Abstract
Phylogenetic analysis of about 200 strains of Salmonella, Shigella, and Escherichia coli was carried out using the nucleotide sequence of the gene for DNA gyrase B (gyrB), which was determined by directly sequencing PCR fragments. The results establish a new phylogenetic tree for the classification of Salmonella, Shigella, and Escherichia coli in which Salmonella forms a cluster separate from but closely related to Shigella and E. coli. In comparison with 16S rRNA analysis, the gyrB sequences indicated a greater evolutionary divergence for the bacteria. Thus, in screening for the presence of bacteria, the gyrB gene might be a useful tool for differentiating between closely related species of bacteria such as Shigella spp. and E. coli. At present, 16S rRNA sequence analysis is an accurate and rapid method for identifying most unknown bacteria to the genus level because the highly conserved 16S rRNA region is easy to amplify; however, analysis of the more variable gyrB sequence region can identify unknown bacteria to the species level. In summary, we have shown that gyrB sequence analysis is a useful alternative to 16S rRNA analysis for constructing the phylogenetic relationships of bacteria, in particular for the classification of closely related bacterial species.
Shigella and Salmonella are pathogens that cause gastroenteropathy in humans (22). Alimentary infections are mostly caused by Salmonella, which has a broad distribution throughout the natural world and a widespread occurrence in animals, especially in poultry and swine (10). Shigella spp. are, in fact, metabolically inactive biogroups of Escherichia coli, and some E. coli strains can cause diarrhea similar to that caused by Shigella. Brenner considered Shigella and E. coli to be a single species, based on DNA homology (4).
In general, the chromosomes of Salmonella, Shigella, and E. coli comprise a single circular DNA molecule consisting of about 4 × 106 bp, with a relative molecular mass of 4 × 109 and a total length of about 1.4 mm. E. coli, Shigella, and Salmonella all belong to the family Enterobacteriaceae.
There are over 2,000 Salmonella serotypes, based on antigenic differences associated with gastroenteritis and enteric (typhoid) fever in humans. The majority of these serotypes belong to a single Salmonella species, Salmonella enterica (23). S. enterica includes six subspecies (Salmonella enterica subsp. enterica, Salmonella enterica subsp. salamae, Salmonella enterica subsp. arizonae, Salmonella enterica subsp. diarizonae, Salmonella enterica subsp. houtenae, and Salmonella enterica subspecies indica).
For Shigella, there are four species (Shigella dysenteriae, Shigella flexneri, Shigella boydii, and Shigella sonnei) (3). Any of the four Shigella species can cause bacillary dysentery.
Finally, for E. coli, there are at least five pathotypes: enteroinvasive E. coli, enterotoxigenic E. coli, enteropathogenic E. coli (EPEC), enterohemorrhagic E. coli (EHEC), and enteroaggregative E. coli (12, 24, 29, 35). Symptoms caused by EPEC resemble salmonellosis, those of enteroinvasive E. coli resemble Shigella, and those of enterotoxigenic E. coli resemble cholera.
Shigella and E. coli strains are often extremely difficult to separate biochemically because there are aerogenic (gas-producing) shigellae and lactose-negative, anaerogenic, nonmotile E. coli strains. E. coli strains can cause a shigella-like diarrhea, and Shigella species are regarded as metabolically inactive biogroups of E. coli (4). Therefore, it would be useful to classify these types of bacteria to aid the treatment of bacterial infections.
PCR has been used to determine the evolutionary relationships of bacteria by analyzing nucleotide sequences of various genes, including 16S/23S rRNA, housekeeping genes, and invasion genes (1, 2, 16, 19, 20, 26). In particular, 16S rRNA sequences have been widely used to construct bacterial phylogenetic relationships (6, 34) or to detect pathogenic bacteria (15). Bacterial analysis by 16S rRNA has become popular because these sections of RNA are conserved and easy to sequence. However, the classification of closely related species of bacteria, for example, Shigella spp. and E. coli, is difficult to achieve through 16S rRNA analysis (7).
As an alternative to 16S rRNA analysis, Yamamoto and Harayama (36, 37, 38) designed a set of PCR primers that allowed both the amplification of the gyrB gene, which encodes the subunit B protein of DNA gyrase (topoisomerase type II), from a large variety of bacteria and the rapid nucleotide sequencing of the amplified gyrB fragments. They then used the gyrB gene in the taxonomic classification of Pseudomonas putida and Acetinobacter strains. The sequences of gyrB genes imply that the rate of molecular evolution is higher than that determined by 16S rRNA sequences (9, 36, 37, 38). We thought that detailed analysis of bacterial phylogenetic relationships might be possible through the gyrB region, as this genetic region can classify bacteria that cannot be classified by their 16S rRNA regions. In particular, the gyrB gene region might be useful to analyze the phylogenetic relationship among Shigella, Salmonella, and E. coli. Using PCR, we amplified the gyrB regions (about 1.2 kb) of bacterial strains isolated from clinical specimens. This region was sequenced directly, and the results were used to compile phylogenetic relationships for the bacterial strains. In this study, we present a new phylogenetic analysis of Shigella, Salmonella, and E. coli determined from the gyrB gene region and compare the results with those of phylogenetic analysis by 16S rRNA.
MATERIALS AND METHODS
Bacterial strains and clinical specimens.
