly (P = 0.0001) less likely than other DR isolates to be in DR clusters.
Isolates from residents of Mexico were not significantly more or less associated with DR clusters than isolates from residents of Texas. In Texas, the significant independent predictors of clustering within DR clusters as opposed to genotype clusters were found to be race, age, country of birth, HIV infection status, and resistance to more than one drug. Specifically, isolates from African Americans, individuals under age 65, individuals born in the United States, and HIV-positive individuals were each more likely to be associated with a DR cluster. Country of origin was not found to be a significant independent predictor of clustering (Table 4).
TABLE 4.
Significant independent predictors of inclusion in clusters of two or more isolatesa
| Analysis subgroup | Characteristic | No. (%) of isolates
|
OR (adjusted CIb) | P valuec | |
|---|---|---|---|---|---|
| DR match clustered (n = 250) | Not DR match clustered (n = 163) | ||||
| All subjectsd | Resistant to more than one drug | 109 (44) | 111 (69) | 0.4 (0.2, 0.6) | <0.0001 |
| HIV positivee | 46 (18) | 11 (7) | 2.5 (1.2, 5.0) | 0.01 | |
| Texas residentsf | Resistant to more than one drug | 45 (29) | 51 (59) | 0.5 (0.2, 1.0) | 0.04 |
| Race | 0.02 | ||||
| White | 24 (14) | 9 (11) | 1.0 | ||
| Hispanic | 76 (44) | 61 (72) | 0.9 (0.4, 2.3) | ||
| Black | 61 (36) | 9 (11) | 3.4 (1.2, 10.0) | ||
| Asian and other | 10 (4) | 6 (7) | 1.0 (0.3, 3.7) | ||
| Female gender | 41 (24) | 34 (40) | 0.5 (0.3, 0.9) | 0.0007 | |
| MDR | 17 (10) | 27 (32) | 0.4 (0.2, 1.0) | 0.04 | |
Significant independent predictors of inclusion in clusters of two or more isolates with identical genotypes and identical resistance to INH, RIF, STR, and/or EMB (DR clustered) versus inclusion in clusters of two or more isolates with identical genotypes and resistance to various drugs (genotype, DR clustered). For Mexican residents, white race was combined with Asian race and other because of the small numbers of isolates. None of the factors was significant for residents of Mexico.
All variables were modeled simultaneously.
Global test of effect.
Variables analyzed were United States versus Mexico, gender, age, HIV infection status, resistance to more than one one drug, multiple-drug resistance (resistance to INH and RIF), and resistance to all four drugs. The Interactions between drug resistance and residence were nonsignificant.
Subjects with unknown HIV infection status were considered negative; unknown HIV infection status was unrelated to clustering.
Variables analyzed were gender, age, HIV infection status, race, foreign born, birth country, resistance to more than one drug, multiple-drug resistance, and resistance to all four drugs.
Geographic distribution of DR cluster strains.
The proportions of strains in DR clusters varied significantly by geographic locality. The area with the highest percentage (55%) of patients whose isolates were included in DR clusters was found to be the adjacent cities of Laredo, Texas, and Nuevo Laredo, Tamaulipas. The Dallas-Fort Worth metropolitan area also had a high percentage (36%) of patients whose patients were included in DR clusters, and that percentage was not significantly different from that for the Laredo-Nuevo Laredo area (P = 0.066). In contrast, the percentages of patients whose isolates were in DR clusters from the southernmost portion of the United States-Mexico border, which includes Hidalgo County, Cameron County, Reynosa, and Matamoros (25%), and from Houston (27%) were significantly lower than the percentage from either the Laredo-Nuevo Laredo area (P < 0.0001 and P = 0.004, respectively) or the Dallas-Fort Worth area (P = 0.004 and P = 0.033, respectively). When the percentages for adjoining geographic areas were compared, there was no significant difference between those for Nuevo Laredo and Laredo, Hidalgo County and Reynosa, or Cameron County and Matamoros. By contrast, DR clustering was significantly more common in Dallas County than in its adjoining county, Tarrant County (OR, 3.4; CI, 1.6, 7.2; P = 0.002).
Of the 78 DR clusters, only 4 were restricted to Mexico alone, while 22 DR clusters were found in both Mexico and Texas, which establishes the spread of strains across the border. Four of these 22 DR clusters included Mexican patients residing in the interior of that country, and 13 included Texan patients residing in counties not adjoining the border (Table 2). One of the DR clusters (cluster 11.008) included isolates from patients from both the interior of Mexico and the interior of Texas. The isolates from this DR cluster included an INH-resistant isolate collected from a patient in Tampico, Tamaulipas, in 1998 with a genotype identical to that of an INH-resistant isolate collected from a Mexican-born patient in Nacogdoches, Texas, in 1999. Tampico and Nacogdoches are separated by approximately 1,100 km (660 mi).
There is also evidence of the interstate spread of the strains that were found to be associated with clusters in this study. Interstate comparisons based on IS6110-based RFLP patterns shared with CDC National Fingerprint Patterns and spoligotype designations showed that isolates from 157 individuals in 5 other CDC surveillance areas (Arkansas, California, Maryland, Massachusetts, and New Jersey) shared genotypes associated with the DR clusters in Texas and Tamaulipas (Table 5). Notably, 7 of these 13 interstate genotype clusters included isolates from individuals residing in Mexico. Two patients from Massachusetts had drug resistance profiles matching the respective genotypes of isolates from Texas (A. C. Miller, personal communication). In addition, both individuals were known to have previously resided in Texas. One individual, a homosexual male, was infected with a RIF-resistant strain matching a strain (Texas pattern 13.121) associated with a gay bar in Dallas. The other, an Asian individual, was infected with an STR-resistant strain (Texas pattern 12.004). The STR-resistant strain with Texas pattern 12.004 is found almost exclusively in the vicinity of the sister cities of Laredo, Texas, and Nuevo Laredo, Tamaulipas. No epidemiological link has yet been identified between the former Houston resident diagnosed with TB in Massachusetts and the Laredo-Nuevo Laredo STR-resistant cluster with pattern 12.004. However, most of the isolates from other states either were drug susceptible or had dissimilar drug resistance profiles.
TABLE 5.
Detection of Texas isolates with the same IS6110-based RFLP patterns as isolates from other states in the United Statesa
| IS6110-based RFLP analysis
|
No. of isolatesb
|
|||||||
|---|---|---|---|---|---|---|---|---|
| No. of bands | Pattern | Spoligotypec | Ariz. | Calif. | Mass. | Md. | Mich. | N.J. |
| 2 | 2.001 | 777776777760771 | 2, 0 | 2, 1 | 6, 0 | 1, 0 | ||
| 2 | 2.001 | 777776777760601 | 11, 0 | 33, 3 | 19, 12 | 28, 14 | ||
| 2 | 2.006 | 400037777413771 | 4, 1 | |||||
| 3 | 3.010 | 777776777760601 | 1, 0 | 1, 0 | ||||
| 4 | 4.001 | 777776777760771 | 0, 1 | |||||
| 4 | 4.002 | 777776777760771 | 6, 1 | 3, 0 | 2, 1 | 15, 1 | 8, 1 | |
| 6 | 6.006 | 3, 0 | ||||||
| 10 | 10.003 | 1, 0 | 1, 0 | 1, 0 | ||||
| 10 | 10.012 | 5, 0 | 2, 0 | 1, 0 | ||||
| 13 | 12.004 | 0, 1 | ||||||
| 13 | 13.121 | 0, 1 | 1, 0 | |||||
| 13 | 13.011 | 3, 0 | 1, 0 | |||||
| 15 | 15.024 | 1, 0 | 1, 0 | |||||
Boldface indicates that the drug resistance panel for the isolate matches that for isolates from states other than Texas. Underlining indicates clusters that include individuals residing in Tamaulipas. Spoligotype designations were determined by using the nomenclature of Dale et al. (7).
The data represent the number of drug-susceptible isolates, number of DR isolates.
Spoligotype data for strains with more than five IS6110 RFLP bands were not available from all of the other genotyping laboratories.
DISCUSSION
Twenty-seven percent of DR M. tuberculosis isolates collected from 919 individuals residing in the U.S. state of Texas and the bordering Mexican state of Tamaulipas were grouped into clusters of two or more isolates on the basis of matching genotypes and drug resistance patterns. Nine percent of the individuals with DR TB were in clusters that included people residing on both sides of the border. Seven percent were in clusters that not only spanned the border but that were also spread beyond the immediate confines of the border.