The strains used in this study originated from reference collections (American Type Culture Collection and Deutsche Sammlung von Mikroorganismen und Zellkulturen GmbH) or were clinical isolates. Two hundred strains were cultured from stool specimens from patients with diarrhea collected from different regions in Japan during the period between April 1999 and May 2000. The serogroups of these 200 strains were identified by using an agglutination kit (Denka Seiken Co., Ltd., Tokyo, Japan), and the strains were numbered successively within each serogroup, for example, S. sonnei P1 (P for patient), P2, etc. Screening for E. coli O157:H7 was performed by culture on sorbitol MacConkey agar (Becton Dickinson and Company, Sparks, Md.).
Preparation of chromosomal DNA.
The bacterial strains used in this study were obtained from clinical specimens and were cultured by standard methods. To isolate chromosomal DNA, one or two freshly grown colonies of bacteria were scraped into a 1.5-ml Eppendorf tube and resuspended in 500 μl of sterile water. The bacterial suspension was then boiled (at 100°C for 10 min) to release the DNA.
PCR.
Chromosomal DNAs were amplified by PCR in a thermocycler 480 (Perkin-Elmer Co., Norwalk, Conn.). PCR was performed in a total volume of 100 μl with 5 U of Taq DNA polymerase (AmpliTaq; Perkin-Elmer), 50 mM KCl, 10 mM Tris-HCl (pH 8.3), 1.5 mM MgCl2, 0.001% (wt/vol) gelatin, 200 mM (each) deoxynucleoside triphosphate (dATP, dCTP, dGTP, and dTTP), 10 pM primer UP1 (5′-GAAGTCATCATGACCGTTCTGCAYGCNGGNGGNAARTTYGA-3′), and 10 pM primer UP2r (5′-AGCAGGGTACGGATGTGCGAGCCRTCNACRTCNGCRTCNGTCAT-3′) (36). A 5-μl bacterial sample was added to the PCR solution, which underwent an initial denaturation step of 95°C for 5 min before 30 cycles of 96°C for 1 min, 63°C for 1 min, 72°C for 1 min, and then a final step of 72°C for 7 min for the last cycle. The PCR products were analyzed by 3% agarose gel electrophoresis.
Sequencing of gyrB genes.
DNA sequencing was performed by the dideoxy chain termination method using a Taq DyeDeoxy Terminator cycle-sequencing kit (Applied Biosystems, Foster City, Calif.) according to the manufacturer's instructions. PCR fragments were determined by using the sequencing primers UP1S (5′-GAAGTCATCATGACCGTTCTGCA-3′) and UP2Sr (5′-AGCAGGGTACGGATGTGCGAGCC-3′) (36). Sequence reactions were analyzed on a PRISM 310 genetic analyzer (Applied Biosystems).
Phylogenetic analysis.
The phylogenetic data described below were obtained by alignment and phylogenetic analysis of the bacterial sequences. The nucleotide sequences of 16S rRNA and gyrB were aligned by using the CLUSTAL V computer program (13). A neighbor-joining analysis (25) was used to reconstruct phylogenetic trees with the DNASTAR computer program (DNASTAR Inc., Madison, Wis.).
Nucleotide sequence accession numbers.
The nucleotide sequence data reported in this paper appear in the GenBank and DDBJ nucleotide sequence databases with the following accession numbers: 16S rRNA genes, X80724, U90318, U88546, AF057362, Z47544, X80681, X96965, X96963, X96964, M59292, AJ251468, AJ251469, AF129440, and AF130981; gyrB genes, AB083821 to AB084027.
RESULTS
Phylogenetic analysis and genetic distance of 16S rRNA.
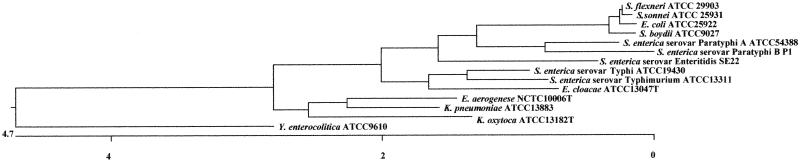
Data for the phylogenetic analysis were obtained from sequences contained in the GenBank nucleotide sequence database (17). The following strains were examined: E. coli (ATCC 25922), S. enterica subsp. enterica serovar Enteritidis (SE22), S. enterica subsp. enterica serovar Paratyphi A (ATCC 54388), S. enterica subsp. enterica serovar Paratyphi B, S. enterica subsp. enterica serovar Typhi (ATCC 19430), S. enterica subsp. enterica serovar Typhimurium (ATCC 13311), S. boydii (ATCC 9027), S. flexneri (ATCC 29903), S. sonnei (ATCC 25931), Yersinia enterocolitica (ATCC 9610), Enterobacter aerogenes (NCTC10006T), Enterobacter cloacae (ATCC 13047T), Klebsiella oxytoca (ATCC 13182T), and Klebsiella pneumoniae (ATCC 13883). Alignment of the 16S rRNA nucleotide sequence, adjusted to 1,435 bases, was performed by the computer program MegAlign (DNASTAR Inc.). Table 1 shows the percent nucleotide divergence and similarity of 16S rRNA for E. coli, Shigella, Salmonella, Enterobacter, and Klebsiella, with Yersinia as an outgroup. S. sonnei and S. flexneri have 99.9% similarity to each other; 99.9 and 99.8%, respectively, to E. coli; and 99.7% to S. boydii. These results indicate that S. flexneri and S. sonnei are more closely related to E. coli than to S. boydii. Figure 1 shows the phylogenetic tree for these strains on the basis of their 16S rRNAs. In this tree, Salmonella subspecies strains were grouped into two clusters. The first cluster contained S. enterica serovar Paratyphi A, S. enterica serovar Paratyphi B, and S. enterica serovar Enteritidis, while the second cluster contained S. enterica serovar Typhi and S. enterica serovar Typhimurium.
TABLE 1.
16S rRNA gene sequence similarity and divergencea of each pair for E. coli, Salmonella, Shigella, Enterobacter, Klebsiella, and Yersinia calculated by DNASTAR
| Strain no. | Strain name | % Similarity with strain no.
|
|||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | 14 | ||
| 1 | E. coli ATCC 25922 | 99.6 | 97.4 | 97.2 | 96.5 | 95.4 | 99.6 | 99.9 | 99.8 | 95.0 | 95.9 | 95.4 | 95.1 | 93.0 | |
| 2 | S. enterica serovar Enteritidis ES22 | 2.6 | 97.6 | 97.4 | 97.4 | 97.1 | 96.7 | 96.9 | 96.9 | 95.1 | 96.9 | 95.1 | 95.5 | 91.6 | |
| 3 | S. enterica serovar Paratyphi A ATCC 54388 | 2.3 | 2.3 | 98.6 | 97.6 | 96.7 | 97.5 | 97.4 | 97.3 | 94.6 | 96.2 | 94.8 | 94.8 | 91.5 | |
| 4 | S. enterica serovar Paratyphi B P1 | 2.5 | 2.6 | 1.3 | 97.1 | 96.3 | 97.2 | 97.2 | 97.1 | 94.5 | 96.1 | 94.5 | 94.6 | 91.3 | |
| 5 | S. enterica serovar Typhi ATCC 19430 | 3.4 | 2.3 | 2.1 | 2.6 | 97.8 | 96.8 | 96.7 | 96.6 | 95.6 | 97.4 | 95.7 | 95.9 | 92.0 | |
| 6 | S. enterica serovar Typhimurium ATCC 13311 | 3.4 | 2.1 | 2.4 | 2.9 | 1.1 | 95.4 | 95.4 | 95.3 | 95.0 | 96.6 | 95.3 | 95.4 | 91.1 | |
| 7 | S. boydii ATCC 9207 | 0.4 | 2.8 | 2.1 | 2.4 | 3.1 | 3.4 | 99.7 | 99.7 | 94.6 | 95.5 | 95.1 | 94.7 | 92.8 | |
| 8 | S. flexneri ATCC 29903 | 0.1 | 2.6 | 2.2 | 2.4 | 3.3 | 3.4 | 0.3 | 99.9 | 94.9 | 95.7 | 95.3 | 95.0 | 93.0 | |
| 9 | S. sonnei ATCC 25931 | 0.2 | 2.6 | 2.3 | 2.5 | 3.4 | 3.4 | 0.3 | 0.1 | 94.8 | 95.6 | 95.1 | 94.8 | 93.0 | |
| 10 | E. aerogenes NCTC10006T | 4.0 | 4.2 | 4.8 | 5.0 | 3.4 | 3.6 | 4.3 | 4.0 | 4.1 | 97.5 | 97.8 | 98.1 | 93.2 | |
| 11 | E. cloacae ATCC 13047T | 3.3 | 2.6 | 3.1 | 3.4 | 1.6 | 1.9 | 3.6 | 3.3 | 3.4 | 2.2 | 97.2 | 98.3 | 92.1 | |
| 12 | K. oxytoca ATCC 13182T | 3.8 | 4.4 | 4.8 | 5.1 | 3.4 | 3.4 | 3.9 | 3.8 | 3.9 | 1.8 | 2.6 | 97.4 | 92.3 | |
| 13 | K. pneumoniae ATCC 13883 | 4.0 | 3.9 | 4.5 | 4.8 | 3.1 | 3.1 | 4.2 | 4.0 | 4.0 | 1.5 | 1.5 | 2.6 | 93.0 | |
| 14 | Y. enterocolitica ATCC 9610 | 5.6 | 6.7 | 6.9 | 7.1 | 6.8 | 6.6 | 5.9 | 5.6 | 5.7 | 4.4 | 6.2 | 5.7 | 4.9 | |
Percent divergence is calculated by comparing sequence pairs in relation to the phylogeny reconstructed by MegAlign (DNASTAR). Percent similarity compares sequences directly without accounting for phylogenetic relationships.
FIG. 1.
Phylogenetic tree based on the nucleotide sequences of 16S rRNA genes. The 16S rRNA sequences were adjusted to 1,435 bases, and the tree was constructed by the neighbor-joining method, using the computer program MegAlign (DNASTAR Inc.). The scale indicates the percentage of base difference (percent divergence). The sequence data for phylogenetic analysis were taken from the GenBank nucleotide sequence database for the following strains: E. coli (ATCC 25922), S. enterica serovar Enteritidis (SE22), S. enterica serovar Paratyphi A (ATCC 54388), S. enterica serovar Paratyphi B, S. enterica serovar Typhi (ATCC 19430), S. enterica serovar Typhimurium (ATCC 13311), S. boydii (ATCC 9027), S. flexneri (ATCC 29903), S. sonnei (ATCC 25931), Y. enterocolitica (ATCC 9610), E. aerogenes (NCTC10006T), E. cloacae (ATCC 13047T), K. oxytoca (ATCC 13182T), and K. pneumoniae (ATCC 13883).
Phylogenetic analysis and genetic distance of gyrB genes.