In a previous genotyping study of DR M. tuberculosis isolates from Texas by Wilson et al. (24), only 20% of the isolates were associated with genotype clusters and less than 1% had both identical genotypes and identical drug resistance panels. In contrast, this study demonstrates that significantly more isolates from patients residing in Texas and Mexico are associated with genotype clusters than the proportion reported previously (45 versus 20% [P < 0.0001]). This is despite the fact that the cluster definition used here was restricted to isolates demonstrating exact rather than 90% IS6110-based RFLP matches in order to more accurately reflect recent transmission (9, 16). In so doing, the sizes of clusters of isolates with six or more IS6110 bands are probably underestimated, as strains have been demonstrated to acquire additional insertions of the IS6110 element in the genome during the course of an outbreak (2, 13, 22). In addition, significantly more isolates from patients in the present study were genotypically identical in terms of identical drug resistance panels compared with the proportion in the previous study (27 versus <1% [P < 0.0001]), even though the requirement for matching drug susceptibility patterns excludes patients who may have initially been infected with a DR strain that then subsequently acquired resistance to an additional drug(s). Therefore, this study most likely underestimates the extent of transmission of the progenitor strains over time in the interest of focusing on more recent transmission. Furthermore, even if all strains with less than six IS6110 bands and matching spoligotype patterns were removed from this study on the basis of the fact that the matched genotypes may not adequately reflect recent transmission (6), the proportion of isolates in clusters with matching genotypes and matching drug resistance patterns still remains significantly higher than that reported previously (21 versus <1% [P < 0.0001]). While the sequencing of genes to detect mutations related to drug resistance would provide additional indications of which isolates were most likely connected to instances of direct transmission, such an undertaking was beyond the scope of this study.
The question of whether the transmission of these strains occurred before or after drug resistance was acquired can be addressed for Texan patients. The corresponding genotypes for 30 (32%) of the 78 DR clusters identified were not associated with those for any of the 1,938 drug-susceptible isolates concurrently genotyped by our laboratory (Fig. 1). The fact that the isolates in these 30 clusters share drug resistance patterns (Table 3) suggests that the initial transmission of drug resistance may have occurred with the DR strain. For instance, the isolates in the border-spanning clusters sharing fingerprint 9.001 are all resistant to INH and STR. Initial epidemiological investigation of the clusters with fingerprint 9.001 showed that half of the individuals had a history of substance abuse and incarceration in Texan and Mexican jails along the border for drug trafficking. One of these individuals is a family relation of two additional patients, including a child, with TB caused by the same strain. Another child in Dallas has yet to be associated with the other patients in any way other than a history of frequent travel to Mexico and a mother with a history of incarceration for illicit drug-related activities. Recent identification of additional patients in Dallas with a history of movement between the United States and Mexico provides evidence of ongoing transmission of the family of strains with the 9.001 fingerprint (data not shown).
The genotypes associated with isolates in the other 48 clusters were also described for drug-susceptible isolates that have been genotyped by our laboratory in conjunction with the CDC National Tuberculosis Genotyping and Surveillance Network. Considering the larger number of susceptible isolates with fingerprints matching those of isolates in smaller DR clusters and the variabilities of the resistance patterns, some of these DR clusters appear to consist of isolates that acquired drug resistance following transmission of a susceptible strain. However, the possibility of the subsequent transmission of acquired drug resistance cannot be excluded and may account for clusters in which there are more DR isolates than drug-susceptible isolates. For example, an epidemiological investigation of one cluster in which the number of drug-sensitive isolates exceeds the number of DR isolates has verified the transmission of acquired resistance. The 13.121 fingerprint cluster, which is associated with 13 individuals infected with RIF-resistant isolates as well as 19 individuals infected with drug-susceptible isolates, represents isolates responsible for an evolving outbreak focused in a predominantly black, male, homosexual population frequenting a particular nightclub in Dallas. Isolates from individuals initially infected were drug susceptible. However, isolates from one individual with a history of noncompliance with directly observed therapy acquired RIF resistance. The subsequent infection of a transgendered entertainer who performed at the establishment on a regular basis resulted in more individuals infected with the RIF-resistant strain. Interstate transmission of an M. tuberculosis strain with an 11-band fingerprint pattern, as opposed to the 13-band pattern described here, via networks of transgendered performers has been documented previously (5). Similarly, a RIF-resistant isolate with the 13-band pattern was recovered from a patient in Massachusetts who had recently moved from Texas. It is of interest that while the gay bar-associated TB strain in Dallas also differs from the well-documented gay bar-associated TB strain in Houston (25), the Houston strain is now being found in Dallas, and vice versa (E. A. Graviss, personal communication).