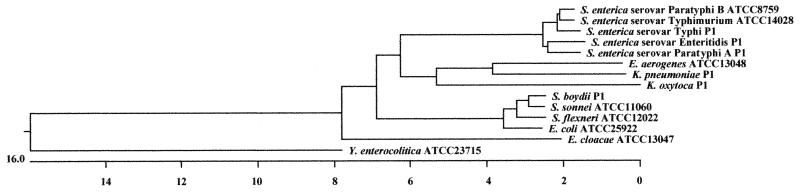
Bacterial samples were subjected to PCR amplification of the gyrB gene region with degenerate primers. The amplified gyrB gene, a region of 1,171 bp, was sequenced from five different strains of Salmonella, three different strains of Shigella, two different strains of Enterobacter, one strain of E. coli, two different strains of Klebsiella, and a strain Yersinia enterocolitica as a control. The following strains were examined: E. coli (ATCC 25922), S. enterica serovar Enteritidis (isolate), S. enterica serovar Paratyphi A (isolate), S. enterica serovar Paratyphi B (ATCC 8759), S. enterica serovar Typhi (isolate), S. enterica serovar Typhimurium (ATCC 14028), S. boydii (isolate), S. flexneri (ATCC 12022), S. sonnei (ATCC 11060), Y. enterocolitica (ATCC 23715), E. aerogenes (ATCC 13048), E. cloacae (ATCC13047), K. oxytoca (isolate), and K. pneumoniae (isolate). Alignment of the gyrB nucleotide sequences, adjusted to 1,171 bases, was performed by the computer program MegAlign. Table 2 shows the percent nucleotide divergence and similarity of the gyrB gene for E. coli, Shigella, Salmonella, Enterobacter, and Klebsiella, with Yersinia as an outgroup. S. sonnei and S. flexneri have 98.4% similarity to each other; 98.1 and 97.8%, respectively, to E. coli; and 99.1 and 98.7%, respectively, to S. boydii. The percent divergence of E. coli from Salmonella is greater in the gyrB gene than in 16S rRNA. Figure 2 shows the phylogenetic tree for these species based on the gyrB gene sequence. As indicated in Fig. 2, bacteria of the same genus are located in the same cluster. In other words, S. enterica serovar Enteritidis, S. enterica serovar Paratyphi A, S. enterica serovar Paratyphi B, S. enterica serovar Typhi, and S. enterica serovar Typhimurium form a cluster with Salmonella in contrast to the result with 16S rRNA (Fig. 1); similarly, S. boydii, S. flexneri, and S. sonnei form a cluster with Shigella.
TABLE 2.
gyrB gene sequence similarity and divergencea of each pair for E. coli, Salmonella, Shigella, Enterobacter, Klebsiella, and Yersinia calculated by DNASTAR
| Strain no. | Strain name | % Similarity with strain no.
|
|||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | 14 | ||
| 1 | E. coli ATCC25922 | 91.3 | 91.0 | 91.0 | 91.1 | 90.9 | 98.0 | 97.8 | 98.1 | 89.2 | 89.2 | 89.2 | 89.3 | 80.4 | |
| 2 | S. enterica serovar Enteritidis P1 | 9.2 | 98.2 | 98.1 | 97.9 | 98.0 | 91.4 | 91.1 | 91.2 | 89.7 | 89.1 | 89.5 | 90.4 | 79.4 | |
| 3 | S. enterica serovar Paratyphi A P 1 | 9.5 | 1.8 | 98.5 | 97.9 | 98.0 | 90.9 | 90.7 | 90.9 | 89.8 | 88.9 | 89.6 | 90.2 | 79.2 | |
| 4 | S. enterica serovar Paratyphi B ATCC 8759 | 9.5 | 1.9 | 1.5 | 98.9 | 99.2 | 90.9 | 91.1 | 90.9 | 90.3 | 89.1 | 89.7 | 90.6 | 79.3 | |
| 5 | S. enterica serovar Typhi P1 | 9.4 | 2.2 | 2.1 | 1.1 | 99.0 | 90.7 | 90.9 | 90.7 | 90.2 | 88.8 | 89.7 | 90.7 | 79.8 | |
| 6 | S. enterica serovar Typhimurium ATCC 14028 | 9.6 | 2.0 | 1.9 | 0.8 | 1.0 | 90.9 | 91.1 | 90.9 | 90.3 | 89.5 | 89.5 | 90.8 | 79.7 | |
| 7 | S. boydii P1 | 2.0 | 9.1 | 9.6 | 9.6 | 9.9 | 9.7 | 98.7 | 99.1 | 89.5 | 89.2 | 89.0 | 89.1 | 80.4 | |
| 8 | S. flexneri ATCC 12022 | 2.3 | 9.5 | 10.0 | 9.5 | 9.7 | 9.5 | 1.3 | 98.4 | 89.5 | 89.2 | 89.3 | 89.0 | 80.8 | |
| 9 | S. sonnei ATCC 11060 | 1.9 | 9.4 | 9.6 | 9.6 | 9.9 | 9.7 | 0.9 | 1.6 | 89.5 | 89.4 | 88.9 | 89.2 | 80.5 | |
| 10 | E. aerogenes ATCC 13048 | 11.7 | 11.1 | 10.8 | 10.4 | 10.4 | 10.3 | 11.4 | 11.3 | 11.4 | 88.3 | 91.3 | 93.4 | 79.2 | |
| 11 | E. cloacae ATCC 13047T | 11.4 | 11.9 | 12.0 | 11.9 | 12.2 | 11.4 | 11.4 | 11.5 | 11.2 | 12.7 | 87.8 | 87.6 | 80.0 | |
| 12 | K. oxytoca P1 | 11.4 | 11.2 | 11.0 | 11.0 | 11.0 | 11.2 | 11.6 | 11.3 | 11.6 | 9.3 | 13.3 | 89.7 | 79.5 | |
| 13 | K. pneumoniae P1 | 11.6 | 10.4 | 10.5 | 10.1 | 10.0 | 9.9 | 11.9 | 11.8 | 11.7 | 6.8 | 13.5 | 11.2 | 79.0 | |
| 14 | Y. enterocolitica ATCC 23715 | 21.2 | 22.9 | 23.3 | 23.3 | 22.4 | 22.5 | 21.6 | 21.1 | 21.3 | 23.3 | 22.1 | 22.7 | 23.7 | |
Percent divergence is calculated by comparing sequence pairs in relation to the phylogeny reconstructed by MegAlign (DNASTAR). Percent similarity compares sequences directly without accounting for phylogenetic relationships.