DR strains of M. tuberculosis were found to be geographically distributed within and between Texas and Mexico. Sixty-four percent of patients in DR clusters were found in nonadjoining counties or jurisdictions. Individuals in DR clusters of two patients were not significantly more or less likely to be in adjoining counties than individuals in DR clusters of more than two patients. Thirty-four percent of the patients in DR clusters were in 1 of 22 binational DR clusters. Of the residents of Texas in these binational DR clusters, 90% were Hispanic and 47% were born in Mexico. While specimens from Texas were collected from throughout the state, specimens from Mexico were collected primarily in the Lower Rio Grande-Rio Bravo Valley border state of Tamaulipas. Despite this, 24% of the patients in binational clusters were found to reside in counties of Texas or jurisdictions of Mexican states not directly adjoining the U.S.-Mexico border. Residents of Mexico were not more or less likely than Texans to be associated with DR clusters. These findings are consistent with the observation that in a clinic-based study in Monterrey, Mexico, a high degree of clustering of M. tuberculosis genotypes suggested extensive recent transmission (26). While Yang et al. (26) did not find similar evidence of clustering of DR isolates in their study, they hypothesized that the sample size was too small to find similar evidence of clustering of DR isolates, as it has previously been demonstrated that as the sample size increases, the level of genotype clustering increases (21).
It is interesting to compare the prevalence of DR clustering in different areas of Texas and Mexico. Even though the number of DR isolates collected in the Dallas-Fort Worth area was comparable to the number of DR isolates collected in the southernmost border area (Hidalgo County, Cameron County, Reynosa, Matamoros) or in the Houston area, the percentage of patients in Dallas-Fort Worth in DR clusters is significantly higher than the percentage in the southernmost border area (P = 0.004) or the Houston metroplex (P = 0.033). Even more striking is the finding that the clusters in which the Dallas patients are found are focused primarily in Dallas, but the same is not true for Fort Worth patients. Individuals in Fort Worth are generally included in clusters focused primarily in either Dallas or Houston or in clusters distributed throughout Texas and Mexico. In fact, Dallas County is the only county in Texas in which the number of patients in DR clusters consisting of patients within the county exceeds the number of patients in DR clusters consisting of patients from outside that county. Dallas and Reynosa are the only cities in the study for which this is true. The significantly disproportionate concentration of clustered patients in Dallas compared with that in Fort Worth (P = 0.0018) suggests that there may be differences in patient migration. Alternatively, local differences in public health policy may be playing a role in identifying DR cases or reducing the rates of transmission of DR TB.
Improved understanding of the effects of immigration on the epidemiology of TB in the United States will be necessary for the elimination of TB from the United States. The results of this study indicate that DR M. tuberculosis isolates are transmitted between Mexico and the United States. Control of DR TB in the United States therefore requires the control of TB in Mexico.
Acknowledgments
We gratefully acknowledge the support of the Robert J. Kleberg, Jr., and Helen C. Kleberg Foundation. This work was also supported in part by a cooperative agreement with the CDC National Tuberculosis Genotyping and Surveillance Network.
We thank Bruce Elliot and the staff of the Texas Department of Health Bureau of Laboratories Mycobacteriology Section for generously performing drug susceptibility testing and for assistance with isolate collection. We also thank Donald Cave, Jack Crawford, Wendy Cronin, Edward Desmond, Jeff Driscoll, Barry Kreiswirth, Michael Kucab, Jeffery Massey, Ann Miller, Leonard Mukasa, and Sumi Sun for contributions to the investigation of the interstate distribution of strains, Janice Pagoda for assistance with the statistical analyses, and Yvonne Camarce for database management.