FIG. 2.
Phylogenetic tree based on the nucleotide sequences of gyrB genes. The gyrB nucleotide sequences were adjusted to 1,171 bases, and the tree was constructed by the neighbor-joining method, using the computer program MegAlign (DNASTAR Inc.). The scale indicates the percentage of base difference (percent divergence). The mean sequence obtained from direct sequencing of PCR products was used for the following strains: E. coli (ATCC 25922), S. enterica serovar Enteritidis (isolate), S. enterica serovar Paratyphi A (isolate), S. enterica serovar Paratyphi B (ATCC 8759), S. enterica serovar Typhi (isolate), S. enterica serovar Typhimurium (ATCC 14028), S. boydii (isolate), S. flexneri (ATCC 12022), S. sonnei (ATCC 11060), Y. enterocolitica (ATCC 23715), E. aerogenes (ATCC 13048), E. cloacae (ATCC 13047), K. oxytoca (isolate), and K. pneumoniae (isolate).
Comparison of the genetic distance and the phylogenetic tree determined by 16S rRNA and gyrB.
A direct comparison of the genetic distance and the phylogenetic tree determined by the 16S rRNA sequence with those determined by the gyrB sequence was not possible because the bacterial strains analyzed were not the same. However, the following observations could be made. First, the rate of genetic divergence of the gyrB sequence differed greatly from that of the 16S rRNA sequence. For example, compared with 16S rRNA, the gyrB analysis shows there is a 4- to 10-fold increase in the length of branches between closely related species of Shigella and E. coli. Moreover, the topology of the phylogenetic tree based on the 16S rRNA sequence was quite different from that based on the gyrB sequence. Second, phylogenetic analysis using the gyrB nucleotide sequence determined a classification of the bacteria different from that determined by 16S rRNA analysis.
Phylogenetic analysis of gyrB genes from clinical specimens.
For this study, data were gathered from the genes of 200 clinical specimens and the genes were sequenced. Alignment of the gyrB nucleotide sequences, adjusted to 1,171 bases, was done by the computer program MegAlign. Figure 3 shows the phylogenetic tree for these species based on the gyrB gene sequence. The numbers of bacterial strains sharing the same sequence in each group are indicated. For example, S. enterica serovar Enteritidis P1 to P5, five strains of S. enterica serovar Enteritidis, have identical gyrB sequences, which were isolated from five patients. On one hand, it has been reported that it is impossible to differentiate between E. coli and Shigella on the basis of 16S rRNA sequence analysis (7). However, even if some Shigella strains are distributed among E. coli strains, our results clearly show that Shigella spp. are not identical to E. coli. On the other hand, it was not possible to classify serogroups of Salmonella and E. coli by phylogenetic analysis of either the 16S rRNA or the gyrB gene. E. coli isolates with the same O serogroups appear in several different clusters. For example, E. coli O157 isolates are found in three different clusters. It is interesting that E. coli O157 BBL63644 was identified as a sorbitol-fermenting EHEC isolate, representing the H− serogroup (5, 23). In general, serogroup does not correspond to genotype. Indeed, our analysis showed that serogroup does not correspond to genotype.
FIG. 3.
Phylogenetic tree based on the nucleotide sequences of gyrB genes taken from 200 clinical specimens. The gyrB nucleotide sequences were adjusted to 1,171 bases, and the tree was constructed by the neighbor-joining method, using the computer program MegAlign (DNASTAR Inc.). The scale indicates the percentage of base difference (percent divergence). Amplified gyrB genes from five subspecies of Salmonella, three species of Shigella, two species of Enterobacter, one species of E. coli, two species of Klebsiella, and Yersinia species used as an outgroup were sequenced. Bacterial strains of the same species with the same gene sequence are displayed in one group; for example, the cluster labeled S. enterica serovar Enteritidis P1-5 includes five strains of S. enterica serovar Enteritidis with identical gyrB sequences which were isolated from patients.
DISCUSSION
Phylogenetic-tree analysis is often used as a method to classify organisms. Various genes have been examined for the analysis of phylogenetic relationships of Salmonella, Shigella, and E. coli (1, 2, 16, 21). In general, 16S rRNA is most frequently used for such analyses; however, phylogenetic relationships between closely related species of Shigella and E. coli are weakly defined by this approach, as the 16S rRNA sequences of bacteria contain highly conserved regions. In terms of cell biology, Shigella spp. are similar to E. coli. Shigella strains are in reality clones of E. coli (14, 28) and are believed to have emerged relatively recently (21, 32). In our phylogenetic-tree analysis of 16S rRNA, S. sonnei and S. flexneri were found to have 99.9% similarity to each other and 99.9 and 99.8% similarity, respectively, to E. coli but only 99.7% similarity to S. boydii (Table 1). The percent divergences of E. coli from S. sonnei, S. flexneri, and S. boydii were 0.2, 0.1, and 0.4%, respectively. These data are in accordance with the previous study using the 16S rRNA gene sequence (34), indicating close relationships among these bacteria. In Fig. 3., S. sonnei (15 specimens) and S. flexneri (17 specimens) are grouped together, but the groups are scattered among E. coli strains. This result gives results comparable to those of studies with other genes (14, 20, 28), supporting the idea that Shigella strains are actually clones of E. coli. The divergence values within or between species are smaller in 16S rRNA analysis than in previous studies using several housekeeping and invasion genes (1, 2, 16, 19, 20, 26). For example, in the nucleotide sequence of the isocitrate dehydrogenase gene, pairs of E. coli strains differed by 5.6% on average and pairs of E. coli and S. enterica strains differed by 13.3% (30). Although the 16S rRNA gene is preferred for phylogenetic studies because of its attributes, such as little evidence for its lateral transfer (27), the sequence variation of 16S rRNA is not as high as those of other genes in numerous studies.
In the phylogenetic-tree analysis of gyrB, S. sonnei and S. flexneri were found to have 98.4% similarity to each other, 98.1 and 97.8% similarity to E. coli, and 99.1 and 98.7% similarity to S. boydii, respectively (Table 2). The percent divergences of E. coli from S. sonnei, S. flexneri, and S. boydii were 1.9, 2.3, and 2.0%, respectively. The divergence values are significantly improved in gyrB analysis compared with 16S rRNA analysis. It is reported that when 20 Pseudomonas strains were analyzed by using the nucleotide sequences of gyrB and the genes for 16S rRNA and RNA polymerase σ70 factor (rpoD), the percent divergences of gyrB were larger than those of the other genes (38). Like the 16S rRNA gene, the gyrB gene does not appear to be frequently horizontally transmitted and can be found in most, if not all, bacterial species (36, 38). Many genes, especially those for catabolism, are known to spread horizontally among different bacterial species, and they cannot be used to trace the evolutionary records of host bacteria (37).
A comparison of Fig. 2 with Fig. 1 highlights the different patterns of bacterial divergence determined through analysis of gyrB genes and 16S rRNA. In the phylogenetic analysis using 16S rRNA, bacteria belonging to the same genus were not always located in the same cluster; by contrast, in the phylogenetic analysis using the gyrB gene, bacteria of the same genus were clustered together. In other words, S. enterica serovar Enteritidis, S. enterica serovar Paratyphi A, S. enterica serovar Paratyphi B, S. enterica serovar Typhi, and S. enterica serovar Typhimurium were located in a cluster with Salmonella. Similarly, S. boydii, S. flexneri, and S. sonnei were located in a cluster with Shigella. In addition, the rate of base substitution was greater for the gyrB sequence than for the 16S rRNA sequence (9, 36, 38). Thus, certain bacterial strains were classified differently under the two phylogenetic analyses.
It seems likely that phylogenetic analysis using the gyrB gene sequence will be able to classify some bacteria that cannot be classified by their 16S rRNA sequences. Cilia et al. (8) have reported that 16S rRNA sequences cannot be used to derive phylogenetic-tree analyses among closely related bacteria, for example, Shigella and E. coli, owing to the similarity in these gene regions. However, our results indicate that such closely related bacteria might be classified by gyrB analysis. 23S rRNA sequence analysis (7) could provide phylogenetic information at the subspecies level for Salmonella, but gyrB analysis generally shows sharper separations.
Approximately 2,000 serotypes of S. enterica can cause sickness in humans, such as S. enterica serovar Typhi, which causes enteric (typhoid) fever and is pathogenic only in humans, and S. enterica serovar Typhimurium, which causes gastroenteritis and is pathogenic for several mammalian species. Currently, salmonellosis is classified clinically according to the symptoms (typhoid fever and paratyphoid) and/or according to the serotype, especially for non-typhoid fever types of salmonellosis. However, as serotype does not necessarily agree with genotype, it would be useful to have another means, such as gyrB gene analysis, to classify the subspecies of S. enterica. In contrast to serotype assays, genotype assays for pathogenic bacteria may be expensive and require technical expertise, but they could detect a wider range of bacterial strains and increase specificity (18), which is useful when cross-reaction is observed with serotyping reagents.
As shown in Fig. 3, E. coli isolates with the same O serogroups appear in several different clusters. Our results also show a mosaic relationship among pathotypes classified by O serogroup and genotypes classified by gyrB region. For example, E. coli isolates with O serogroups associated with EPEC and EHEC appear in different clusters or different pathotypes are grouped in one cluster. These results indicate lateral transfer of genes for O antigens (20, 31), as well as genes for H antigens (23), among E. coli strains.
The rate of evolution of the gyrB genetic region is higher than that of the 16S rRNA region, and the gyrB genetic region is found in all bacterial species. We believe that the gyrB region will have high reliability for identifying pathogenic bacteria. Although the 16S rRNA sequence method is a highly accurate and rapid method for identifying most bacteria to the genus level (33), the gyrB sequence method might be more useful for identifying bacteria to the species level. In practical terms, this means that whereas it is easy to amplify the 16S rRNA sequence because it is highly conserved across bacteria, it is harder to amplify the gyrB region because of its variability. However, the primer set developed by Yamamoto and Harayama (36) for amplification of the bacterial gyrB region by PCR simplifies the rapid amplification of this region. It is known that the amplified gyrB region we used includes the region involving quinolone resistance (11, 39). However, the 200 clinical specimens in our study do not include quinolone-resistant strains (data not shown), indicating that this resistance does not influence the interpretation of our results.