REFERENCES
- 1.Asch, S., B. Leake, and L. Gelberg. 1994. Does fear of immigration authorities deter tuberculosis patients from seeking care? West. J. Med. 161:373-376. [PMC free article] [PubMed] [Google Scholar]
- 2.Bifani, P. J., B. Mathema, Z. Liu, S. L. Moghazeh, B. Shopsin, B. Tempalski, J. Driscol, R. Frothingham, J. M. Musser, P. Alcabes, and B. N. Kreiswirth. 1999. Identification of a W variant outbreak of Mycobacterium tuberculosis via population-based molecular epidemiology. JAMA 282:2321-2327. [DOI] [PubMed] [Google Scholar]
- 3.Centers for Disease Control and Prevention. 1998. Population-based survey for drug resistance of tuberculosis—Mexico. Morb. Mortal. Wkly. Rep. 47:371-375. [PubMed] [Google Scholar]
- 4.Centers for Disease Control and Prevention. 1995. Tuberculosis among foreign-born persons who had recently arrived in the United States—Hawaii, 1992-1993, and Los Angeles County, 1993. Morb. Mortal. Wkly. Rep. 44:703-707. [PubMed] [Google Scholar]
- 5.Centers for Disease Control and Prevention. 2000. HIV-related tuberculosis in a transgender network—Baltimore, Maryland, and New York City area, 1998-2000. Morb. Mortal. Wkly. Rep. 49:317-320. [PubMed] [Google Scholar]
- 6.Cronin, W. A., J. E. Golub, L. S. Magder, N. G. Baruch, M. J. Lathan, L. N. Mukasa, N. Hooper, J. H. Razeq, D. Mulcahy, W. H. Benjamin, and W. R. Bishai. 2001. Epidemiologic usefulness of spoligotyping for secondary typing of Mycobacterium tuberculosis isolates with low copy numbers of IS6110. J. Clin. Microbiol. 39:3709-3711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dale, J. W., D. Brittain, A. A. Cataldi, D. Cousins. J. T. Crawford, J. Driscoll, H. Heersma, T. Lillebaek, T. N. Quitugua, N. Rastogi, D. van Soolingen, and V. Wright. 2001. Spacer oligonucleotide typing of Mycobacterium tuberculosis: recommendations for standardized nomenclature. Int. J. Tuberc. Lung Dis. 5:216-219. [PubMed] [Google Scholar]
- 8.Escobedo, M., and F. G. Cosio. 1997. Tuberculosis and the United States-Mexico border. J. Border Health 2:40-47. [Google Scholar]
- 9.Gillespie, S. H., A. Dickens, and T. D. McHugh. 2000. False molecular clusters due to nonrandom association of IS6110 with Mycobacterium tuberculosis. J. Clin. Microbiol. 38:2081-2086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jasmer, R. M., A. Ponce de Leon, P. C. Hopewell, R. G. Alarcon, A. R. Moss, E. A. Paz, G. F. Schecter, and P. M. Small. 1997. Tuberculosis in Mexican-born persons in San Francisco: reactivation, acquired infection and transmission. Int. J. Tuberc. Lung Dis. 1:536-541. [PubMed] [Google Scholar]
- 11.Kamerbeek, J., L. S., A. Kolk, M. van Agterveld, D. van Soolingen, S. Kuijper, A. Bunschoten, H. Molhuizen, R. Shaw, M. Goyal, and J. D. A van Embden. 1997. Simultaneous detection and strain differentiation of Mycobacterium tuberculosis for diagnosis and epidemiology. J. Clin. Microbiol. 35:907-914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kremer, K., D. van Soolingen, R. Frothingham, W. H. Haas, P. W. Hermans, C. Martin, P. Palittapongarnpim, B. B. Plikaytis, L. W. Riley, M. A. Yakrus, J. M. Musser, and J. D. van Embden. 1999. Comparison of methods based on different molecular epidemiological markers for typing of Mycobacterium tuberculosis complex strains: interlaboratory study of discriminatory power and reproducibility. J. Clin. Microbiol. 37:2607-2618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mazurek, G. H., M. D. Cave, K. D. Eisenach, R. J. Wallace, J. H. Bates, and J. T. Crawford. 1991. Chromosomal DNA fingerprint patterns produced with IS6110 as strain-specific markers for epidemiologic study of tuberculosis. J. Clin. Microbiol. 29:2030-2033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.McKenna, M. T., E. McCray, and I. Onorato. 1995. The epidemiology of tuberculosis among foreign born persons in the United States, 1986 to 1993. N. Engl. J. Med. 332:1071-1076. [DOI] [PubMed] [Google Scholar]