In summary, we have shown that gyrB sequence analysis is a fruitful approach to determine the phylogenetic relationships of bacteria and may be an alternative to 16S rRNA analysis. In particular, gyrB analysis of bacteria is an effective means to classify closely related species. Further research on gyrB sequence analysis will clarify in more detail the classification of bacterial species.
Acknowledgments
We thank Yoshimi Katow and Kazunori Hochido for assistance in culture. We also thank Noboru Fujinami for stimulating discussions.
This study was supported by the New Energy and Industrial Technology Development Organization (NEDO).
REFERENCES
- 1.Boyd, E. F., K. Nelson, F. S. Wang, T. S. Whittam, and R. K. Selander. 1994. Molecular genetic basis of allelic polymorphism in malate dehydrogenase (mdh) from natural populations of Escherichia coli and Salmonella enterica. Proc. Natl. Acad. Sci. USA 91:1280-1284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Boyd, E. F., J. Li, H. Ochman, and R. K. Selander. 1997. Comparative genetics of the inv-spa invasion gene complex of Salmonella enterica. J. Bacteriol. 179:1985-1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brenner, D. J., G. R. Fanning, G. V. Miklos, and A. G. Steigerwalt. 1973. Polynucleotide sequence relatedness among Shigella species. Int. J. Syst. Bacteriol. 23:1-7. [Google Scholar]
- 4.Brenner, D. J. 1984. Family I. Enterobacteriaceae, p. 408-420. In N. R. Krieg and J. G. Holt (ed.), Bergey's manual of systematic bacteriology, vol. 1. Williams & Wilkins, Baltimore, Md.
- 5.Brunder, W., A. S. Khan, J. Hacker, and H. Karch. 2001. Novel type of fimbriae encoded by the large plasmid of sorbitol-fermenting enterohemorrhagic Escherichia coli O157:H(−). Infect. Immun. 69:4447-4457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chang, H. R., L. H. Loo, K. Jeyaseelan, L. Earnest, and E. Stackebrandt. 1997. Phylogenetic relationships of Salmonella typhi and Salmonella typhimurium based on 16S rRNA sequence analysis. Int. J. Syst. Bacteriol. 47:1253-1254. [DOI] [PubMed] [Google Scholar]
- 7.Christense, H., S. Nordentoft, and J. E. Olsen. 1998. Phylogenetic relationships of Salmonella based on rRNA sequences. Int. J. Syst. Bacteriol. 48:605-610. [DOI] [PubMed] [Google Scholar]
- 8.Cilia, V., B. Lafay, and R. Christen. 1996. Sequence heterogeneities among 16S ribosomal RNA sequences, and their effect on phylogenetic analyses at the species level. Mol. Biol. Evol. 13:451-461. [DOI] [PubMed] [Google Scholar]
- 9.Dams, E., L. Hendriks, Y. Van der Peer, J. M. Neefs, G. Smits, I. Vandenbempt, and R. De Wachter. 1988. Compilation of small ribosomal subunit RNA sequences. Nucleic Acids Res. 16:87-173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Davies, R. H., and C. Wray. 1996. Determination of an effective sampling regime to detect Salmonella enteritidis in the environment of poultry units. Vet. Microbiol. 50:117-127. [DOI] [PubMed] [Google Scholar]
- 11.Gensberg, K., Y. F. Jin, and L. J. Piddock. 1995. A novel gyrB mutation in a fluoroquinolone-resistant clinical isolate of Salmonella typhimurium. FEMS Microbiol. Lett. 132:57-60. [DOI] [PubMed] [Google Scholar]
- 12.Gilligan, P. H. 1999. Escherichia coli. EAEC, EHEC, EIEC, ETEC. Clin. Lab. Med. 19:505-521. [PubMed] [Google Scholar]
- 13.Higgins, D. G., A. J. Bleasby, and R. Fuchs. 1992. CLUSTAL V: improved software for multiple sequence alignment. Comput. Appl. Biosci. 8:189-191. [DOI] [PubMed] [Google Scholar]
- 14.Karaolis, D. K., R. Lan, and P. R. Reeves. 1994. Sequence variation in Shigella sonnei (Sonnei), a pathogenic clone of Escherichia coli, over four continents and 41 years. J. Clin. Microbiol. 32:796-802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lampel, K. A., J. A. Jagow, M. Trucksess, and W. E. Hill. 1990. Polymerase chain reaction for detection of invasive Shigella flexneri in food. [DOI] [PMC free article] [PubMed]
- 16.Li, J., H. Ochman, E. A. Groisman, E. F. Boyed, F. Solomon, K. Nelson, and R. K. Selander. 1995. Relationship between evolutionary rate and cellular location among the Inv/Spa invasion proteins of Salmonella enterica. Proc. Natl. Acad. Sci. USA 92:7252-7256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Maidak, B. L., N. Larsen, M. J. McCaughey, R. Overbeek, G. J. Olsen, K. Fogel, J. Blandy, and C. R. Woese. 1994. The Ribosomal Database Project. Nucleic Acids Res. 22:3485-3487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.McDaniels, A. E., E. W. Rice, A. L. Reyes, C. H. Johnson, R. A. Haugland, and G. N. Stelma, Jr. 1996. Confirmational identification of Escherichia coli, a comparison of genotypic and phenotypic assays for glutamate decarboxylase and β-d-glucuronidase. Appl. Environ. Microbiol. 62:3350-3354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nelson, K., T. S. Whittam, and R. K. Selander. 1991. Nucleotide polymorphism and evolution in the glyceraldehyde-3-phosphate dehydrogenase gene (gapA) in natural populations of Salmonella and Escherichia coli. Proc. Natl. Acad. Sci. USA 88:6667-6671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nelson, K., and R. K. Selander. 1994. Intergeneric transfer and recombination of the 6-phosphogluconate dehydrogenase gene (gnd) in enteric bacteria. Proc. Natl. Acad. Sci. USA 91:10227-10231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pupo, G. M., R. Lan, and P. R. Reeves. 2000. Multiple independent origins of Shigella clones of Escherichia coli and convergent evolution of many of their characteristics. Proc. Natl. Acad. Sci. USA 97:10567-10572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Reeves, M. W., G. M. Evins, A. A. Heiba, B. D. Plikaytis, and J. J. Farmer III. 1989. Clonal nature of Salmonella typhi and its genetic relatedness to other salmonellae as shown by multilocus enzyme electrophoresis, and proposal of Salmonella bongori comb. nov. J. Clin. Microbiol. 27:313-320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Reid, S. D., R. K. Selander, and T. S. Whittam. 1999. Sequence diversity of flagellin (fliC) alleles in pathogenic Escherichia coli. J. Bacteriol. 181:153-160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rich, C., S. Favre-Bonte, F. Sapena, B. Joly, and C. Forestier. 1999. Characterization of enteroaggregative Escherichia coli isolates. FEMS Microbiol. Lett. 173:55-61. [DOI] [PubMed] [Google Scholar]
- 25.Saitou, N., and M. Nei. 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4:406-425. [DOI] [PubMed] [Google Scholar]
- 26.Selander, R. K., J. Li, and K. Nelson. 1996. Evolutionary genetics of Salmonella enterica, p. 2691-2707. In F. C. Neidhardt et al. (ed.), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed., vol. 2. American Society for Microbiology, Washington, D.C.
- 27.Sneath, P. H.1993. Evidence from Aeromonas for genetic crossing-over in ribosomal sequences. Int. J. Syst. Bacteriol. 43:626-629. [DOI] [PubMed] [Google Scholar]
- 28.Stevenson. G., B. Neal, D. Liu, M. Hobbs, N. H. Packer, M. Batley, J. W. Redmond, L. Lindquist, and P. Reeves. 1994. Structure of the O antigen of Escherichia coli K-12 and the sequence of its rfb gene cluster. J. Bacteriol. 176:4144-4156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sunabe, T., and Y. Honma. 1998. Relationship between O-serogroup and presence of pathogenic factor genes in Escherichia coli. Microbiol. Immunol. 42:845-849. [DOI] [PubMed] [Google Scholar]
- 30.Wang, F. S., T. S. Whittam, and R. K. Selander. 1997. Evolutionary genetics of the isocitrate dehydrogenase gene (icd) in Escherichia coli and Salmonella enterica. J. Bacteriol. 179:6551-6559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang, L., and P. R. Reeves. 2000. The Escherichia coli O111 and Salmonella enterica O35 gene clusters: gene clusters encoding the same colitose-containing O antigen are highly conserved. J. Bacteriol. 182:5256-5261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang, L., W. Qu, and P. R. Reeves. 2001. Sequence analysis of four Shigella boydii O-antigen loci: implication for Escherichia coli and Shigella relationships. Infect. Immun. 69:6923-6930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang, R. F., W. W. Cao, and C. E. Cerniglia. 1994. A 16S rDNA-based PCR method for rapid and specific detection of Clostridium perfringens in food. Mol. Cell. Probes 8:131-137. [DOI] [PubMed] [Google Scholar]
- 34.Wang, R. F., W. W. Cao, and C. E. Cerniglia. 1997. Phylogenetic analysis and identification of Shigella spp. by molecular probe. Mol. Cell. Probes 11:427-432. [DOI] [PubMed] [Google Scholar]
- 35.Willshaw, G. A., T. Cheasty, H. R. Smith, S. J. O'Brien, and K. Adak. 2001. Verocytotoxin-producing Escherichia coli (VTEC) O157 and other VTEC from human infections in England and Wales: 1995-1998. J. Med. Microbiol. 50:135-142. [DOI] [PubMed] [Google Scholar]
- 36.Yamamoto, S., and S. Harayama. 1995. PCR amplification and direct sequencing of gyrB genes with universal primers and their application to the detection and taxonomic analysis of Pseudomonas putida strains. Appl. Environ. Microbiol. 61:1104-1109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yamamoto, S., and S. Harayama. 1996. Phylogenetic analysis of Acinetobacter strains based on the nucleotide sequences of gyrB genes and on the amino acid sequences of their products. Int. J. Syst. Bacteriol. 46:506-511. [DOI] [PubMed] [Google Scholar]
- 38.Yamamoto, S., and S. Harayama. 1998. Phylogenetic relationships of Pseudomonas putida strains deduced from the nucleotide sequences of gyrB, rpoD and 16S rRNA genes. Int. J. Syst. Bacteriol. 3:813-819. [DOI] [PubMed] [Google Scholar]
- 39.Yoshida, H., M. Bogaki, M. Nakamura, L. M. Yamanaka, and S. Nakamura. 1991. Quinolone resistance-determining region in the DNA gyrase gyrB gene of Escherichia coli. Antimicrob. Agents Chemother. 35:1647-1650. [DOI] [PMC free article] [PubMed] [Google Scholar]