- 15.NCCLS. 2000. Susceptibility testing of mycobacteria, nocardia, and other aerobic actinomycetes; tentative standard, 2nd ed. NCCLS, Wayne, Pa. [PubMed]
- 16.Niemann, S., S. Rusch-Gerdes, E. Richter, H. Thielen, H. Heykes-Uden, and R. Diel. 2000. Stability of IS6110 restriction fragment length polymorphism patterns of Mycobacterium tuberculosis strains in actual chains of transmission. J. Clin. Microbiol. 38:2563-2567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rubel, A. J., and L. C. Garro. 1992. Social and cultural factors in the successful control of tuberculosis. Public Health Rep. 107:626-636. [PMC free article] [PubMed] [Google Scholar]
- 18.Small, P. M., P. C. Hopewell, S. P. Singh, A. Paz, J. Parsonnet, D. C. Ruston, G. F. Schecter, C. L. Daley, and G. K. Schoolnik. 1994. The epidemiology of tuberculosis in San Francisco. A population-based study using conventional and molecular methods. N. Engl. J. Med. 330:1703-1709. [DOI] [PubMed] [Google Scholar]
- 19.Taylor, J. P., D. Bergmire-Sweat, and L. Suarez. 1999. Epidemiology of drug-resistant tuberculosis in Texas. Am. J. Epidemiol. 149:359-365. [DOI] [PubMed] [Google Scholar]
- 20.van Embden, J. D., M. D. Cave, J. T. Crawford, J. W. Dale, K. D. Eisenach, B. Gicquel, P. Hermans, C. Martin, R. McAdam, and T. M. Shinnick. 1993. Strain identification of Mycobacterium tuberculosis by DNA fingerprinting: recommendations for a standardized methodology. J. Clin. Microbiol. 31:406-409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.van Soolingen, D., M. W. Borgdorff, P. E. de Haas, M. M. Sebek, J. Veen, M. Dessens, K. Kremer, and J. D. van Embden. 1999. Molecular epidemiology of tuberculosis in The Netherlands: a nationwide study from 1993 through 1997. J. Infect. Dis. 180:726-736. [DOI] [PubMed] [Google Scholar]
- 22.van Soolingen, D., P. W. Hermans, P. E. de Haas, D. R. Soll, and J. D. van Embden. 1991. Occurrence and stability of insertion sequences in Mycobacterium tuberculosis complex strains: evaluation of an insertion sequence-dependent DNA polymorphism as a tool in the epidemiology of tuberculosis. J. Clin. Microbiol. 29:2578-2586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wells, C. D., M. Ocana, K. Moser, D. Bergmire-Sweat, J. C. Mohle-Boetani, and N. J. Binkin. 1999. A study of tuberculosis among foreign-born Hispanic persons in the U.S. states bordering Mexico. Am. J. Respir. Crit. Care Med. 159:834-837. [DOI] [PubMed] [Google Scholar]
- 24.Wilson, R. W., Z. Yang, M. Kelley, M. D. Cave, J. M. Pogoda, R. J. Wallace, Jr., J. P. Cegielski, D. F. Dunbar, D. Bergmire-Sweat, L. B. Elliott, and P. F. Barnes. 1999. Evidence from molecular fingerprinting of limited spread of drug-resistant tuberculosis in Texas. J. Clin. Microbiol. 37:3255-3259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yaganehdoost, A., E. A. Graviss, M. W. Ross, G. J. Adams, S. Ramaswamy, A. Wanger, R. Frothingham, H. Soini, and J. M. Musser. 1999. Complex transmission dynamics of clonally related virulent Mycobacterium tuberculosis associated with barhopping by predominantly human immunodeficiency virus-positive gay men. J. Infect. Dis. 180:1245-1251. [DOI] [PubMed] [Google Scholar]
- 26.Yang, Z. H., A. Rendon, A. Flores, R. Medina, K. Ijaz, J. Llaca, K. D. Eisenach, J. H. Bates, A. Villarreal, and M. D. Cave. 2001. A clinic-based molecular epidemiologic study of tuberculosis in Monterrey, Mexico. Int. J. Tuberc. Lung Dis. 5:313-320. [PubMed] [Google Scholar]